Association Mapping of Yellow Rust (Puccinia striiformis F. sp. tritici) Resistance in Bread Wheat (Triticum aestivum L.)
Received: 12-Jul-2021 / Accepted Date: 26-Jul-2021 / Published Date: 02-Aug-2021 DOI: 10.4172/2329-8863.1000472
Abstract
Genetic resistance is the most economic and effective means of reducing yield losses caused by the disease. Marker-trait associations in germplasm relevant to breeding program via Association Mapping (AM) can be an effective way to identify loci useful for selection. In present study an accession penal of 123 spring bread wheat genotypes have been assembled and genotyped with 10263 SNPs markers. Further, AM analysis using a General linear model (GLM) identified three genomic region located on wheat chromosome 2A, 3A and 5B; contain 33 common significant markers at three locations which are significantly associated with genes conferring resistance to yellow rust. Mixed Linear Model (MLM), corrected for population structure and kinship relatedness and identified one common genomic regions located on chromosome 3A which are significantly associated with genes conferring resistance to yellow rust at all locations. For this reason, a constant search for new genes for resistance is required, and wild relatives of wheat may be a rich resource for identifying novel resistance genes for stripe rust. Additionally, our study highlighted the presence of valuable genetic variation that could be exploited to sustainably enhance yellow rust resistance in bread wheat, but further characterization and successful validation, diagnostic markers linked to yellow rust resistance genes can be used for breeding wheat varieties with resistance to the wheat yellow rusts.
Keywords: Kubsa; Puccinia stiiriformis tritici; Single nucleotide polymorphism; Yellow rust; Germplasm
Introduction
Wheat (Triticum aestivum L.) is grown globally and is the world’s second most important cereal crop [1]. It is considered as one of the first domesticated food crops and for more than 80 countries has been the primary food staple of major civilizations of world and it‘s the most widely adapted major cereal crop that is cultivated on larger land area than any other crop worldwide. Globally wheat is grown on 220.4 million hectares with a total production 750 million Metric Tons (Mt) annually, which makes it the second important grain crop after maize. China is currently the world‘s leading wheat producer, accounting for approximately 15% of the world‘s total production [2].
Area under wheat production in Africa is estimated approximately 8.8 million ha (2.5million ha east and South Africa and 6.3 million ha in North Africa). Within east and North Africa, Ethiopia is the top producer, next to Egypt, volume produced and the numbers of farmers engaged in its production are huge [3]. In the period of 2009-2011, the country ranked first both in area and production of wheat in sub- Saharan Africa with a share of 55% and 47.8%, respectively [4]. In Ethiopia wheat is cultivated on about 4.7 million hectares of land accounting for 13.5% of the total grain crop area, with an annual production of 4.54 million tons, contributing about 15.63% of the total grain production [5]. According to CAS, (2017) report wheat ranks fourth after tef, maize and sorghum. The Bale, Arsi and Shewa areas of Oromia region are the highest wheat producing areas of Ethiopia and are considered as-wheat belt areas, produce about 52.83% of Ethiopian wheat [5]. Wheat is largely grown in the mid and highland areas of Ethiopia spanning at altitudes of 1500 to 3000 m. a. s. l. However, it is mainly grown between 1800 to 2500 m. a. s. l in the country [6].
With the global population increasing and food security expected to become more important, wheat will continue to play a fundamental role as an important staple food crop for the vast majority of the global human population. Nationally, wheat contributes an estimated 12% to the daily per capital calorie intake and source of carbohydrates, making it the third most important contributor to national calorie intake, after maize and sorghum [7,8]. Wheat is the world leading cereal grain serving as a staple food for more than one-third of the global population [6,9].
To address the rising demand for wheat and improve global food security, an annual increase in wheat production by up to 1.7% is necessary [10]. Global wheat breeding efforts have made significant contributions to the improvement of wheat yield potential. However, annual growth rate of wheat yield has been declining or static in the recent decade [11,12]. The sustainable production and supply of wheat for future generation is threatened and challenged by the world population growth rate, global climate changes, and various biotic and abiotic stresses [11]. Of biotic stress; yellow rust is one of the most devastating diseases [13-15]. Over 45 million tons of wheat (valued at $9 billion) is lost due to wheat diseases and other pests annually, among which yellow rust has become a serious threat to wheat, causing 50-100% yield losses [16]. This is mainly due to the breakdown of existing resistance genes and gradual adaptation of new strains in warmer regions, particularly the Central and West Asia and North Africa (CWANA) region [17]. Yellow rust cause significant yield and quality losses and challenge the achievement of wheat productivity for gains needed to supply the growing demand [18]. In Ethiopia majorities of commercial bread wheat cultivars have become susceptible to stem rust and/or yellow rust [19]. This is mainly due to the pathogen's ability to mutate, multiply rapidly and to use its airborne dispersal mechanism from one field to another and even over long distances [20-24]. As new races are continually produced in the 86 pathogen population, most Yr genes have become ineffective [25].
