Cytokine-Dependent Expression Regulation of ALOX15
Received: 31-Mar-2016 / Accepted Date: 26-May-2016 / Published Date: 31-May-2016 DOI: 10.4172/2576-3881.1000106
Abstract
Lipoxygenases (LOX) are lipid-peroxidizing enzymes that play a role in cell differentiation, but have also been implicated in the pathogenesis of inflammatory, hyperproliferative and neurological disorders. They are widely distributed in plants and mammals but also occur sporadically in lower organisms. The human genome involves six functional LOX genes and a corrupted pseudogene. 20 years ago it was reported that expression of ALOX15 was specifically induced in human peripheral monocytes by the classical Th2 cytokine interleukin 4 and later expression array profiles indicated that this enzyme is the most strongly upregulated gene product in human monocytes. Although the molecular basis for this IL4-dependent expression regulation has extensively been studied during the past 20 years, there are still a number of unsolved questions. This review is aimed at summarizing the current knowledge on the cytokine-dependent expression regulation of ALOX15 with particular focus on the Th2 cytokines interleukin-4 and interleukin-13 in various cells and tissues and at critically evaluating the potential biological implication of this effect.
Keywords: Eicosanoids; Lipoxygenase; Leukotrienes; Inflammation; Energy metabolism
6246Introduction
According to the IUPAC recommendations lipoxygenases (LOXs) are classified as fatty acid dioxygenases since they introduce one molecule of atmospheric oxygen into the hydrocarbon tail of fatty acids [1,2]. LOXs are widely distributed in plants and mammals but they only occur sporadically in bacteria, fungi and lower marine organisms [3]. The first mammalian LOXs have been described in the mid 1970 in human blood platelets [4,5] and rabbit reticulocytes [6]. Since then a large number of LOX-isoforms have been described in various animal species. Complete sequencing of the human genome indicated the existence of six different LOX genes (ALOX5, ALOX12, ALOX12B, ALOX15, ALOX15B, ALOXE3), which encode for functionally distinct LOX-isoforms [7]. In addition, a corrupted pseudogene (7) has been detected in the human genome (ALOXE12 ). Free polyenoic fatty acids, such as linoleic acid (9,12-octadecadienoic acid), alpha- (9,12,15-octadecatrienoic acid) and gamma- (6,9,12- octadecatrienoic acid) linolenic acid, arachidonic acid (5,8,11,14- eicosatetraenoic acid), 5,8,11,14,17-eicosapentaenoic acid and 4,7,10,13,16,19-docosahexaenoic acid are preferred substrates of mammalian LOXs. Since free fatty acids do not abundantly occur in most mammalian cells the LOX-pathway is initiated by enzymatic hydrolysis of membrane ester lipids [1]. The primary products of the LOX reaction, the hydroperoxy fatty acids, are subsequently converted to a large array of bioactive mediators, which include leukotrienes [1], lipoxins [8], hepoxilins [9], eoxins [10], resolvins [11], protectins [12] and others. However, LOXs exhibit their biological functions not only by producing bioactive mediators. As oxidizing enzymes they are also involved in the regulation of the cellular redox homeostasis, which strongly impacts the gene expression pattern. Moreover, since ALOX15 orthologs are capable of oxidizing complex ester lipids such as phospholipids [13] and cholesterol esters [14] they modify the functional characteristics of biomembranes and lipoproteins.
The expression of LOX-isoforms in general and of ALOX15 in particular is highly regulated on transcriptional and translational levels [15]. In 1992 it was first reported that expression of ALOX15 can be specifically induced in isolated human monocytes by the Th2-cytokine interleukin-4 (IL4) [16]. Two years later [17] another classical Th2 cytokine, interleukin 13 (IL13), was shown to upregulate ALOX15 expression. Although the mechanistic basis for the IL4/IL13- dependent expression regulation has not extensively been studied in these early reports these papers mark the beginning of a new chapter in LOX research. A quick PubMed search with the key words “lipoxygenase” and “interleukin” revealed some 720 entries over the past 25 years. This review is aimed at summarizing the current knowledge on the cytokine-dependent expression regulation of ALOX15 with particular emphasis on the Th2 cytokines IL4 and IL13. We will also critically evaluate the suggested mechanistic scenarios and the applicability of the robust in vitro effects for the in vivo situation.
