A Survey of Viral-bacterial Co-infection in Respiratory Samples Using Multiplex Real Time-PCR
Received: 01-Apr-2019 / Accepted Date: 21-Apr-2019 / Published Date: 26-Apr-2019 DOI: 10.4172/2332-0877.1000400
Abstract
Influenza and non-Influenza virus related respiratory tract infections are amongst the most common reasons reported for physician visits and hospitalizations, globally. The economic burden associated with these viral infections, within the United States, are upwards of $50 billion annually. Concomitant, or subsequent, co-infections of the respiratory tract by pneumonia-causing pathogenic bacteria, like Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis and Staphylococcus aureus, are often observed in patients suffering from these respiratory viral infections and contribute to significant co-morbidity and mortality. In the present study, respiratory swabs from patients were tested for the presence of viral and bacterial pathogens, by Real-Time PCR, employing the Thermo-Fisher Open Array® platform. Amongst the 5793 samples tested (Dec 2017-Dec 2018), 2357 (40.68%) tested positive for the presence of Influenza and Non-Influenza viral infections. Out of these positive patient samples, 1175 (49.85%) also tested positive for the presence of one or more pneumonia-causing bacterial species. The co-infection data obtained was distributed on the basis of age. Interestingly, nearly 30% of the cases from the younger patient subset (<1 year old and 1-15 years old) were found to be co-infected with S. pneumoniae, H. influenzae and M. catarrhalis within the same sample. In addition, infant (<1 year old) patient viral positive samples, displayed higher than previously reported levels of M. catarrhalis co-infections (72.08%). With the ever-increasing popularity of point of care testing, this study demonstrates the importance and efficacy of a rapid, comprehensive multiplex-PCR, syndromic-panel approach to respiratory disease diagnostics, assisting clinicians to make better, and timely, decisions, potentially leading to improved patient care and outcomes.
Keywords: Influenza; Co-infections; Influenza like illness; Respiratory virus; Respiratory infection
Keywords
Influenza; Co-infections; Influenza like illness; Respiratory virus; Respiratory infection
Introduction
Respiratory viral disease caused by Influenza A, B (Influenza) and other non-influenza viruses [Respiratory Syncytial Virus (RSV), Rhinovirus (RV), Coronavirus (CV) Parainfluenza virus (PIV), Human metapneumovirus (HMPV), Adenovirus (AV)], can be complicated by secondary bacterial infection, which has significant impact on disease presentation and the associated morbidity and mortality rates. Bacterial co-infections assert important implications on clinical and patient care decisions [1-3]. Influenza and non- influenza viral infections of the respiratory tract renders the very young, and people over the age of 65 years, highly susceptible to bacterial pneumonia. The total economic burden of Influenza, calculated as both direct medical care and indirect costs, in the United States, is $11.2 billion [4]. Non-influenza viral respiratory tract infections have an even larger economic impact, with the economic burden of these infections, within the United States, estimated at approximately $40 billion annually [5]. The comorbidity associated with concurrent bacterial pneumonia and influenza infection, has been well documented in the four Influenza pandemics of the last century [6]. It is now believed that the high rate of mortality and disease severity reported during these pandemics, was due in part to concurrent or secondary bacterial co-infections with Streptococcus pneumoniae, Hemophilus influenzae and Staphylococcus aureus [3]. These same bacterial pathogens are also responsible for the increased morbidity of other non-influenza respiratory viral infections [7]. A number of cellular and molecular mechanisms have been elucidated, that demonstrate how viral infections of the respiratory tract predispose to bacterial infection. Viral infections modify the respiratory tract epithelial cell surface, resulting in enhanced adherence of the bacteria to the tissue [8]. Studies using mice models have shown that Influenza viral infection promote superinfections by S. pneumoniae [9] and S. aureus [10]. Similar observations have been reported with respect to RSV and AV infection, presenting increased adhesion of non-typeable H. influenzae in cultured human respiratory tract epithelial cells [11]. In addition, viral infections of the respiratory tract have an adverse effect on the host immune response against bacteria [12]. Influenza infection resulted in upregulated apoptosis of alveolar macrophages, which