Yellow rust is an important disease of spring bread wheat in the highlands of Ethiopia at altitudes ranging from 2150 to 2850 m. a. s. l and the major production bottleneck in the major wheat producing regions of Ethiopia [26]. Arsi and Bale zones are the major producing areas of wheat and are the known hotspots for the epidemics of yellow rust of wheat [19]. Even though there is seasonal variability in the occurrence of yellow rust in Bale highlands, the main and long rainy season is ideal for yellow rust development [27]. In south-eastern part of Ethiopia, yellow rust is a major threat to wheat production resulting in high yield and quality losses [28]. The importance of stripe rust in the highlands of Ethiopia has been described previously [27-30]. Various management have been recommended like chemical, biological, cultural and other management approaches are not more effective in controlling yellow rust, due to long distance movement of spores, able to mutate and form new races [22,31].
The most effective strategy for control of yellow rust is breeding and growing resistant cultivars, as this approach has no additional cost to farmers and is environmentally desirable. Using DNA-based molecular markers have several advantages over the traditional phenotypic selection and their potential benefits as Marker-Assisted Selection (MAS) have been widely discussed, especially to provide solutions to overcome some of the problems faced by classical phenotypic screening approaches in plant breeding programs [32,33]. The genetic information, wheat germplasm and molecular markers generated here provide resource that could be used to help design and develop wheat varieties with improved adult plant genetic resistance to emerging races of yellow rust [34,35]. Molecular markers can be used to tag rust resistance genes and further their use can serve for the improvement of the efficiency of selection in plant breeding by MAS, with the number of available robust genetic markers such as SSRs and SNPs increasing and the cost of genotyping decreasing. Single Nucleotide Polymorphisms (SNPs) are also recent type of molecular markers and are becoming popular to be widely used for mapping of associated resistance to yellow rust [36,37].
Therefore, searching for new source of resistance to yellow rust from new bread wheat genotypes with the help of molecular markers is necessary to cope up with the emerging virulent races of the pathogen [38].
Materials and Methods
Plant materials and experimental design
In the field experiments 240 spring bread wheat genotypes and 7 check varieties of known and varying host response were used (Appendix: 1). These genotypes was obtained from the ICARDA and the genotypes where evaluated for their resistance to yellow rust at adult stages.
For the field experiment, the genotypes were conditioned in nonreplicated trials, using an augmented design evaluated for their resistance to yellow rust disease. To facilitate uniform disease build-up within the nursery, continuous yellow rust susceptible spreader rows (using a mixture of susceptible cultivars Morocco and Kubsa in 1:2 proportion) was planted perpendicular to all entries on both sides of the plots by 20 cm.
DNA extraction
Genomic DNA was extracted from 2-week-old pooled leaf samples collected from five plants per accession. The samples were frozen in liquid nitrogen and stored at -80˚C until free dry. DNA extraction was carried out following the method, 10 μL of a 100 ng μl-1 DNA of each sample was sent to traits genetics, Germany as commercial service provider for genotyping using nucleotide polymorphism (SNPs) markers [39]. The initial genotypic matrix contained 15K (15000SNPs) reduced to 10263 markers after filtrations (markers with missing data>10% and Minor Allele Frequency (MAF)<5% were removed. The 10263 markers were used to run GWAS and we set 0.15 minimal allele frequency and each markers has to cover at least 80 genotypes.
Population structure
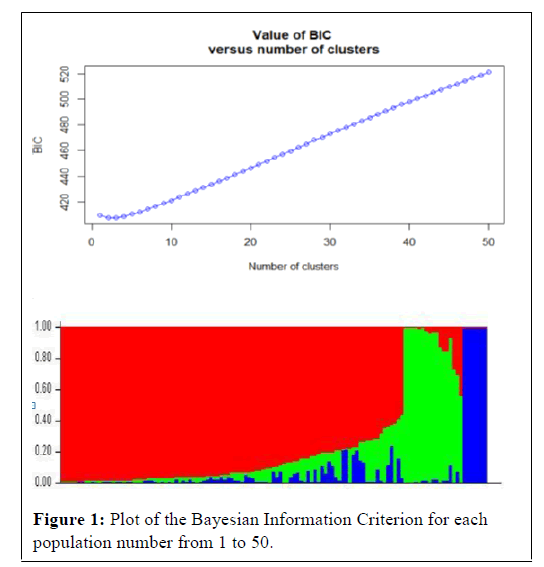
The genetic structure of the 123 genotypes was investigated using 101 highly polymorphic markers spread in the whole genomes [40]. Genetic distance between pair of chosen markers on the same chromosome was more than 50cm to minimize LD was by tightly linked markers. The Discriminant Analysis of Principal Components (DAPC) was performed using the =adegenet’ package 1.4-1 in Rstudio and a Bayesian clustering method was applied to identify clusters of genetically similar individuals using the software STRUCTURE version 2.3 [40]. To infer population structure among the wheat genotypes, 10 independent run for each K (k=the number of sub populations) was performed based on an admixture model and correlated allele frequency. Both the length of burn-in period and the number of iterations were set at 100,000. To reach the appropriate k value, the estimated normal logarithm of the probability of fit [LnP (D)] provided in the STRUCTURE output was plotted against k. This value reaches a plateau when the minimal number of groups that best describe the population substructure has been reached.
