Chemical Modification of CRISPR gRNAs Eliminate type I Interferon Responses in Human Peripheral Blood Mononuclear Cells
Received: 10-Jan-2018 / Accepted Date: 22-Jan-2018 / Published Date: 29-Jan-2018 DOI: 10.4172/2576-3881.1000121
Abstract
Objectives: CRISPR/Cas9 is currently the primary tool used for genome editing in mammalian cells. To cleave and alter genomic DNA, both the Cas9 nuclease and a guide RNA (gRNA) must be present in the nucleus. One preferred method of introducing these reagents is direct transfection of a recombinant Cas9 protein complexed with a synthetic gRNA as a ribonucleoprotein (RNP) complex. It is well established from prior work in RNA interference that synthetic RNAs can induce a type I interferon (IFN) response that can limit the application of such methods both in vitro and in vivo. While the immunological properties of short siRNAs are well understood, little is known about the immune recognition of longer CRISPR gRNAs. The objective of our in vitro study was to investigate how the composition of the gRNA influences its recognition by human immune cells.
Methods: The study was performed in vitro in human peripheral blood mononuclear cells (PBMCs). The PBMCs from healthy donor volunteers were treated with gRNA for 24 h, and the levels of type I IFNs in culture supernatants were measured by a multiplex enzyme-linked immunosorbent chemiluminescent assay. Prior to the analysis in PBMCs, the physicochemical parameters and functionality of all nucleic acid constructs were confirmed by electrospray-ionization mass spectrometry and CRISPR/Cas9 gene editing assessment in HEK293-Cas9 cells, respectively.
Results: We found that unmodified synthetic CRISPR gRNAs triggered a strong IFN response in PBMC cultures in vitro that could be prevented with chemical modification. Likewise, in vitro–transcribed single-guide RNAs (sgRNAs) also triggered a strong IFN response that could only be partially suppressed by phosphatase removal of the 5’-triphosphate group. However, the process by which the gRNA is prepared (i.e., chemically synthesized as a two-part crRNA:tracrRNA complex or in vitro–transcribed as an sgRNA) does not directly influence the immune response to an unmodified gRNA. When experiments were performed in the HEK293 cells, only in vitro–transcribed sgRNA containing 5’-triphosphate induced IFN secretion.
Conclusion: The results of our structure–activity relationship study, therefore, suggest that chemical modifications commonly used to reduce the immunostimulation of traditional RNA therapeutics can also be used as effective tools to eliminate undesirable IFN responses to gRNAs.
Keywords: CRISPR/Cas9; sgRNA; gRNA; crRNA; tracrRNA; Interferons; Immunotoxicity
Introduction
Type I interferons (IFNs) are a group of many proteins that play a vital role in mammalian antiviral and antitumoral host defense [1-3]. The most prominent members of this family are IFN-a, IFN-b, and IFN-w, which in turn include several proteins. For example, there are 13 proteins in the IFN-a group and two in the IFN-b group. The IFN proteins are produced by many cell types, including both immune cells (lymphocytes, macrophages, and dendritic cells) and non-immune cells (fibroblasts, endothelial cells, and osteoblasts) [1,2]. Among the immune cells, the most prominent producers of type I IFNs are the plasmacytoid dendritic cells. Type I IFNs play a key role in antiviral defense by activating intrinsic mechanisms of the infected and neighboring cells to limit the spread of viral pathogens. They also modulate innate immune responses by promoting antigen presentation and activating natural killer–cell functions [1,2]. Furthermore, type I IFNs activate the adaptive immune system by triggering the development of high-affinity antigen-specific lymphocyte responses and immunological memory [3]. In addition to their protective role, type I IFNs can have deleterious consequences for the host by triggering pyrogenic (fever) reactions and contributing to autoimmune diseases [4].
Agonists inducing type I IFN responses are not limited to tumor cells or bacterial and viral pathogens. Certain pharmaceutical products can also trigger an IFN response [5,6]. Therapeutic nucleic acids, such as siRNAs, mRNAs, and antisense oligodeoxynucleotides (ODNs), are among such products [5-7]. Type I IFN induction by these products is often associated with safety concerns due to their pyrogenic activity. Therefore, the pharmaceutical industry has made many attempts to identify the mechanisms of IFN response to therapeutic nucleic acids and ways to overcome such responses. Modifying the backbone; adding 2’-modified ribose backbones, such as 2’-O-Methyl RNA (2’OMe) residues; and removing 5’-triphosphates from in vitro–transcribed (IVT) RNAs are among the approaches used in the field of therapeutic nucleic acids to reduce the risk of fever and fever-like reactions triggered by type I IFN responses as well as improve compound stability and efficacy [7-11].
The clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology has attracted much attention due to its efficacy in genome editing, and its therapeutic application is a rapidly developing field. Since this technology involves RNA components, the immunological response to guide RNA (gRNA) is among the many safety questions that have yet to be addressed. Unlike other therapeutic nucleic acids, little is known about the immune recognition of gRNAs. We therefore conducted an in vitro study using human peripheral blood mononuclear cells (PBMCs) to understand whether gRNAs can induce type I IFN production and to identify methods to prevent this reaction. Prior to the in vitro analysis in PBMCs, we performed physicochemical characterization of various gRNA constructs and confirmed their functionality in the model cell line HEK293, commonly used for proof-of-concept gene-editing studies.