compromised the innate pulmonary immunity, leading to superinfection by S. pneumonia [13]. Similarly, production of anti-microbial peptides can be deregulated by pulmonary viral infections, leading to increased bacterial co-infection [14]. Conversely, there is evidence that bacterial infection of the respiratory tract, can also lead to enhanced viral infection [15,16]. Respiratory disease symptoms due to Influenza and non-influenza viruses, are very similar, and are termed Influenza-like illnesses (ILI). This necessitates a rapid and reliable diagnosis of the causative organism [17,18]. The growing popularity of low complexity point of care (POC)/near POC testing for respiratory disease diagnosis, is based on the rapid turnaround time and ease of operation, with minimal required technical expertise [19]. However, these tests can suffer from lower sensitivity and a limited test menu of organisms, especially with respect to respiratory tract bacterial pathogens. In many cases, these POC tests are restricted to one or only a few respiratory viruses [20-22]. Polymerase Chain Reaction (PCR) based molecular diagnosis is the new “gold standard” of laboratory-based testing for infectious diseases [23]. It has been asserted that employment of a comprehensive multiplex-PCR, syndromic-panel approach to the diagnosis of respiratory tract infectious diseases, can help achieve reliable results in a quick time frame, thus reducing the overall direct and indirect medical costs associated with these infections [24]. There is a large body of work detailing the association of bacterial co-infections during Influenza pandemics. However, the available data for seasonal variations of Influenza and other respiratory viruses, and the associated bacterial co-infections, is sparse [25]. Recommendations by the Center for Disease Control and Prevention (CDC) and other regulating bodies focus on good antibiotic stewardship, warn against treating these known pneumonia causing bacteria in out-patient settings, making clinical decisions on treatment tougher for physicians. The work presented in this paper was undertaken with the aim to determine the rate of viral-bacterial coinfection, in patients presenting with symptoms of ILI. We have developed a comprehensive multiplex-PCR, syndromic-panel test for respiratory tract infectious disease diagnosis. The test menu covers Influenza and several non-influenza respiratory viruses. In addition, bacterial pathogens that are responsible for co-infections, or are commensals of the respiratory tract, but can act as opportunistic pathogens, are also covered in our test menu. The co-infection data presented in this paper allows for a more comprehensive understanding of co-infection rates, distributed over an age range, facilitating better clinical stewardship, when dealing with patients with ILI.
Materials and Methods
Nucleic Acid Extraction Respiratory samples (Nasopharyngeal/ Throat swabs) were received from patients in the HealthTrackRx laboratory in Denton, Texas, United States. Total nucleic acid was isolated, following manufacturers’ instructions, using the Mag-Bind Universal Pathogen Nucleic Acid purification kit (Omega Bio-Tek, Norcross, Georgia, USA), integrated on an automated liquid handling platform (Firefly Nimbus 96; Hamilton Robotics, Reno, Nevada, USA). Reverse Transcription and Pre-Amplification RNA isolated from the patient samples was reverse transcribed using the SuperScript IV reverse transcriptase system (ThermoFisher, California, USA) according to manufacturer’s instructions. Rare targets within the sample were subjected to pre-amplification, as per manufacturer’s instructions, before real-time PCR analysis using the TaqManTM Pre- Amp Mastermix and custom primers provided by the manufacturer (ThermoFisher, California, USA).
Real-Time PCR analysis
Real-Time PCR analysis was performed using the QuantStudioTM 12K Flex Real-Time PCR system (ThermoFisher, California, USA). Diluted pre-amplified nucleic acid, isolated from patient samples, was placed on Open Array TM plates containing the target primers and probes, using the AccuFillTM system. Real-Time PCR analysis was performed on the QuantStudioTM 12K Flex, using the Gene Expression program according to the manufacturer’s instructions (ThermoFisher, California, USA). The PCR primers and probes were designed, validated and manufactured by a ThermoFisher proprietary process.
Data analysis
All statistical analysis was performed using Microsoft Excel. While collating patient test results, no personal identifying data was accessed. Each patient sample was assigned a randomly generated sample identification number and only the gender and age of the sample were used for data analysis.