Linkage disequilibrium
From the complete set of 8127 polymorphic markers, only 7590 markers with known position, only marker with known position were selected to perform the linkage disequilibrium analysis, using TASSEL V 5.2 software [41]. For estimating Linkage Disequilibrium (LD), SNPs alleles with Minor Allele Frequency (MAF) higher than 0.05 were used. Pair-wise LD was measured using the squared allelefrequency correlations (R2) [42].
Association mapping and analysis
The CI and molecular data were used for the association mapping [43]. TASSEL version 5.2 was used to perform association mapping analysis using both the General Linear Model (GLM) and Mixed Linear Model (MLM) methods which takes into consideration Kinship matrix (K) [44]. The MLM was again used but after including population structure (Q) as a covariate to control both Type I and Type II errors. The genetic positions of the SNPs markers were based on the wheat 10K SNP consensus map [45]. Manhattan plot used to indicate the significance level of the markers.
Results
Population structure
As indicated in Figure 1, the K values steadily kept on increasing until K=3 indicating that in the 123 spring bread wheat genotypes in this study are clustered into three sub populations, the number on the x axis that corresponds the first peak before it drops, is considered as the number of clusters. The Discriminant Analysis of Principal Components (DAPC) was performed using the =adegenet‘package 1.4-1 in Rstudio and then we run the structure based on the results we got (R-plot attached; 3 subpopulations) then, estimated from pair wise comparisons as a measure of genetic distance between subpopulations. In the second graph, it is indicated by colors; clearly we have 3 major colors covering distinct big areas. Cluster one is the largest with 99 genotypes accounting for approximately 84.48% of the total genotypes. Cluster 2 and 3 consisted of 17 and 7 genotypes respectively.
The proportion of the genome of each individual originating from each inferred population (a total of 3 and each color represent a single population). Structure analysis of the 123 spring bread wheat genotypes in this study grouped them into 3 clusters indicating the presence of significant genetic variation among the population.
Linkage disequilibrium
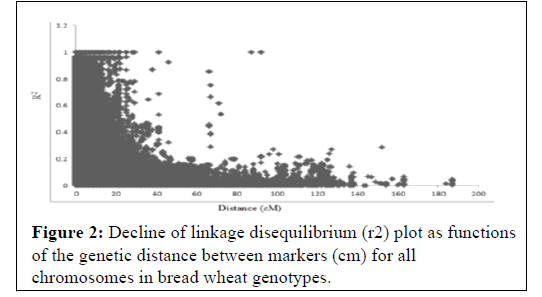
In this study the extent of LD and average rate of LD decay were estimated by squared correlation coefficient (r2) for all pairs of SNPs along each chromosome. When r2 is zero, alleles at two loci do not co-occur more frequently than would be expected under random sampling. R-square approaches its maximum of 1 as alleles at two loci show more frequent co-occurrence within the population sample examined (Figure 2). In contrast, the chromosomes where significant markers linked to yellow rust resistance had good marker coverage and therefore reliable LD decay estimates.
The extent of LD was assessed among all 511876 pairs of SNPs loci for all accessions. Across all accessions as many as 40.5% of the total marker pairs were in LD (based on P <0.001). A scatter plot of r2 values versus genetic distances between all markers across the genome abruptly declined to 0.2 within ̴25 cm when all mapped SNPs markers with chromosome position were analyzed.
SNPs markers statistics
All genotypes were tested with 10263 SNPs markers. A total 8127 SNPs markers were selected for analysis due to their polymorphism used for the AM analysis. Of these 7590 (93.4%) were of known map position in the consensus map in which, 2915, 3722,and 953 were specific to A,B, and D genomes, respectively. A position of 537 polymorphic markers was unknown. In the present study, several markers associated with yellow rust resistance were identified. Chromosomes with the largest markers are 2B (728 markers) followed by 6B (644 markers). Chromosomes 4D and 7D showed the least number of loci, 34 and 79 markers, respectively in Tables 1 and 2.
| Chromosome | No of loci | No of position | Average distance |
|---|---|---|---|
| 1A | 364 | 78 | 6.53 |
| 1B | 534 | 82 | 5.26 |
| 1D | 190 | 42 | 17.32 |
| 2A | 429 | 68 | 7.23 |
| 2B | 728 | 99 | 4.29 |
| 2D | 345 | 54 | 6.71 |
| 3A | 355 | 73 | 10.23 |
| 3B | 466 | 80 | 4.98 |
| 3D | 82 | 26 | 45.06 |
| 4A | 305 | 74 | 9.93 |
| 4B | 273 | 61 | 6.79 |
| 4D | 34 | 15 | 32.74 |
| 5A | 532 | 86 | 5.59 |
| 5B | 615 | 95 | 6.42 |
| 5D | 104 | 28 | 9.33 |
| 6A | 475 | 71 | 8.44 |
| 6B | 644 | 77 | 4.24 |
| 6D | 119 | 33 | 11.57 |
| 7A | 455 | 89 | 8.86 |
| 7B | 462 | 93 | 11.08 |
| 7D | 79 | 44 | 44.12 |
Table 1: Genetic markers statistics, chromosomes, number of marker, number of markers with position on the consensus SNPs map and the average distance between each two adjacent markers for each chromosome.