Materials And Methods
Reagents
Lithium heparin vacutainers were purchased from BD Biosciences (San Jose, CA). RPMI, fetal bovine serum (FBS), penicillin-streptomycin, Dulbecco’s phosphate-buffered saline (DPBS; Ca2+/Mg2+-free), Hank’s balanced salt solution (HBSS), MEGAclear™ Transcription Clean-Up Kit, Geneticin, Opti-MEM®, and Lipofectamine® RNAiMAX transfection reagent were purchased from Thermo Fisher Scientific (Waltham, MA). ODN2216 and chemically synthesized RNAs were synthesized by Integrated DNA Technologies, Inc. (IDT; Coralville, IA). A HiScribe™ T7 High Yield RNA Synthesis Kit, DNase I, and Antarctic Phosphatase were purchased from New England Biolabs (Ipswitch, MA). Multiplex chemiluminescence plates for the detection of type I IFNs were custom-manufactured by Quansys Biosciences (Logan, UT). Dulbecco’s Modified Eagle Medium (DMEM) was purchased from ATCC (Manassas, VA). QuickExtract™ DNA Extraction Solution was purchased from Epicentre (Madison, WI). KAPA HiFi HotStart DNA Polymerase was purchased from Kapa Biosystems (Wilmington, MA). A Mutation Discovery Kit for the Fragment Analyzer™ was purchased from Advanced Analytical Technologies, Inc. (Ames, IA).
Preparation of gRNAs
Chemically synthesized oligoribonucleotides were manufactured by IDT using standard phosphoramidite chemistry. Short CRISPR RNAs (crRNAs) were synthesized as standard desalt RNAs, whereas long trans-activating crRNAs (tracrRNAs) were purified by reversed-phase high-performance liquid chromatography. Chemically-modified RNAs were the Alt-R® CRISPR-Cas9 crRNA and tracrRNA products that employ a proprietary modification pattern that includes end-blocking groups, 2’OMe RNA, and phosphorothioate (PS) linkages as a two-part system where synthetic crRNA and tracrRNA are annealed to form an active gRNA complex [12]. IVT single-guide RNAs (sgRNAs) were synthesized from gBlocks® Gene Fragments (IDT) templates using the HiScribe™ T7 High Yield RNA Synthesis Kit (New England Biolabs) following the manufacturer’s protocol, including a DNase I treatment to remove residual template DNA. IVTs were purified using the MEGAclear™ Transcription Clean-Up Kit (Thermo Fisher Scientific). Where indicated, Antarctic Phosphatase was used to remove the 5’ triphosphate following the manufacturer’s recommended protocol. To reduce the final volume and remove residual phosphatase, IVT RNA was phenol-chloroform-isoamyl alcohol and chloroform-extracted, which was followed by ethanol precipitation. The correct products pre- and post-phosphatase treatment were verified by electrospray-ionization mass spectrometry (ESI-MS) for all gRNAs used. No remaining triphosphate containing IVTs were detected after phosphatase treatment. Prior to testing, all gRNAs were normalized to 100 µM in Duplex Buffer (30 mM HEPES, pH 7.5, 100 mM potassium acetate).
Research donor blood
Healthy volunteer blood specimens were drawn under National Cancer Institute at Frederick Protocol OH99-C-N046. Blood was obtained from different donors to account for potential inter-donor variability and it was collected in BD vacutainer tubes containing lithium heparin as an anticoagulant.
Endotoxin analysis
To study potential particle contamination with endotoxin, the test samples were analyzed by a turbidity Limulus amoebocyte lysate (LAL) assay according to the protocol [13,14]. No endotoxin was detected in any test sample at concentrations used in the in vitro cytokine assay.
Cytokine response in PBMC cultures
Experiments were performed according to Nanotechnology Characterization Laboratory protocol ITA-10 [15]. Briefly, whole blood anticoagulated with lithium heparin was diluted in PBS, and PBMCs were isolated using Ficoll-Paque gradient-density centrifugation. Purified PBMCs were incubated with controls and gRNA samples complexed with RNAiMAX transfection reagent (Thermo Fisher Scientific). The complexation was performed according to the manufacturer’s instructions. The RNAiMAX transfection reagent alone was added to both the negative and positive control samples to establish the baseline relevant to gRNA-treated samples. The final concentration of gRNA was 1 μM. The incubation of cell cultures continued for 24 h. At the end of incubation, the supernatants were collected and centrifuged for five minutes at 18,000 g before they were analyzed for the presence of type I IFNs (IFN-a, IFN-b and IFN-w) by multiplex assay (Quansys Biosciences).