Results
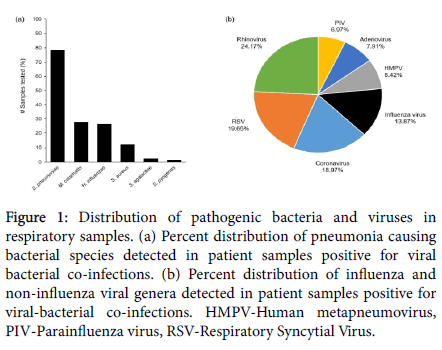
In the present study, spanning a period of one year (December 2017 to December 2018), a total of 5793 respiratory samples were tested for the presence of viruses and bacteria, causing influenza like illness (ILI). Amongst these, 2357 (40.68%) tested positive for the presence of Influenza and Non-Influenza viral respiratory infections. Out of the patient samples that tested positive for the presence of these respiratory viruses, 1175 (49.85%) also tested positive for the presence of one or more bacterial species responsible for causing bacterial pneumonia. The main viral genera detected were Rhinovirus (24.17%), RSV (19.65%), Coronavirus (18.97%), Influenza virus (13.87%), HMPV (8.42%), Adenovirus (7.91%) and PIV (6.97%) (Figure 1a). The main bacterial pathogens, detected as viral co-infections, were Streptococcus pneumoniae (78.38%), Moraxella catarrhalis (27.74%), Haemophilus influenzae (26.38%) and Staphylococcus aureus (12.08%). Amongst the total cases, S. agalactiae (Group B Strep) and S. pyogenes (Group A Strep) were detected at 2.38% and 1.61% respectively (Figure 1b). In addition, Chlamydophila pneumoniae, Klebsiella pneumoniae, Mycoplasma pneumoniae and Bordetella pertussis were detected in less than 1% of the patient samples tested (Data not presented).
Figure 1: Distribution of pathogenic bacteria and viruses in respiratory samples. (a) Percent distribution of pneumonia causing bacterial species detected in patient samples positive for viral bacterial co-infections. (b) Percent distribution of influenza and non-influenza viral genera detected in patient samples positive for viral-bacterial co-infections. HMPV-Human metapneumovirus, PIV-Parainfluenza virus, RSV-Respiratory Syncytial Virus.
Age distribution of bacterial co-infection in Influenza positive samples
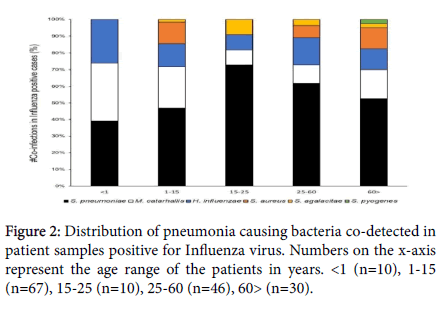
Influenza virus (A and B) were detected in 315 cases. Out of those, 163 (51.74%) cases were found to have at least one bacterial co- infection. Distribution of the Influenza virus-bacterial co-infection data by age, shows S. pneumoniae as the dominant co-infecting species present across all age ranges. S. pneumoniae, M. catarrhalis and H. influenzae were the only bacterial co-infections detected in the infant samples (<1 year of age), with 40% of the samples testing simultaneously positive for all three bacterial species. In addition, this trio of upper respiratory bacterial pathogens, co-detected together in a sample, was observed only in the younger population. No S. aureus co- infections were detected in the <1 years and 15-25 years age groups with S. pneumoniae being detected in 90% and 80% of the samples in these age groups, respectively. M. catarrhalis co-infections were detected at an elevated rate in the younger populations (80% and 43% in <1 and 1-15 years of age respectively) compared to the 15-25 years (10%) and 25-60 years (13%) age groups. The pathogen re-emerges at comparatively higher rate (23%) in the >60 years age group. Only one case of Streptococcus pyogenes was detected as a co-infection in the older population (>60 years of age) (Figures 2 and 3).
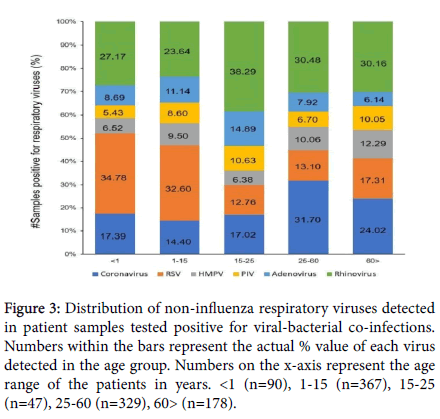
Figure 3: Distribution of non-influenza respiratory viruses detected in patient samples tested positive for viral-bacterial co-infections. Numbers within the bars represent the actual % value of each virus detected in the age group. Numbers on the x-axis represent the age range of the patients in years. <1 (n=90), 1-15 (n=367), 15-25 (n=47), 25-60 (n=329), 60> (n=178).