The multiplicity of published and unpublished experimental data on this topic makes it impossible to consider all reports for this review. Although we did our best to balance the list of cited references, we might have overlooked important contributions. We apologize to those distinguished colleagues who significantly contributed to the field but whose work has not been referenced.
Lipoxygenases (LOX) As Lipid Peroxidizing Enzymes
LOXs are fatty acid dioxygenases, which introduce molecular dioxygen into the hydrocarbon chain of polyunsaturated fatty acids. The LOX-reaction consists of four elementary reactions (hydrogen abstraction, radical rearrangement, oxygen insertion, radical reduction) and leads to the formation of specific lipid hydroperoxides [18]. The stereochemistry of all four elementary reactions is tightly controlled so that from the mixture of theoretically possible oxygenation products only one isomer is selectively formed. The product specificity depends on the chemical identity of the LOXisoform. For instance, human ALOX5 [19] converts arachidonic acid predominantly to 5S- hydroperoxy-6E,8Z,11Z,14Z-eicosatetraenoic acid (5S-HpETE), whereas other product isomers such as 5R-HpETE, 15S-HpETE or 11S/R-HpETE are not formed. In contrast, human ALOX12 [20] oxygenates arachidonic acid almost exclusively to 12SHpETE. The corresponding 12R-enantiomer and other isomers of hydroperoxy arachidonic acid are absent.
Diversity of LOX family and occurrence of LOX sequences in the domains of terrestrial life
LOXs form a very diverse family of enzymes. They occur in two (bacteria, eucarya) of the three domains of terrestrial life but have not been identified in archaea. Although a number of LOX-like sequences are present in various archaea there is no report on the expression of a functional LOX enzyme in this domain. Similarly, there are a number of LOX-like sequences in viral genomes but so far no functional viral LOX has been identified. We recently cloned a LOX-like sequence from a giant virus (mimivirus), which infects Acanthamoeba polyphaga, and expressed the recombinant enzyme in E. coli . This protein consists of 565 amino acids and thus, is some 100 amino acids smaller than typical mammalian LOXs. Interestingly, the amino acid sequence of the mimivirus protein (Figure 1) involves two potential iron-ligand clusters: Cluster 1: His-Met-Arg-Lys-Thr, cluster 2: His-Ser-Lys-Asn- His (the putative iron liganding amino acids are marked in bold). Most functional LOXs contain two of such iron ligand clusters. Although the distance between the two putative iron clusters is somewhat bigger as in most mammalian LOX they may still adopt a 3D conformation suitable for iron liganding.
Figure 1: Amino acid sequence of the putative LOX from the Acanthamoeba polyphaga mimivirus. The cDNA sequence of the putative LOX from the Acanthamoeba polyphaga mimivirus was retrieved from the NCBI database (ADO18315.1). The open reading frame encodes for a 565 amino acid protein that involves two potential iron ligand clusters and a C-terminal Ile (black background). The amino acids given in capital letters might constitute the potential proteinogenic iron ligands. The one letter code for amino acids is employed.
In addition, in most mammalian LOXs the C-terminal amino acid is an Ile, which functions a 5th proteinogenic iron ligand. The mimivirus protein does also carry C-terminal Ile and thus, should have a fully functional iron ligand sphere. We cloned the putative mimivirus LOX, expressed the corresponding protein as N-terminal his-tag fusion construct in E. coli and purified it by affinity chromatography on Niagarose. Unfortunately, when we incubated the purified enzyme with arachidonic acid or linoleic acid we could not detect the formation of typical LOX products. Moreover, when we determined the iron content of the purified enzyme we only quantified substoichiometric amounts of iron in the enzyme preparation (less than 10% of the expected value based on a 1:1 enzyme:iron stoichiometry). These data suggest that this LOX-like sequence of the Acanthamoeba polyphaga mimivirus does not encode a functional LOX and thus, there is still no report on the occurrence of a functional LOX in viruses.