Preparation and characterization of nucleic acid constructs
HEK293 cells that constitutively express the Cas9 nuclease (“HEK293-Cas9” cells) were used to verify the functionality of gRNAs. HEK293 cells were also used to test IFN responses to the different RNAs following lipofection. HEK293-Cas9 and HEK293 were maintained in DMEM supplemented with 10% FBS, 1% penicillin-streptomycin. The DMEM used for HEK293-Cas9 was also supplemented with 500 µg/mL G418. For lipoplex formation, 1.5 µL of the gRNA at a working concentration of 3 µM was mixed with 0.75 µL Lipofectamine® RNAiMAX in the presence of Opti-MEM® at a final volume of 50 µL and incubated at room temperature for 10 minutes. Next, 40,000 HEK293-Cas9 cells were plated on top of the lipoplex mixture in 100 µL of DMEM with 10% FBS in a 96-well plate (reverse transfection). The final volume was 150 µL with a gRNA concentration at 30 nM. Cells were incubated at 37°C and 5% CO2.
Forty-eight hours post-transfection, cells were washed with 100 µL PBS, lysed using 50 µL QuickExtract™ DNA Extraction Solution, and heated to 65°C for 15 min, which was followed by another 15 min at 95°C. Genomic DNA was diluted three-fold with nuclease-free water, and 1.5 µL (~15 ng) was amplified using 0.15 U KAPA HiFi HotStart DNA Polymerase in a final volume of 10 µL. Total gene editing was measured using an Alt-R® Genome Editing Detection Kit (T7EI) (IDT). Polymerase chain reaction primers (IDT) were designed to be at least 100 bp distant from the gRNA cut site. Cleavage products were separated on the Fragment Analyzer™ using the Mutation Discovery Kit. Editing frequencies were calculated using the following formula: average molar concentration of the cut products/(average molar concentration of the cut products+molar concentration of the uncut product) × 100. Supernatants from HEK293 cells were collected 24 h after the delivery of nucleic acid constructs and analyzed for the presence of IFNs by multiplex assay (Quansys Biosciences).
Results
Characterization of nucleic acid constructs
Endotoxin is a known immunostimulatory contaminant that can be introduced into reagents during their preparation and carried over to the final products [16]. Since we intended to analyze nucleic acid constructs in primary human blood cells, we verified that all materials did not contain endotoxin at levels sufficient to affect the experiments. According to the kinetic turbidity LAL assay, endotoxin was undetectable in all tested constructs (Table 1).
| Sample Number | HPRT target site | Material Type | Sample Description | Endotoxin EU/mL* |
|---|---|---|---|---|
| A | 38087 | gRNA | Synthesized, unmodified RNA, 2-part complex | <0.1 |
| B | 38087 | gRNA | Synthesized, modified RNA (end-blocked, 2’OMe, PS linkages), 2-part complex | <0.1 |
| C | 38087 | sgRNA | In vitro–transcribed RNA (with 5’-triphosphate) | <0.1 |
| D | 38087 | sgRNA | In vitro–transcribed RNA, phosphatase-treated | <0.1 |
| E | 38285 | gRNA | Synthesized, unmodified RNA, 2-part complex | <0.1 |
| F | 38285 | gRNA | Synthesized, modified RNA (end-blocked, 2’OMe, PS linkages), 2-part complex | <0.1 |
| G | 38285 | sgRNA | In vitro–transcribedRNA (with 5’-triphosphate) | <0.1 |
| H | 38285 | sgRNA | In vitro–transcribedRNA, phosphatase-treated | <0.1 |
| I | NA | ssDNA | Alt-R® Cas9 Electroporation Enhancer | <0.1 |
| J | NA | - | Duplex Buffer (100 mM potassium acetate, 30 mM HEPES, pH 7.5) | <0.1 |
Table 1: Nucleic acid constructs: The study included nine nucleic acid (NA) constructs (eight gRNA and one ssDNA) and a control buffer used for the NA preparation and storage. gRNA were prepared to target two sites of the HPRT gene. The gRNAs differed by 20-base protospacer guide domains targeting the 38285 site with sequence 5’-CUUAUAUCCAACACUUCGUG-3’ and the 38087 site with sequence 5’-AAUUAUGGGGAUUACUAGGA-3’. For each site, the gRNA were either chemically synthesized or in vitro–transcribed. The two parts of the synthetic gRNA were separately synthesized (crRNA and tracrRNA), then annealed to form a complete gRNA complex. These constructs were either unmodified or included end-blocking groups, 2’-OMe RNA, and PS linkages (Alt-R® CRISPR-Cas9 crRNA and tracrRNA). The in vitro–transcribed sgRNA was prepared as a single RNA strand and either directly used (with a 5’-triphosphate moiety) or treated with phosphatase to instead leave a 5’-hydroxyl group. All samples were assessed for endotoxin contamination by the kinetic turbidity LAL assay. * - of 100 μM stock; sgRNA–single-guide RNA; ssDNA–single-stranded DNA; gRNA–guide RNA; HPRT-hypoxanthine-guanine phosphoribosyltransferase; 2’-OMe-oxy-methyl modification added to the 2’ hydroxyl of the ribose moiety; PS-phosphorothioate modification of the backbone; HEPES - (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) is a zwitterionic organic chemical buffering agent; LAL-Limulus amoebocyte lysate; EU–endotoxin units; mL-milliliter.