Age distribution of non-Influenza respiratory viral infections
Given the number of viral species responsible for respiratory tract infections in the current study, and for ease of data visualization, the distribution of non-Influenza viral infections was evaluated across the same age ranges (Figure 3), as employed for the Influenza-bacteria co- infections. Amongst the patient samples positive for viral-bacterial co- infections, approximately 25% tested positive for Rhinovirus infections. A higher proportion of Rhinovirus infection was detected in the 15-25 years age group (38.29%). Similarly, Coronavirus infections accounted for approximately 19% of the co-infection samples and were observed in 31.7% of the 25-60 years age group. RSV infections were over-represented in the younger population (34.78% in <1 year of age and 32.6% in 1-15 years of age), as opposed to the 19.65% detection in the total co-infection samples subset. AV, PIV and HMPV infections displayed smaller variations in distribution across all age ranges.
Age distribution of bacterial co-infections in respiratory viral positive samples
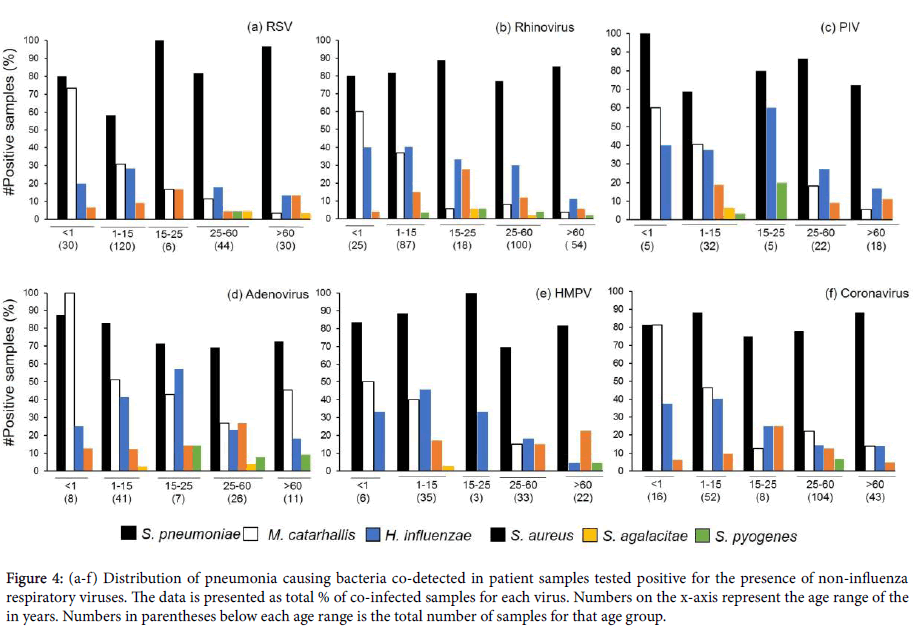
Distribution of bacterial pathogen co-infections in non-influenza virus positive samples reflected the trend observed for Influenza positive patient samples. S. pneumoniae co-infection was the highest reported in these samples, across all age ranges (Figures 4a and 4b). The next highest instance of co-infection was M. catarrhalis, followed by H. influenzae. M. catarrhalis co-infections displayed a downward trend with the progression of age, with no co-infections of the pathogen reported for the 15-25 years old age group sample subset which were also positive for PIV or HMPV infections (Figures 4c-4f). Sporadic instances of S. agalactiae and S. pyogenes co-infections were reported for all viral infections with no observable distribution trend.
Figure 4: (a-f) Distribution of pneumonia causing bacteria co-detected in patient samples tested positive for the presence of non-influenza respiratory viruses. The data is presented as total % of co-infected samples for each virus. Numbers on the x-axis represent the age range of the in years. Numbers in parentheses below each age range is the total number of samples for that age group.