| Location | Marker | Chromosome location | Position | p-value | R2 | Observed proportion (%) |
|---|---|---|---|---|---|---|
| KARC | Kukri_rep_c102953_304 | 3A | 83 | 8.24E+04 | 9.8 | 60.27 |
| Ku_c35823_743 | 2A | 122 | 9.24E+04 | 9.4 | 99.63 | |
| MWU | BS00049403_51 | 5B | 121 | 1.11E-04 | 14.2 | 83.6 |
| IAAV9068 | 5B | 121 | 1.11E-04 | 14.2 | 83.64 | |
| Ra_c111671_555 | 2B | 103 | 4.91E-04 | 10.7 | 56.58 | |
| Kukri_rep_c102953_304 | 3A | 83 | 6.44E-04 | 10.3 | 60.27 | |
| SARC | Kukri_rep_c102953_304 | 3A | 83 | 7.07E-05 | 13.9 | 60.27 |
| IAAV9068 | 3A | 85 | 2.64E-04 | 11.6 | 56.58 |
Association mapping for yellow rust response
Table 2: Association analysis for yellow rust severity of the wheat association mapping panel using MLM model at KARC ,MWU and SARC Significant markers at p˂0.0001.
Association mapping has been reported as an effective strategy to identify linked markers with disease resistance for possible marker assisted selection. Genome-wide association analysis was performed for yellow rust CI for each of the three environments. In view of the strong genotype by location interaction, marker-phenotype association tests were conducted separately for each location as well as for the responses averaged over the three locations. The AM analysis was conducted by performing using both the General Linear Model (population structure correction; Q GLM) and the mixed linear model (population structure and familiar relatedness correction K+Q MLM), efficiency of the two model is determined by using QQ plot. Introducing the experiment-wise correction, 33 common markers at all location showed significant (P ≤ 0.0001) representing three genomic regions and the others number of position effects in the Q GLM model. These markers was located on chromosome 3A, 2A, 5A and NP while in the Q+K MLM model the significance was limited to one region on chromosome 3A which showed the strongest association with yellow rust response at three locations .These results clearly show that how MLM is restrictive. The chromosome with the highest estimate of polymorphic information content was 2B. Manhattan plots depicting association between significant markers greater than 3.022 and yellow response in different environments were displayed.
The association analysis conducted at KARC using GLM detected 37 highly significant SNP markers. These SNPs markers were located on chromosomes 1A, 2A, 3A, 4D, 5B and 7A. On chromosome 1A, one highly significant marker was detected (IACX5994). On chromosome 2A, five highly significant markers were detected in the association analysis. On chromosome 3A, twenty six highly significant markers were detected. On chromosome 4D only one significant SNP marker (RFL_Contig2119_607) was detected at 18 cm. In the same way, only one marker (wsnp_Ku_c38713_47298856) was detected on chromosome 5B at 168 cm. On chromosome 7B there were three SNP markers located at 209 cm. In contrast, the chromosomes where significant markers linked to yellow rust resistance had good marker coverage and reliable LD decay estimates.
The individual analysis at MWU using the GLM method detected significant markers on chromosomes 2A, 3A, 3B, 5B, 6B, 7A and 7B. On chromosome 2A the highly significant SNP markers were distributed from 20 to 27 cm. On chromosome 3A there were 35 highly significant SNP markers were detected. On chromosome 3B, two highly significant SNP markers were detected at 14 and 21 cm. On chromosome 5B, the highly significant markers were located at 121 cm. On chromosome 6B, the highly significant markers were located at 4 and 92 cm. On chromosome 7A, the highly significant markers were located at 113,114 and 121cm. Finally, chromosome 7B contained significant marker at 131 cm. The association analysis conducted with the data collected from SARC for yellow rust severity with the GLM method detected SNP markers significantly associated with resistance to yellow rust located on chromosomes 1A, 2A, 3A, 4A, 5B and 7B. On chromosome 1A, one SNP markers significantly associated with yellow rust located at 144 cm. On chromosome 2A, two SNP markers significantly associated with yellow rust located at 20 and 47 cm were detected. On chromosome 3A, thirty one SNPs marker were detected. On chromosome 4A, one SNP markers significantly associated with yellow rust located at 144 cm. On chromosome 5B, three markers highly significant associated with yellow rust located at 121 and 146 cm. Finally, on chromosome 7B, two SNP markers were detected at 134 cm.
The association analysis conducted with combined data set and individually data set on all location in the wheat AM using the MLM method detected SNP markers significantly linked to yellow rust resistance on chromosomes 2A, 3A, 2B, and 5B. On chromosome 3A, the SNP highly significant markers were located at 83 cm at all locations. At KARC, on chromosome 2A, one marker highly significant associated with yellow rust located at 122 cm. At MWU, on chromosome 2B and 5B one, one highly significant marker are located at 113 and 121 cm respectively. In addition to 3A common for all location SARC have another 3A, which contain one marker highly significant at 85 cm.