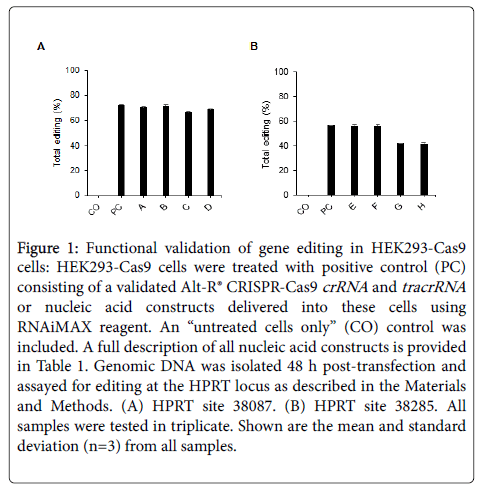
To verify that all nucleic acid constructs were correct, we performed ESI-MS for all gRNAs used. All samples showed the correct, expected mass (data not shown), including the IVT sgRNAs pre-and post-phosphatase treatment. All gRNAs were normalized to 100 µM in IDT Duplex Buffer prior to use. The buffer was included as a negative control in all bioassays. The gRNAs were all tested for functional genome-editing activity in HEK293-Cas9 cells and resulted in the expected cleavage events in the genomic HPRT locus at sites 38087 and 38285 (Figures 1A and 1B). Toxicity was visually observed for cultures transfected with IVT sgRNAs, both with and without 5’-triphosphate 5’-ends (constructs C, D, G, and H). The IVT sgRNAs (constructs G and H) targeting HPRT site 38285 had editing frequencies slightly lower than the chemically synthesized gRNAs and control gRNA as validated by Alt-R® CRISPR-Cas9 crRNA and tracrRNA (Figure 1). However, gene editing was still observed with these constructs, indicating that the correct gRNA product was present to enable CRISPR/Cas9–mediated cleavage. No editing was detected in the “untreated cells only” controls.
Figure 1: Functional validation of gene editing in HEK293-Cas9 cells: HEK293-Cas9 cells were treated with positive control (PC) consisting of a validated Alt-R® CRISPR-Cas9 crRNA and tracrRNA or nucleic acid constructs delivered into these cells using RNAiMAX reagent. An “untreated cells only” (CO) control was included. A full description of all nucleic acid constructs is provided in Table 1. Genomic DNA was isolated 48 h post-transfection and assayed for editing at the HPRT locus as described in the Materials and Methods. (A) HPRT site 38087. (B) HPRT site 38285. All samples were tested in triplicate. Shown are the mean and standard deviation (n=3) from all samples.
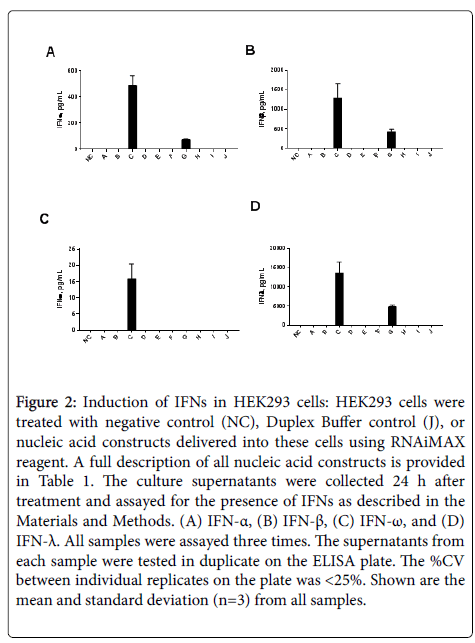
When supernatants from HEK293 cells were analyzed for the presence of IFNs, both type I and type III IFNs were found only in samples exposed to IVT sgRNAs containing 5’-triphosphates (constructs C and G). In contrast, IVT sgRNAs without 5’-triphosphates (D and H) did not induce an IFN response (Figures 2A-2D). Construct C was a more potent IFN inducer than construct G, despite both constructs containing a 5’-triphosphate group (Figure 2).
Figure 2: Induction of IFNs in HEK293 cells: HEK293 cells were treated with negative control (NC), Duplex Buffer control (J), or nucleic acid constructs delivered into these cells using RNAiMAX reagent. A full description of all nucleic acid constructs is provided in Table 1. The culture supernatants were collected 24 h after treatment and assayed for the presence of IFNs as described in the Materials and Methods. (A) IFN-a, (B) IFN-b, (C) IFN-w, and (D) IFN-l. All samples were assayed three times. The supernatants from each sample were tested in duplicate on the ELISA plate. The %CV between individual replicates on the plate was <25%. Shown are the mean and standard deviation (n=3) from all samples.