Discussion and Conclusion
The present study is a survey of viral and bacterial co-infections detected in patient samples displaying symptoms of Influenza-like illness. The data presented in this paper is, to a large extent, in agreement with previously published studies. However, a few novel and interesting observations were made with respect to the current state of our understanding of viral-bacterial co-infections in respiratory illnesses. S. pneumoniae was found to be the most common bacterial infection associated with the viral infections detected in this study, irrespective of the viral species or the age group.
Pneumonia and other related diseases caused by S. pneumoniae, are responsible for millions of hospitalizations and deaths, both in the United States and globally [26,27]. Previous studies have reported S. pneumoniae to be the most prevalent co-infection associated with Influenza infection, ranging from 20% to 54.8% of the total cases [28-30]. Prior viral infection of the airways facilitates the colonization of the respiratory tract by S. pneumoniae [31] as the bacterium is able to catabolize sialic acid that is released from the host cell surface by viral neuraminidases [32] giving it a nutritional advantage and increasing its pathogenic potential. Additionally, pneumococcal resistance to traditional antibiotics like Augmentin, have made S. pneumonia harder to treat empirically [33] with 27.47% of the co- infection cases testing positive for M. catarrhalis, this pathogen was more prevalent than H. influenzae and S. aureus in our study. To our knowledge, this is the first study reporting such high instances of M. catarrhalis co-infection rate within the same data set. M. catarrhalis is a common causal agent of otitis media [34] and complications (bronchitis, pneumonia) associated with chronic obstructive pulmonary disease (COPD) [35].
The emerging importance of the pathogen was reported in the 2009 Influenza pandemic, where 17.64% of the Influenza positive cases were co-infected by M. catarrhalis [36]. Two reasons that can explain the under-reporting in previous studies and, by contrast, the high levels of M. catarrhalis co-infections in our study are: (a) the use of a sensitive detection technique and (b) prior to the 1980’s, M. catarrhalis was listed as a non-pathogenic bacterium. In fact, a meta-analysis of 27 studies on Influenza co-infections lists M. catarrhalis as a frequent source of infection since the 1980’s but it is still regarded by some physicians as normal colonizers that do not require treatment [37].
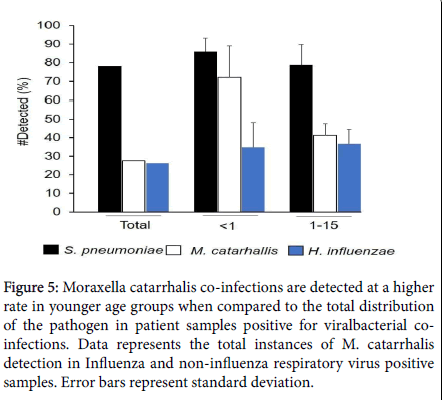
A particularly interesting observation was made with respect to the detection of M. catarrhalis in the younger population (M. catarrhalis in the S. pneumoniae and H. influenzae are not significantly different than the total co-infection rates (Figure 5). Similar infection rates of M. catarrhalis have been reported in infants up to 2 years of age with respect to the development of otitis media [34]. M. catarrhalis displays increased cell adhesion and proinflammatory responses, with exposure to cold ambient conditions (26°C for 3 hours). This may occur with prolonged exposure to cold air temperatures and can result in Influenza-like/viral cold-like symptoms [38]. Humoral immune responses against M. catarrhalis seem to be age-dependent, with the level of immunoglobulin G (IgG) gradually increasing in children. A significant decrease in antibody concentration, during the first 3 months of life, from healthy children is seen, and the M. catarrhalis- specific IgG antibodies remain at a low level in children below 1 year of age. From the age of 1 year, the immune response of children increases slowly, to reach maternal levels at the age of 10 years [39]. Our data, along with the above prior immune-system observations, suggest M. catarrhalis as a strong candidate for childhood vaccination programs [40].
Figure 5: Moraxella catarrhalis co-infections are detected at a higher rate in younger age groups when compared to the total distribution of the pathogen in patient samples positive for viralbacterial coinfections. Data represents the total instances of M. catarrhalis detection in Influenza and non-influenza respiratory virus positive samples. Error bars represent standard deviation.