Discussion
As indicated in Figure 1, the K values steadily kept on increasing until K=3 indicating that in the 123 spring bread wheat genotypes in this study are clustered into three main sub populations. The existence of such clusters to imply genetic variation based on molecular data might be attributed to the utilization of diverse parents, originated from different sources, in the breeding program of ICARDA reported that wheat breeding program at ICARDA utilizes parents originated from ICARDA, CIMMYT, and from a wide range of genetically unrelated winter wheat from Turkey, Iran, Russia, Ukraine, Romania, Bulgaria, Hungary, and the United States of America [46]. The utilization of such diverse parents in the breeding program has contributed to the reported genetic variation. The significant SNP markers associated with spring bread wheat to yellow rust reported in this study provide several resistance loci to fight the disease, of which some are likely novel. The extent of LD was assessed among all 511876 pairs of SNPs loci for all accessions. Across all accessions as many as 40.5% of the total marker pairs were in LD (based on P <0.001). A scatter plot of R2 values versus genetic distances between all markers across the genome abruptly declined to 0.2 within ̴25cm when all mapped SNPs markers with chromosome position were analyzed. When compared with cultivated durum wheat and bread wheat, LD decayed not more rapidly in this spring bread wheat genotypes. Previous studies reported that the genome-wide LD r2 decayed to 0.2 within a distance of 5-20 cm in the elite bread wheat populations [47-50]. This result is expected for selfpollinated crop species such as wheat reported that rapid rate of inbreeding with selfing results in a low recombination frequency in self-pollinated species. In the previous studies, the estimated LD decay of wheat was 0.5 to 40 cm, which is relatively high when compared with cross-pollinated crops such as maize (200 to 2000 bp) [47,51-53].
In our study we used 123 spring bread wheat genotypes were tested with 10263 SNPs markers. A total 8127 of SNPs markers were selected for analysis due to their polymorphism used for the AM analysis, in which, 2915, 3722, and 953 were specific to A, B, and D genomes, respectively and others no positions. In our finding higher marker density were found on chromosome B. Similar with our finding. Chromosomes with the largest markers are 2B (728 markers) followed by 6B (644 markers). Several yellow rust QTLs and Yr genes are mapped on these chromosomes including Yr5 on chromosome 2B [54]. Chromosomes D showed the least number of loci, also reported large non- polymorphic chromosomal sections in the D genome, especially on 4D and 7D (>30cm) [55].
Association mapping has been reported as an effective strategy to identify linked markers with disease resistance for possible marker assisted selection. The discovery of significant SNPs can allow for tagging of lines that are enriched for alleles associated with the trait, and their use in gene introgression for resistance breeding. Reported that ,discovery of resistant sources in existing breeding programs can speed up the process of gene introgression into elite lines, gene pyramiding for elevated resistance to the disease, and possible identification of diagnostic markers that can be used in marker assisted resistance breeding [56]. The association mapping analysis was conducted by performing using both the General Linear Model and the mixed linear model. The association analysis conducted at KARC using GLM detected 37 highly significant SNP markers. These SNPs markers were located on chromosomes 1A, 2A, 3A, 4D, 5B and 7A. On chromosome 4D only one significant SNP marker (RFL_Contig 2119_607) was detected at 18cm, reported a significant association of IWA5375 and other linked SNP (IWA5766) with yellow rust resistances, which were mapped in close proximity to the RFL_Contig2119_607 locus [57]. In the same way, only one marker (wsnp_Ku_c38713_47298856) was detected on chromosome 5B at 168 cm. Other QTL including QYr-5B_Oligoculm and YrEXP2 were previously mapped close to the region of wsnp_Ku_c38713_47298856. On chromosome 7B there were three SNP markers located at 209 cm [58,59]. At MWU using the GLM method detected significant markers on chromosomes 2A, 3A, 3B, 5B, 6B, 7A and 7B. Chromosome 7B contained significant marker at 131cm. It could be related to the previously mapped yellow rust QTL QYr-7B_Oligoculm on the same chromosome [58]. The association analysis conducted with the data collected from SARC for yellow rust severity with the GLM method detected SNP markers significantly associated with resistance to yellow rust located on chromosomes 1A, 2A, 3A, 4A, 5B and 7B.
The association analysis in the wheat detected markers significantly associated with yellow rust resistance in each location using GLM and MLM methods. Reported that genes for yellow rust resistance have been found in almost every chromosome of the wheat genome [60]. But in our finding, analyses conducted with data collected from all sites detected significant SNP markers only on chromosome 1A, 2A, 3A, 2B, 5B and 7B. Marker IAAV9068 was present on 73 genotypes (59.34%), of which 64(87.6%) showed resistance/moderately resistant response to yellow rust. This suggests that genetic regions around these loci may be useful for choosing parents and incorporating new yellow rust resistance genes into adapted wheat cultivars after testing validity of this marker, these agree with [61]. Marker Kukri_rep_c102953_304 was present on 70 genotypes (56.9%), of which 62(88.57%) resistance/moderately resistant response to yellow rust. Of 33common significant markers, the markers with the highest proportion of presence in the genotypes evaluated were IAAV9068 on chromosome 3A and Kukri_rep_c102953_304 also on Chromosome 3A..