Induction of type I IFNs in PBMCs: a structure–activity relationship
PBMCs are the primary immune responders to therapeutic nucleic acids when these materials enter into systemic circulation [17]. Many studies demonstrated that in vitro assays utilizing healthy donor PBMC cultures are accurate and predictive of cytokine storm and pyrogenic reactions to drug products in humans [18-22]. We therefore used the freshly drawn blood of three healthy donor volunteers to isolate PBMCs and exposed these cells to various gRNA samples and controls. Since the biomedical applications of gRNA commonly include a delivery agent, we used an RNAiMAX lipid-based carrier in our in vitro experiments. We added gRNA-RNAiMAX complexes to PBMC cultures and monitored the levels of type I IFNs (IFN-a, IFN-w and IFN-b) at 24 h post-treatment. In a preliminary experiment, the RNAiMAX reagent alone was tested to verify that it does not affect cell viability and responses to the assay positive control ODN2216, a CpG oligonucleotide, a known TLR9 agonist and a potent inducer of type I IFNs. Since no adverse effects on the assay performance were observed (data not shown), RNAiMAX at the same concentration as that used to form complexes with gRNA was also added to the negative and positive control samples. This experimental design allowed us to compare the effects of various gRNA constructs against a baseline that was equivalent across all tested samples.
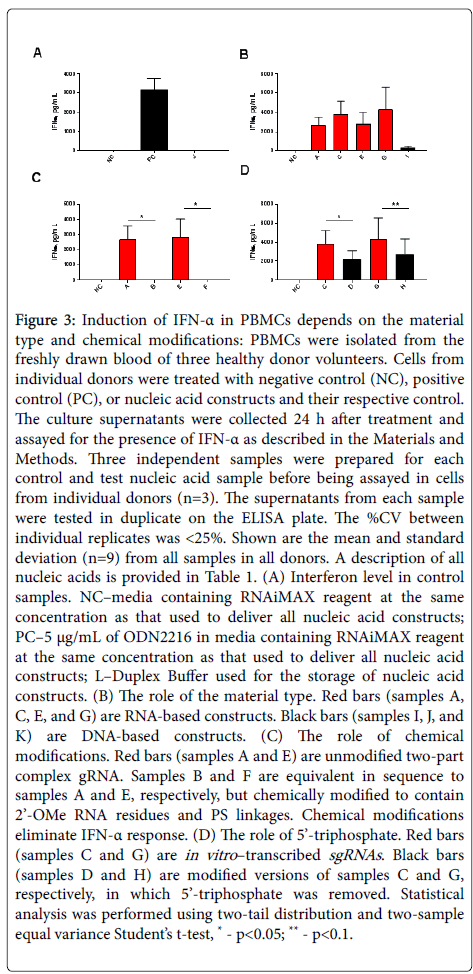
No IFN-a was detected in the negative control and Duplex Buffer control samples, while ODN2216 resulted in high IFN-a levels (Figure 3A). All unmodified gRNA constructs—regardless of their sequence, target site, and origin (i.e., a chemically synthesized annealed two-part complex or IVT sgRNA)—induced higher levels of IFN-a than single-stranded DNA (ssDNA) construct (Figure 3B, compare samples A, C, E, and G vs. sample I). Substitution of 2’OMe RNA residues for RNA at multiple locations in the two-part gRNA complex eliminated the IFN-a response (Figure 3C, compare sample A vs. B and sample E vs. F). In contrast to the unmodified IVT sgRNA samples, their counterparts without 5’-triphosphate moiety resulted in lower levels of IFN-a secretion by the cells (Figure 3D, compare samples C vs. D and G vs. H).
Figure 3: Induction of IFN-a in PBMCs depends on the material type and chemical modifications: PBMCs were isolated from the freshly drawn blood of three healthy donor volunteers. Cells from individual donors were treated with negative control (NC), positive control (PC), or nucleic acid constructs and their respective control. The culture supernatants were collected 24 h after treatment and assayed for the presence of IFN-a as described in the Materials and Methods. Three independent samples were prepared for each control and test nucleic acid sample before being assayed in cells from individual donors (n=3). The supernatants from each sample were tested in duplicate on the ELISA plate. The %CV between individual replicates was <25%. Shown are the mean and standard deviation (n=9) from all samples in all donors. A description of all nucleic acids is provided in Table 1. (A) Interferon level in control samples. NC–media containing RNAiMAX reagent at the same concentration as that used to deliver all nucleic acid constructs; PC–5 μg/mL of ODN2216 in media containing RNAiMAX reagent at the same concentration as that used to deliver all nucleic acid constructs; J–Duplex Buffer used for the storage of nucleic acid constructs. (B) The role of the material type. Red bars (samples A, C, E, and G) are RNA-based constructs. Black bars (sample I) are DNA-based constructs. (C) The role of chemical modifications. Red bars (samples A and E) are unmodified two-part complex gRNA. Samples B and F are equivalent in sequence to samples A and E, respectively, but chemically modified to contain 2’-OMe RNA residues and PS linkages. Chemical modifications eliminate IFN-a response. (D) The role of 5’-triphosphate. Red bars (samples C and G) are in vitro–transcribed sgRNAs. Black bars (samples D and H) are modified versions of samples C and G, respectively, in which 5’-triphosphate was removed. Statistical analysis was performed using two-tail distribution and two-sample equal variance Student’s t-test, * - p<0.05; ** - p<0.1.