The pathogen’s role in otitis media and COPD is well established [41] but given the higher rate of co-infection associated with respiratory viruses, especially in the younger population, M. catarrhalis can emerge as a serious pathogen. A possible explanation for the high rate of M. catarrhalis co-infection rate observed in this study could be the use of pneumococcal (Streptococcus pneumoniae) vaccines. It has been demonstrated that use of pneumococcal vaccines led to a decrease in nasopharyngeal colonization of vaccine-covered serotypes, with a concomitant increase in colonization by the non-vaccine pneumococcal serotypes, H. influenzae and M. catarrhalis [42]. Along with S. pneumoniae and H. influenzae, S. aureus has been reported to be one of the most common co-infecting pathogens (28% of all co- infection instances) during, or after, Influenza infection [37]. In the present survey, S. aureus co-infection rate was the fourth most abundant (12.08%), which is significantly lower than the co-infection rate previously reported for the pathogen. S. aureus is a common cause of pneumonia in humans and a frequently associated co-infection with Influenza. The potential pathogen, especially methicillin resistant strains (MRSA), can prove to be fatal in many cases [43]. Our patient population resided mainly on the out-patient population, which could provide insight on the lower detection rate of S. aureus in the current study. Globally pneumonia is one of the leading causes of mortality in children aged 5 and under [44]. As suggested by our study, the expected trends of pneumonia causing bacterial co-infections can change over time. Thus, a robust detection technique, that does not suffer from the limitations of classic culture and sensitivity protocols, (i.e. time to final reporting, microbial viability problems during sample transport to the lab, inability to culture some microbes, long length of culture time for some microbes, etc.), is needed. Patient samples, wherein 3 or more bacterial species were detected, comprise 10.46% (123/1175) of the total co-infection cases in the current study. In the majority of these cases (90/123), we detected S. pneumoniae, M. catarrhalis and H. influenza, in conjunction with a viral infection. An overwhelming majority of these cases (81/90) were present in the younger population (
Potentially, in one out of every two patients using a viral-only detecting POC test, clinicians would have missed the diagnosis of a concurrent bacterial infection, likely increasing morbidity and mortality, and certainly increasing “time to successful treatment” and infection-associated costs. Weighing the balance between good antibiotic stewardship and effective treatment is ill defined, and is left to physician discretion, potentially increasing adverse patient outcomes in the present, to avoid the spread of antibiotic resistance in the future. Despite the insight into the co-infection trends of bacterial pathogens with Influenza and other respiratory viruses, our study suffers from some limitations. The data generated is only for the detection of the bacterial and viral pathogens in patients displaying ILI symptoms. We did not have access to their clinical conditions prior to, or after, reporting of the results, nor do we know the clinical outcome. In addition, the use of vaccines and antibiotics prior to the collection of samples was not detailed, precluding their consideration in our discussion. As a future endeavor, the clinical conditions of the patients, along with treatment plans, could be tracked in coordination with the treating physician. Efforts are underway in our laboratory to generate a comprehensive dataset for the associated antibiotic resistance markers, to further assist in the correct initial treatment, and to maximize antibiotic stewardship considerations for bacterial co-infections. We recognize the importance of POC tests in an out-patient setting for quick viral diagnosis, and the need for rapid viral panels on hospitalized patients, but these techniques do not test for the most common pneumonia causing bacteria. Based on the current findings, these bacteria need to be tested as nearly half of post-viral patients will have at least one bacterial co-infection known to cause lower respiratory tract infections, which can lead to adverse patient outcomes, including morbidity and mortality in at risk age groups.
Acknowledgements
The authors would like to thank Dr. Pallavi Upadhyay for critical reading and editing of the manuscript.
References
- Brealey JC, Sly PD, Young PR, Chappell KJ (2015) Viral bacterial co-infection of the respiratory tract during early childhood. FEMS Microbiol Lett 362: 62.
- Rothberg MB, Haessler SD (2010) Complications of seasonal and pandemic influenza. Crit Care med 38: 91-97.
- Gupta RK, George R, Nguyen-Van-Tam JS (2008) Bacterial pneumonia and pandemic influenza planning. Emerg Infect Dis 14:1187-1192.
- Putri WC, Muscatello DJ, Stockwell MS, Newall AT (2018) Economic burden of seasonal influenza in the United States. Vaccine 36: 3960-3966.
- Fendrick AM, Monto AS, Nightengale B, Sarnes M (2003) The economic burden of non-influenza-related viral respiratory tract infection in the United States Ann Intern Med 163: 487-494.