Because of the diversity of the LOX family classification of these enzymes is a critical point. In the early days of LOX research the enzymes have been classified with respect to their reaction specificity of arachidonic acid oxygenation and three distinct enzyme subtypes (5-LOX, 12-LOX, 15-LOX) have been differentiated. Unfortunately, this arachidonic acid based classification is not applicable for all LOXisoforms. Some plant LOXs do not accept arachidonic acid as substrate [21] and LOX-isoforms from lower marine organisms exhibit different reaction specificities [22-25]. Another disadvantage of the specificitybased nomenclature system is that it does not consider evolutionary aspects of phylogenetic relatedness. For instance, human platelet 12- LOX (ALOX12) exhibits the same reaction specificity as mouse ALOX15 but with respect to their evolutionary relatedness these two enzymes are far apart. In contrast, human ALOX15 and mouse ALOX15 share a much higher degree of phylogenetic relatedness but exhibit different positional specificities. Since mouse ALOX15 converts arachidonic acid to 12S-HpETE and the human ortholog produces 15- HpETE, the two orthologous enzymes (functional equivalents in different species) are classified in different groups according to the specificity-based nomenclature system. This is highly confusing. On the other hand, the evolutionary concepts of LOX classification have the disadvantage that the degree of amino acid conservation is rather low when enzymes of evolutionary distant species are compared. For instance, the 12-lipoxygenating LOX1 of the zebrafish shares a similar degree of amino acid conservation with all human LOX-isoforms [26] and thus, it is impossible to assign this enzyme to any of the human isoforms based on the degree of amino acid conservation.
Mammalian ALOX15 orthologs
In mammals the situation is less confusing. As indicated above, the human genome involves 6 functional LOX genes and a corrupted pseudogen. In mice there is an ortholog for each human LOX gene (ALOX15, ALOX15b, ALOX12, ALOX12b, ALOX5, ALOXE3 ) but the human ALOXE12 pseudogen is functional (Aloxe12) in mice. Thus, mice express seven different LOX-isoforms. Functional inactivation of the different genes induced different phenotypes [7] and together with other findings these data suggest that the different LOX-isoforms exhibit different biological functions. Mammalian ALOX15 orthologs have been implicated in cell differentiation [27,28] but also in the pathogenesis of cardio- vascular [29,30], inflammatory [31], hyperproliferative [32] and neurological [33] diseases. Blood platelets of Alox12 knockout mice are more sensitive to agonist stimulation [34] and the water barrier of their epidermis [35] does not develop regularly. Alox5 has also been implicated in cardio-vascular disorders [36] but also plays a role in allergic diseases [37,38]. Alox12b and Aloxe3 are involved in epidermal differentiation and functional inactivation of the corresponding genes leads to dehydration of the newborns because of excessive loss of water [39,40]. In humans, mutations in the ALOX12B and ALOXE3 genes have been related to autosomal recessive congenital ichthyosis, a severe hereditary skin disease [41]. Moreover, Alox12b appears to be involved in adipocyte differentiation [42]. For ALOX15B and ALOXE12 knockout mice are currently not available.