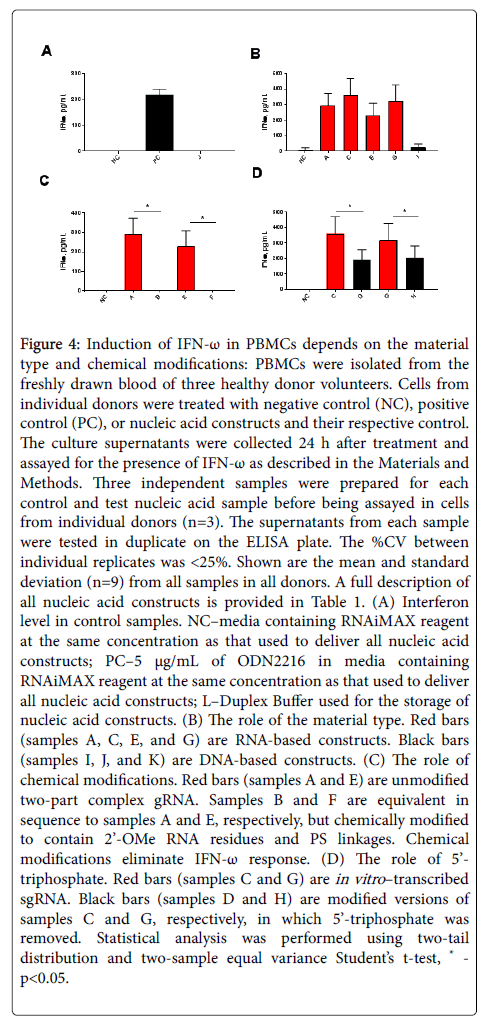
Similar results were observed with IFN-w (Figure 4) and IFN-b (data not shown). High levels of IFN-w were induced by the ODN2216, while neither the negative control nor the Duplex Buffer control showed a detectable IFN induction (Figure 4A). gRNA constructs were more potent IFN-w agonists than ssDNA (Figure 4B, compare samples A, C, E, and G vs. sample I). Chemical modification completely eliminated IFN-w (Figure 4C, compare sample A vs. B and sample E vs. F), while the removal of 5’-triphosphate from unmodified IVT sgRNAs reduced but did not eliminate secretion of this type I IFN (Figure 4D, compare sample C vs. D and G vs. H).
Figure 4: Induction of IFN-w in PBMCs depends on the material type and chemical modifications: PBMCs were isolated from the freshly drawn blood of three healthy donor volunteers. Cells from individual donors were treated with negative control (NC), positive control (PC), or nucleic acid constructs and their respective control. The culture supernatants were collected 24 h after treatment and assayed for the presence of IFN-w as described in the Materials and Methods. Three independent samples were prepared for each control and test nucleic acid sample before being assayed in cells from individual donors (n=3). The supernatants from each sample were tested in duplicate on the ELISA plate. The %CV between individual replicates was <25%. Shown are the mean and standard deviation (n=9) from all samples in all donors. A full description of all nucleic acid constructs is provided in Table 1. (A) Interferon level in control samples. NC–media containing RNAiMAX reagent at the same concentration as that used to deliver all nucleic acid constructs; PC–5 μg/mL of ODN2216 in media containing RNAiMAX reagent at the same concentration as that used to deliver all nucleic acid constructs; J–Duplex Buffer used for the storage of nucleic acid constructs. (B) The role of the material type. Red bars (samples A, C, E, and G) are RNA-based constructs. Black bars (sample I) are DNA-based constructs. (C) The role of chemical modifications. Red bars (samples A and E) are unmodified two-part complex gRNA. Samples B and F are equivalent in sequence to samples A and E, respectively, but chemically modified to contain 2’-OMe RNA residues and PS linkages. Chemical modifications eliminate IFN-w response. (D) The role of 5’-triphosphate. Red bars (samples C and G) are in vitro–transcribed sgRNA. Black bars (samples D and H) are modified versions of samples C and G, respectively, in which 5’-triphosphate was removed. Statistical analysis was performed using two-tail distribution and two-sample equal variance Student’s t-test, * - p<0.05.
The effects of gRNA and ssDNA on IFN-a and IFN-w were consistent between all tested donors. The trends observed in IFN-b were similar to those observed in IFN-a and IFN-w. However, the overall levels of this member of the type I IFN family were lower, and the responses were more pronounced in some but not all donors (data not shown).
Discussion
Macromolecular therapeutic nucleic acids are a large family of materials that includes antisense oligonucleotides, triplex-forming oligodeoxyribonucleotides, immunostimulatory oligonucleotides, splice-switching oligonucleotides, inhibitory RNA (siRNAs and shRNAs), and aptamers [23]. Preclinical and clinical studies of these materials have revealed numerous challenges, including pharmacokinetics, toxicology, and instability in the blood [24-31]. While many of these hurdles have been successfully addressed through chemical modifications of the backbone, changes in sequences, or alterations to dose regimen ([28,29,32], CRISPR/Cas9 is a new technology that relies on a nucleic acid component for function and therefore, may face similar hurdles.