- Joseph C, Togawa Y, Shindo N (2013) Bacterial and viral infections associated with influenza. Influenza Other Respir Viruses 7: 105-113.
- Smith AM, McCullers JA (2014) Secondary bacterial infections in influenza virus infection pathogenesis. Influenza Pathogenesis and Control,Volume I Springer, Cham.
- McCullers JA (2014) The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol 12: 252-262.
- Nakamura S, Davis KM, Weiser JN (2011) Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J Clin Invest 121: 3657-3665.
- Iverson AR, Boyd KL, McAuley JL, Plano LR, Hart ME et al. (2011) Influenza virus primes mice for pneumonia from Staphylococcus aureus. J Infect Dis 203: 880-888.
- Novotny LA, Bakaletz LO (2016) Intercellular adhesion molecule 1 serves as a primary cognate receptor for the Type IV pilus of nontypeable Haemophilus influenzae. Cellular microbiology 18: 1043-1055.
- Mehta D, Petes C, Gee K, Basta S (2015) The role of virus infection in deregulating the cytokine response to secondary bacterial infection. J Interferon Cytokine Res. 35: 925-934.
- Ghoneim HE, Thomas PG, McCullers JA (2013) Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. The J Immunol 191: 1250-1259.
- McGillivary G, Mason KM, Jurcisek JA, Peeples ME, Bakaletz LO (2009) Respiratory syncytial virusâ€induced dysregulation of expression of a mucosal βâ€defensin augments colonization of the upper airway by nonâ€typeable Haemophilus influenzae. Cellular microbiology 11: 1399-1408.
- Nguyen DT, Louwen R, Elberse K, van Amerongen G, Yüksel S et al. (2015) Streptococcus pneumoniae enhances human respiratory syncytial virus infection in vitro and in vivo. PLoS One 10: 127098.
- Gulraiz F, Bellinghausen C, Bruggeman CA, Stassen FR (2014) Haemophilus influenza increases the susceptibility and inflammatory response of airway epithelial cells to viral infections. The FASEB Journal 29: 849-858.
- Hasman H, Pachucki CT, Unal A, Nguyen D, Devlin T et al. (2009) Aetiology of influenza-like illness in adults includes parainfluenza virus type 4. J Med Microbiol 58: 408-413.
- Laguna-Torres VA, Gómez J, Ocaña V, Aguilar P, Saldarriaga T et al. (2009) Influenza-like illness sentinel surveillance in Peru. PloS one 4: 6118.
- Zumla A, Al-Tawfiq JA, Enne VI, Kidd M, Drosten C et al. (2014) Rapid point of care diagnostic tests for viral and bacterial respiratory tract infections-needs, advances, and future prospects. Lancet Infect Dis 14: 1123-1135.
- Mahony JB, Petrich A, Smieja M (2011) Molecular diagnosis of respiratory virus infections. Crit Rev Cl Lab Sci 48: 217-249.
- Kaman WE, Elshout G, Bindels PJ, Mitsakakis K, Hays JP (2016) Current problems associated with the microbiological point-of-care testing of respiratory tract infections in primary care. Future Microbiol 11: 607-610.
- Bruning AH, de Kruijf WB, van Weert HC, Willems WL, de Jong MD et al. (2017) Diagnostic performance and clinical feasibility of a point-of-care test for respiratory viral infections in primary health care. Fam Pract 34: 558-563.
- Basein T, Gardiner B, Andujar-Vazquez GM, Chandranesan AS, Rabson A et al. (2017) Clinical utility of universal PCR and its real-world impact on patient management. Open Forum Infect Dis S627.
- Schreckenberger PC, McAdam AJ (2015) Point-counterpoint: large multiplex PCR panels should be first-line tests for detection of respiratory and intestinal pathogens. J Clin Microbiol 53: 3110-3115.
- Falsey AR, Becker KL, Swinburne AJ, Nylen ES, Formica MA et al. (2013) Bacterial complications of respiratory tract viral illness: a comprehensive evaluation. J Infect Dis 208: 432-441.
- Pittet LF, Posfay-Barbe KM (2012) Pneumococcal vaccines for children: a global public health priority. Clin Microbiol Infect 18: 25-36.