In this finding higher marker densities were observed in the A and B genome chromosomes (2.11 and 3.14 markers per cm, respectively) compared to the D genome chromosomes, which may be due to lower rates of recombination. Similar to these, reported that higher marker densities were observed in A and B chromosomes (8.11 and 6.17 markers per cm, respectively) compared to the D genome chromosomes [62,63]. Used the 90K SNP chip to genotype 726 wheat accessions including landraces and found a similar trend, where only 15% of the reported markers were in the D genome [45]. Also found large non-polymorphic chromosomal sections in the D genome, especially on 4D and 7D (>30cm), But in these finding we report there are few polymorphic markers present on 4D and 7D chromosomes [55]. According the study conducted by the typically low genetic variation in the D genome of modern wheat means that breeding efforts essentially act to manipulate diversity largely in the A and B genomes [55, 64,65].
Accessions from this diversity panel could be used to increase genetic diversity, particularly for the D genome in modern germplasm. In our finding several QTL located on chromosome 3A by using GLM that provide resistance to yellow rust obtained in all locations (around 33 common SNPs are present) and one common QTL were obtained by using MLM, but according to study conducted by using the same markers and the same genotypes with our finding they do not get common markers for different locations on septoria and stem rust respectively, these indicate that our genotypes contain more markers which are resistance to yellow rust [66]. Since the markers (SNPs) we used are not used before many of the significant markers we found are might represent novel yellow rust loci, but they require validation/ further investigations.
Conclusion
It could be concluded that genetic resistance is the most economic and effective means of reducing yield losses caused by the disease. Association mapping in elite germplasm has the potential to accelerate the translation of basic genetic information towards applications in crop improvement and cultivar release. Accessions with high a percentage of yellow rust resistance associated alleles are excellent genetic resources that could serve as parental breeding lines to enable more efficient breeding for yellow rust resistance. Application of the latest genotyping platforms, such as SNPs, could lead to the rapid detection of novel genomic regions underpinning yellow rust resistance. We found 33 common markers for all sites by using GLM and 1common marker by using MLM. This result indicates that the genotypes we used in this finding contain many genes which are resistance to yellow rust. We identified several resistance loci; both confirming previously reported resistance genes and describing novel genomic regions in the bread wheat genotypes. The molecular markers linked to resistance loci identified in the current study can be used to efficiently target the selection of associated QTL to diversify the genetic basis of the spring bread wheat genotypes. Our results suggest that GWAS is an effective method for characterizing genes in cultivated bread wheat and confirm that bread wheat is a rich source of stripe rust resistance loci that can be used for wheat improvement. Gene pyramiding, or combining several resistance genes into one genotype, is one strategy for developing durable resistance that the pathogen may not be able to overcome. For this reason, a constant search for new genes for resistance is required, and wild relatives of wheat may be a rich resource for identifying novel resistance genes for stripe rust.
References
- Pant K, Ojha B, Thapa D, Kharel R, Gautam N, et al. (2020) Evaluation of biofortified spring wheat genotypes for yield and micronutrient contents. Fundam Appl Agric 5: 78-87.
- CSA (Central Statistical Authority) of Ethiopia (2014) Agricultural survey sample. Report on area and production of crops (private peasant holding, meher season). Statistical Bulletin Number 33. Addis Ababa, Ethiopia.
- Negassa A, Shiferaw B, Jawoo K, Sonder K, Smale M, et al. (2013) The potential for wheat production in Africa: Analysis of biophysical suitability and economic profitability. CIMMYT.
- CSA (Central Statistical Agency) of Ethiopia (2017) Agricultural sample survey: Report on area and production of major crops.
- Hei N, Shimelis H, Laing M (2017) Appraisal of farmers’ wheat production constraints and breeding priorities in rust prone agro-ecologies of Ethiopia. Afr J Agri Res 12: 944-952.
- Guush B, Zelekawork P, Kibrom T, Seneshaw T (2011) Foodgrain consumption and calorie intake patterns in Ethiopia.
- Tehseen M, Tonk F, Tosun M, Amri A, Sansaloni C, et al. (2020) Genome-Wide Association Study of Resistance to PstS2 and Warrior Races of Puccinia Striiformis f. Sp.Tritici (Stripe Rust) in Bread Wheat Landraces. Plant Genome 14: e20066.
- Alemu M (2013) Genetic Variability and Association among Agronomic characters in some wheat (T.aestivum) Genotypes in Arsi Zone, Oromia Region, Ethiopia.
- Rosegrant M, Agcaoili M (2010) Global food demand, supply, and price prospects to 2010.
- Dixon J, Braun H, Crouch J (2009) Overview: transitioning wheat research to serve the future needs of the developing world. In: Dixon J, Braun H-J, Kosina P., Crouch, J., editors. Wheat facts and futures Mexico.
- Fischer R, Byerlee D, Edmeades G (2009) Can technology deliver on the yield challenge to 2050?
- Wellings C (2011) Global status of stripe rust: A review of historical and current threats. Euphytica 179: 129-141.
- Wan A, Zhao Z, Chen XM, He Z, Jin S, et al. (2004) Wheat stripe rust epidemics and virulence of Puccinia striiformis f.sp. tritici in China in 2002. Plant Disease 88: 896-904.