In this study, we characterized various nucleic acid constructs relevant to the CRISPR/Cas9 technology. We first confirmed that all of the gRNAs were the correct mass by ESI-MS and that all of the gRNAs directed the correct genome-editing events (i.e., cleaved the correct site in the human genome) by a functional assay in HEK293-Cas9 cells (Figure 1). We found that only IVT sgRNA containing 5’-triphosphate induces all IFN types (Figure 2). Removal of the 5’-triphosphate eliminated IFN induction. Differences in IFN induction between sgRNA targeting different sites of the target HPRT gene (constucts C and G) suggest that the sgRNA sequence may contribute to the IFN induction. These data also suggest that HEK293 cells can respond to sgRNA through the RIG-I pathway, which is reported to depend on the presence of 5’-triphosphate [33,34]. Moreover, the data suggest that HEK293 cells do not contain TLR9, another endosomal nucleic acid–sensing protein, because ODN2216, a known potent TLR9 agonist, does not induce IFNs in the HEK293 cells (data not shown). It is interesting that cells showed visual evidence for cytotoxicity following phosphatase-treated IVT sgRNAs even though IFN secretion was not detected, suggesting that some other mechanism underlies the cytotoxicity.
We demonstrated that the induction of type I IFN responses in the human primary PBMC cultures by various gRNA constructs follows the same trends previously established for other traditional therapeutic oligonucleotides, such as siRNAs (Figures 3 and 4). We showed that IFN induction can be eliminated by 2’-modification of the ribose backbone (Figures 3C and 4C). We also found that removal of 5’-triphosphate from IVT sgRNAs reduced but did not eliminate their ability to stimulate an IFN response in human blood leukocytes (Figures 3D and 4D). Unlike in HEK293 cells, the delivery of gRNAs and sgRNAs into PBMCs did not result in a change in cell viability.
Conclusion
The results of our study suggest that the risk of undesirable pro-inflammatory responses to gRNA can be minimized by optimizing the chemical composition. 2’OMe RNA modification of chemically synthesized gRNA is advisable when one desires to block IFN response. The removal of 5’-triphosphate in otherwise-unmodified IVT sgRNAs can reduce immune stimulation in the PBMCs and eliminate IFN responses in HEK293 cells. As no simple method exists to place 2’-modified residues at select positions within an IVT sgRNA, it is unlikely that this approach will have significant utility for therapeutic applications.
Conflicts Of Interest
MSS and MAB are employed by Integrated DNA Technologies (IDT), which sells reagents similar to some reported in this work. However, IDT is not a publicly traded company, and the authors do not own any shares or equity in IDT.
Acknowledgment
The study was supported in whole or in part by federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- Chin KL, Anis FZ, Sarmiento ME, Norazmi MN, Acosta A, et al. (2017) Role of Interferons in the Development of Diagnostics, Vaccines, and Therapy for Tuberculosis. J Immunol Res 2017: 5212910.
- Negishi H, Taniguchi T, Yanai H (2017) The Interferon (IFN) Class of Cytokines and the IFN Regulatory Factor (IRF) Transcription Factor Family. Cold Spring Harb Perspect Biol pii: a028423.
- Di Franco S, Turdo A, Todaro M, Stassi G (2017) Role of Type I and II Interferons in Colorectal Cancer and Melanoma. Front Immunol 8: 878.
- Picard C, Belot A (2017) Does type-I interferon drive systemic autoimmunity?. Autoimmun Rev 16: 897-902.
- Bessis N, GarciaCozar FJ, Boissier MC (2004) Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther 11: 10-17.
- Sakurai H, Kawabata K, Sakurai F, Nakagawa S, Mizuguchi H, et al. (2008) Innate immune response induced by gene delivery vectors. Int J Pharm 354: 9-15.
- Burel SA, Machemer T, Ragone FL, Kato H, Cauntay P, et al. (2012) Unique O-methoxyethyl ribose-DNA chimeric oligonucleotide induces an atypical melanoma differentiation-associated gene 5-dependent induction of type I interferon response. J Pharmacol Exp Ther 342: 150-162.
- Lennox KA, Behlke MA (2010) A direct comparison of anti-microRNA oligonucleotide potency. Pharm Res 27: 1788-1799.
- Watts JK, Yu D, Charisse K, Montaillier C, Potier P, et al. (2010) Effect of chemical modifications on modulation of gene expression by duplex antigene RNAs that are complementary to non-coding transcripts at gene promoters. Nucleic Acids Res 38: 5242-5259.
- Leeds JM, Henry SP, Geary R, Burckin T, Levin AA, et al. (2000) Comparison of the pharmacokinetics of subcutaneous and intravenous administration of a phosphorothioate oligodeoxynucleotide in cynomolgus monkeys. Antisense Nucleic Acid Drug Dev 10: 435-441.
- Behlke MA (2008) Chemical Modification of siRNAs for in vivo Use. Oligonucleotides 18: 305-319.
- Jacobi AM, Rettig GR, Turk R, Collingwood MA, Zeiner SA, et al. (2017) Simplified CRISPR tools for efficient genome editing and streamlined protocols for their delivery into mammalian cells and mouse zygotes. Methods 121-122: 16-28.