- Huang SS, Johnson KM, Ray GT, Wroe P et al. (2011) Healthcare utilization and cost of pneumococcal disease in the United States. Vaccine 29: 3398-3412.
- MartÃn-Loeches I, Sanchez-Corral A, Diaz E, Granada RM, Zaragoza R et al. (2011) Community-acquired respiratory coinfection in critically ill patients with pandemic 2009 influenza A (H1N1) virus. Chest 139: 555-562.
- Cox CM, Blanton L, Dhara R, Brammer L, Finelli L (2011) 2009 Pandemic influenza A (H1N1) deaths among children-United States, 2009–2010. Clin Infect Dis 52: S69-74.
- Shieh WJ, Blau DM, Denison AM, DeLeon-Carnes M, Adem P et al. (2010) 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am J Pathol 177: 166-175.
- Grijalva CG, Griffin MR, Edwards KM, Williams JV, Gil AI et al. (2014) The role of influenza and parainfluenza infections in nasopharyngeal pneumococcal acquisition among young children. Clin Infect Dis 58: 1369-1376.
- Siegel SJ, Roche AM, Weiser JN (2014) Influenza promotes pneumococcal growth during coinfection by providing host sialylated substrates as a nutrient source. Cell Host Microbe 16: 55-67.
- Kim L, McGee L, Tomczyk S, Beall B (2016) Biological and epidemiological features of antibiotic-resistant Streptococcus pneumoniae in pre-and post-conjugate vaccine eras: a United States perspective. Clin Microbiol Rev 29: 525-552.
- Faden H, Harabuchi Y, Hong JJ (1994) Epidemiology of Moraxella catarrhalis in children during the first 2 years of life: relationship to otitis media (1994) J Infect Dis 169: 1312-1317.
- Murphy TF, Brauer AL, Grant BJ, Sethi S (2005) Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am J Respir Crit Care Med 172: 195-199.
- Nguyen T, Kyle UG, Jaimon N, Tcharmtchi MH, Coss-Bu JA et al. (2012) Coinfection with Staphylococcus aureus increases risk of severe coagulopathy in critically ill children with influenza A (H1N1) virus infection. Crit Care Med 40: 3246-3250.
- Klein EY, Monteforte B, Gupta A, Jiang W, May L et al. (2016) The frequency of influenza and bacterial coinfection: a systematic review and metaâ€analysis. Influenza Other Respir Viruses 10: 394-403.
- Aebi C (2011) Moraxella catarrhalis-pathogen or commensal? Adv Exp Med Biol 697: 107-116.
- Ejlertsen T, Thisted E, Ostergaard PA, Renneberg J (1994) Maternal antibodies and acquired serological response to Moraxella catarrhalis in children determined by an enzyme-linked immunosorbent assay. Clin Diagn Lab Immunol 1: 464-468.
- Ruckdeschel EA, Kirkham C, Lesse AJ, Hu Z, Murphy TF (2008) Mining the Moraxella catarrhalis genome: identification of potential vaccine antigens expressed during human infection. Infect Immun 76: 1599-1607.
- Goldstein EJ, Murphy TF, Parameswaran GI (2009) Moraxella catarrhalis, a human respiratory tract pathogen. Clin Infect Dis 49:124-131.
- Casey JR, Pichichero ME (2004) Changes in frequency and pathogens causing acute otitis media in 1995-2003. Pedia Infect Dis 23: 828-824.
- Mulcahy ME, McLoughlin RM (2016) Staphylococcus aureus and influenza A virus: Partners in coinfection. MBio 7: 02016-02068.
- Liu L, Oza S, Hogan D, Chu Y, Perin J et al. (2016) Global, regional, and national causes of under-5 mortality in 2000-15 an updated systematic analysis with implications for the Sustainable Development Goals. The Lancet 388: 3027-3035.
Citation: Singh V, Reddy J, GrangerJ (2019) A Survey of Viral-bacterial Co-infection in Respiratory Samples Using Multiplex Real Time-PCR . J Infect Dis Ther 7: 400. DOI: 10.4172/2332-0877.1000400
Copyright: © 2019 Singh V, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 4768
- [From(publication date): 0-2019 - Dec 19, 2025]
- Breakdown by view type
- HTML page views: 3820
- PDF downloads: 948