- Milus E, Kristensen K, Hovmøller M (2009) Evidence for increased aggressiveness in a recent widespread strain of Puccinia striiformis f. sp. tritici causing stripe rust of wheat. Phytopathology 99: 89â€94.
- ICARDA (2011) Research to action-strategies to reduce the emerging wheat stripe rust disease. International wheat stripe rust symposium, Aleppo, Syria.
- William H, Trethowan R, Crosby-Galvan E (2007) Wheat breeding assisted by markers: CIMMYT’s experience. Euphytica 157: 307-319.
- SARC (Sinana Agricultural Research Center) (2004) Sinana Agricultural Research Center Pathology Department Progress Reports for the Period 2002-2004.
- Singh R, Huerta J, William H (2005) Genetics and breeding for durable resistance to leaf and stripe rusts in wheat. Turk J Agric For 29: 121-127.
- Chen X, Moore M, Milus E, Long, D, Line R, et al. (2007) Wheat Stripe Rust Epidemics and Races of Puccinia striiformis f. sp. tritici in the United States in 2000. Plant Dis 86: 39-46.
- Chen W, Wellings C, Chen X, Kang, Z ,Liu T (2014) Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici. Mol Plant Pathol 15: 433–446.
- Jia J, Zhao S, Kong X, Li Y, Zhao G, et al. (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496: 91-95.
- Zhao J, Wang M, Chen X, Kang Z (2016) Role of alternate hosts in epidemiology and pathogen variation of cereal rusts. Ann Rev Phytopathol 54: 207-228.
- Sharmaâ€Poudyal D, Chen X, Rupp R (2014) Potential oversummering and overwintering regions for the wheat stripe rust pathogen in the contiguous United States. Int J Biometeorol 58: 987â€997.
- Ayele B, Temesgen K (2008) Review of relative infection of Research on Diseases of small cereals crops. In: Abraham Tadesse (ed). Increasing crop production through improved plant protection. Int Res J Plant Sci 1: 375-418.
- Bekele H, Shambel K, Dereje H (2002) Seasonal variations in the occurrence of wheat stripe rust in Bale highlands. Int J Pest Manag 6: 65-72.
- Dereje H (2003) Effects of yellow rust (Puccinia striiformis) on yield, yield components and quality of improved bread wheat (Triticum aestivum L.) varieties.
- Getaneh W, Andrushenko A, Mozgovoy A (1990) Wheat rust situation in 1987 and 1989 crop season in Ethiopia. In: Proceedings of the Ethiopian Phytopathological Committee 1990.
- Getinet G, Marten G, Temesgen K, Mengistu H, Yeshi A (1990) Wheat disease survey in Ethiopia in 1988.
- Feyissa R, Kudryavtsev E, Chiapparino E, Chiari T (2005) On-farm conservation and enhancement of local durum Wheat genetic resources in Ethiopia.
- Anderson J (2003) Plant genomics and its impact on wheat breeding. In: Plant molecular breeding (H.J.Newbury, Ed.), Blackwell Pub. Boca Raton 184-215.
- McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Xia XC (2017) Catalogue of gene symbols for wheat: 2017 Supplement.
- Bouvet L, Percival-Alwyn L, Berry S, Fenwick P, Mantello C, et al. (2021) Wheat Genetic Loci Conferring Resistance to Yellow Rust in the Face of Recent Epidemics of Genetically Diverse Races of the Fungus Puccinia StriiformisF. Sp.Tritici. Res squ 8: 1-35.
- Rani R, Singh R, Yadav N (2019) Evaluating stripe rust resistance in Indian wheat genotypes and breeding lines using molecular markers. C R Biol 342: 154-174.
- Cuthbert R (2011) Molecular mapping of septoria tritici blotch resistance in hexaploid wheat (Triticum aestivum L.).
- Huang X, Han B (2014) Natural variations and genome-wide association studies in crop plants. Annu Rev Plant Biol 65: 531-551.
- Tadesse W, Ogbonnaya FC, Jighly A, Nazari K, Rajaram S, et al. (2014) Association Mapping of Resistance to Yellow Rust in Winter Wheat Cultivars and Elite Genotypes.
- Ogbonnaya F, Seah I, Delibes A, Jahier J, Lopez-Brana I, et al. (2001) Molecular genetic characterization of nematode resistance from Aegilops ventricosa and its derivatives in wheat. Theor Appl Genet 102: 623-629.
- Pritchard J, Stephens M, Donnelly P (2000) Inference of Population Structure Using Multilocus Genotype Data. Genetics 155: 945-959.
- Huang B, George A, Forrest K (2012) A multi parent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol J 10: 826-39.
- Flint-Garcia S, Thornsberry E, Buckler S (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54: 357-374.
- Ogbonnaya FC, Rasheed A, Okechukwu EC, Jighly A, Makdis F, et al. (2017) Genome wide association study for agronomic and physiological traits in spring wheat evaluated in a range of heat prone environments. Theor Appl Genet 130: 1819-1835.
- Zhu C, Gore M, Buckler SE, Yu J (2008) Status and Prospects of Association Mapping in Plants. Plant Genot 1: 5-2.