- Neun BW, Dobrovolskaia MA (2011) Detection and quantitative evaluation of endotoxin contamination in nanoparticle formulations by LAL-based assays. Methods Mol Biol 697: 121-130.
- Neun BW, Dobrovolskaia MA (2018) Considerations and Some Practical Solutions to Overcome Nanoparticle Interference with LAL Assays and to Avoid Endotoxin Contamination in Nanoformulations. Methods Mol Biol 1682: 23-33.
- Potter TM, Neun BW, Rodriguez JC, Ilinskaya AN, Dobrovolskaia MA, et al. (2018) Analysis of Pro-inflammatory Cytokine and Type II Interferon Induction by Nanoparticles. Methods Mol Biol 1682: 173-187.
- Fennrich S, Hennig U, Toliashvili L, Schlensak C, Wendel HP, et al. (2016) More than 70 years of pyrogen detection: Current state and future perspectives. Altern Lab Anim 44: 239-253.
- Coch C, Luck C, Schwickart A, Putschli B, Renn M, et al. (2013) A human in vitro whole blood assay to predict the systemic cytokine response to therapeutic oligonucleotides including siRNA. PloS one 8: e71057.
- Daneshian M, Von Aulock S, Hartung T (2009) Assessment of pyrogenic contaminations with validated human whole-blood assay. Nature protocols 4: 1709-1721.
- Dobrovolskaia MA (2015) Pre-clinical immunotoxicity studies of nanotechnology-formulated drugs: Challenges, considerations and strategy. J Control Release 220: 571-583.
- Hasiwa N, Daneshian M, Bruegger P, Fennrich S, Hochadel A, et al. (2013) Evidence for the detection of non-endotoxin pyrogens by the whole blood monocyte activation test. ALTEX 30: 169-208.
- Hoffmann S, Peterbauer A, Schindler S, Fennrich S, Poole S, et al. (2005) International validation of novel pyrogen tests based on human monocytoid cells. J Immunol Methods 298: 161-173.
- Vessillier S, Eastwood D, Fox B, Sathish J, Sethu S, et al. (2015) Cytokine release assays for the prediction of therapeutic mAb safety in first-in man trials--Whole blood cytokine release assays are poorly predictive for TGN1412 cytokine storm. J Immunol Methods 424: 43-52.
- Alvarez-Salas LM (2008) Nucleic acids as therapeutic agents. Curr Top Med Chem 8: 1379-1404.
- Henry SP, Geary RS, Yu R, Levin AA (2001) Drug properties of second-generation antisense oligonucleotides: how do they measure up to their predecessors?. Curr Opin Investig Drugs 2: 1444-1449.
- Henry SP, Monteith D, Bennett F, Levin AA (1997) Toxicological and pharmacokinetic properties of chemically modified antisense oligonucleotide inhibitors of PKC-alpha and C-raf kinase. Anticancer Drug Des 12: 409-420.
- Henry SP, Monteith D, Levin AA (1997) Antisense oligonucleotide inhibitors for the treatment of cancer: 2. Toxicological properties of phosphorothioate oligodeoxynucleotides. Anticancer Drug Des 12: 395-408.
- Holmlund JT, Monia BP, Kwoh TJ, Dorr FA (1999) Toward antisense oligonucleotide therapy for cancer: ISIS compounds in clinical development. Curr Opin Mol Ther 1: 372-385.
- Levin AA (1999) A review of the issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim Biophys Acta 1489: 69-84.
- Monteith DK, Henry SP, Howard RB, Flournoy S, Levin AA, et al. (1997) Immune stimulation--a class effect of phosphorothioate oligodeoxynucleotides in rodents. Anticancer Drug Des 12: 421-432.
- Schubert D, Levin AA, Kornbrust D, Berman CL, Cavagnaro J, et al. (2012) The Oligonucleotide Safety Working Group (OSWG). Nucleic Acid Ther 22: 211-212.
- Evans DC, Watt AP, Nicoll-Griffith DA, Baillie TA (2004) Drug-protein adducts: an industry perspective on minimizing the potential for drug bioactivation in drug discovery and development. Chem Res Toxicol 17: 3-16.
- Keefe AD, Pai S, Ellington A (2010) Aptamers as therapeutics. Nat Rev Drug Discov 9: 537-550.
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, et al. (2009) Recognition of 5' triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. J Immunity 31: 25-34.
- Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, et al. (2009) 5'-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A 106: 12067-12072.
Citation: Schubert MS, Cedrone E, Neun B, Behlke MA, Dobrovolskaia MA, et al. (2018) Chemical Modification of CRISPR gRNAs Eliminate type I Interferon Responses in Human Peripheral Blood Mononuclear Cells. J Cytokine Biol 3: 121. DOI: 10.4172/2576-3881.1000121
Copyright: © 2018 Schubert MS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 7286
- [From(publication date): 0-2018 - Dec 22, 2025]
- Breakdown by view type
- HTML page views: 6205
- PDF downloads: 1081