- Wang S, Wong D, Forrest K, Allen A, Chao S, et al. (2014) Characterization of polyploid wheat genomic diversity using a high-density single nucleotide polymorphism array. Plant Biotechnol J 12: 787-796.
- Tadesse W, Ogbonnaya FC, Jighly A, Sanchez-Garcia M, Sohail Q, et al. (2015) Genome-Wide Association Mapping of Yield and Grain Quality Traits in Winter Wheat Genotypes. PLoS ONE 10: e0141339.
- Chao S, Zhang W, Dubcovsky J, Sorrells M (2007) Evaluation of Genetic Diversity and Genome-wide Linkage Disequilibrium among U.S. Wheat (Triticum aestivumL.) Germplasm Representing Different Market Classes. Crop Sci 47: 1018-1030.
- Zhang D, Bai G, Zhu C, Yu J, Carver B (2010) Genetic diversity, population structure, and linkage disequilibrium in U.S. elite winter wheat. Plant Genet 3: 117-127.
- Zegeye H, Rasheed A, Makdis F, Badebo A, Ogbonnaya FC (2014) Genome-wide association mapping for seedling and adult plant resistance to stripe rust in synthetic hexaploid wheat. PLoS One 9: e105593.
- Naruoka Y, Garland-Campbell KA, Carter AH (2015) Genome-wide association mapping for stripe rust (Puccinia striiformis f. sp. tritici) in US Pacific Northwest winter wheat (Triticum aestivum L.). Theor Appl Genet 128: 1083-1101.
- Maccaferri M, Sanguineti M, Natoli E, Araus-Ortega J, Bensalem M, et al. (2007) A panel of elite accessions of durum wheat (Triticum durum Desf.) suitable for association mapping studies. Plant Genet Res 4: 79-85.
- Emebiri L, Oliver R, Mrva K, Mares D (2010) Association mapping of late maturity α-amylase (LMA) activity in a collection of synthetic hexaploid wheat. Mol Breed 26: 39-49.
- Tenaillon M, Sawkins M, Long A, Gaut R, Doebley J, et al. (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays spp. mays. L.). Proc. Natl. Acad. Sci. U. S. A. 98: 9161-9166.
- Marchal C, Zhang J, Zhang P, Fenwick P, Steuernael B, et al. (2018) BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat Plants 4: 662–668.
- Voss-Fels K, Frisch M, Qian L, Kontowski S, Friedt W, et al. (2015) Sub genomic diversity patterns caused by directional selection in bread wheat gene 4 pools. The Plant Genome.
- Bajgain P, Rouse MN, Bulli P, Bhavani S, Gordon T, et al. (2015) Association mapping of North American spring wheat breeding germplasm reveals loci conferring resistance to Ug99 and other African stem rust races. Plant Biotechnol J 15: 249.
- Maccaferri M, Zhang J, Bulli P, Abate Z, Chao S, et al. (2015) A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.) G3 (Bethesda) 5: 449-465.
- Suenaga K, Singh R, Huerta-Espino J, William H (2003) Microsatellite markers for genes Lr34/Yr18 and other quantitative trait loci for leaf rust and stripe rust resistance inbread wheat. Phytopathology 93: 881-890.
- Lin F, Chen M (2008) Molecular mapping of genes for race-specific overall resistance to stripe rust in wheat cultivar Express. Theor Appl Genet 116: 797-806.
- Boyd L (2005) Centenary review: can Robigus defeat an old enemy?-yellow rust of wheat. J Agric Sci 143: 233-243.
- Wang Y, Yu C, Cheng Y, Yao F, Long L, et al. (2021) Genome-wide association mapping reveals potential novel loci controlling stripe rust resistance in a chinese wheat landrace diversity panel from the southern autumn sown spring wheat zone. BMC Genomics 22: 34.
- Cavanagh C, Chao S, Wang S, Huang B, Stephen S, et al.(2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci USA 110: 8057-8062.
- Nielsen N, Backes G, Stougaard J, Andersen S, Jahoor A (2014) Genetic diversity and population structure analysis of European hexaploid bread wheat (Triticum aestivum. L.) varieties. PLoS ONE 9: e94000.
- White J, Law J, MacKay I, Chalmers K, Smith J, et al. (2008) The genetic diversity of UK, US and Australian cultivars of Triticum aestivum measured by DArT markers and considered by genome. Theor Appl Genet 116: 439-453.
- Henry RJ, Nevo E (2014) Exploring natural selection to guide breeding for agriculture. Plant 15 Biotechnol J 12: 655-662.
- Binalf L (2018) Association mapping of septoria Tritici Blotch (Septoria tritici) Resistance in bread wheat (Triticum aestivum L.) in Bale and Arsi highlands, Ethiopia.
Citation: Olana A, Shifa H, Tadesse W (2021) Association Mapping of Yellow Rust (Puccinia striiformis F. sp. tritici) Resistance in Bread Wheat (Triticum aestivum L.). ACST 9: 472. DOI: 10.4172/2329-8863.1000472
Copyright: © 2021 Olana A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 3118
- [From(publication date): 0-2021 - Nov 09, 2025]
- Breakdown by view type
- HTML page views: 2279
- PDF downloads: 839