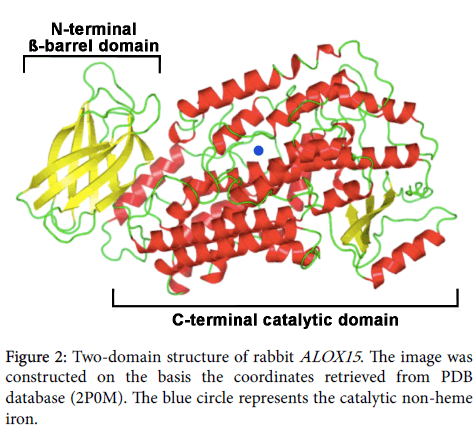
Protein chemical and structural properties of mammalian ALOX15 orthologs: Mammalian ALOX15 orthologs are single polypeptide chain proteins, which consist of some 650 amino acids. The polypeptide chain folds into a two-domain structure with a small (some 110 amino acids) N-terminal ß-barrel domain, which has been implicated in membrane binding and activity regulation [18,43]. The big (some 550 amino acids) alpha helical C-terminal domain harbors the catalytic center, which involves the catalytically active non-heme iron (Figure 2). In SDS PAGE these enzymes migrate in the 75 kDa molecular weight range. The two domains are covalently interconnected by a flexible linker peptide but tight together by non-covalent interactions. For rabbit ALOX15 it has been reported that the two structural subunits may move relatively to each other [44] and the degree of this interdomain movement depends on pH and salt concentration [45]. The N- terminal beta-barrel unit is not essential for catalytic activity, but gene technical deletion of this structural motif lowers the oxygenase activity and impairs membrane binding [46]. In its resting state mammalian ALOX15 orthologs contain ferrous non-heme iron at the active site and the iron ion is liganded by two clusters of amino acids (cluster A: His-A1-A2-A3-A4-His-, cluster B: His-B1-B2-B3- His), in which the His constitute the immediate iron ligands. The 5th proteinogenic iron ligand is the C-terminal Ile and truncation of this amino acid leads to a loss of the catalytic activity. A water molecule or a hydroxyl group completes the octahedral iron ligand sphere and this immediate iron ligand [47,48] is likely to be replaced by the fatty acid substrate. During the catalytic cycle the non-heme iron shuttles between its ferrous and its ferric form [43].
Enzymatic properties of mammalian ALOX15 orthologs: Although mammalian ALOX15 orthologs only have a single catalytic center they are multi- catalytic enzymes and exhibit at least three different catalytic activities [18]: i) lipoxygenase activity, ii) lipohydroperoxidase activity, iii) leukotriene synthasa activity. Which reaction is catalyzed at a given time point depends on the availability of the reaction substrates and on the reaction conditions. For instance, under aerobic conditions and when free polyenoic fatty acids are available, ALOX15 orthologs catalyze the lipoxygenase reaction converting the free polyenoic fatty acids to their corresponding hydroperoxides [49]. If these hydroperoxides still contain bisallylic methylene groups they can be lipoxygenated a second time, which leads to double or even multiple oxygenation products [50]. Such multiple oxygenation of free polyenoic fatty acids by different types of LOXs leads to the formation of lipoxins [51,52], resolvins [53], protectins [11] and maresins [12], which exhibit anti-inflammatory properties.
If no free polyenoic fatty acids are available ALOX15 orthologs can also oxygenate complex ester lipids such as phospholipids [6] and cholesterol esters [14] even if they are incorporated in complex lipidprotein assemblies such as biomembranes [13] and lipoproteins [54]. Although the biomembrane and lipoprotein oxygenase activity of ALOX15 orthologs is at least one order of magnitude lower than their fatty acid oxygenase activity it can clearly be measured and has been suggested to be of biological relevance [27,55].
Hydroperoxy lipids can further be converted in the presence of polyenoic fatty acids to secondary hydroperoxidase products, such as short chain aldehydes, alkanes, ketodienes as well as oxygenated or non-oxygenated fatty acid dimers [56]. This hydroperoxidase activity of ALOX15 orthologs also involves valency shuttling of the non-heme iron. It is initiated by homolytic cleavage of the peroxy group by the ferrous enzyme, which is paralleled by oxidation of the enzyme to its ferric form. To start the next catalytic cycle the enzyme must be reduced back to its ferrous form and free polyenoic fatty acids might serve as reducing agents [57]. The lipohydroperoxidase activity of ALOX15 orthologs is strongly favored under anaerobic conditions but can also be detected during hypoxia [58]. The leukotriene synthase activity of ALOX15 orthologs converts hydroperoxy fatty acids that still contain bisallylic methylenes, such as 15-HpETE, to secondary reaction products containing a conjugated triene system (50). This activity proceeds under aerobic and anaerobic conditions and does not require additional polyenoic fatty acids as reducing agents. The leukotriene synthase activity of ALOX15 orthologs has been implicated in the formation of eoxins, which serve as pro-inflammatory mediators in allergic diseases [10,59].
Among these catalytic activities the lipoxygenase activity with free polyenoic fatty acids as substrate has most comprehensively been characterized. The reaction kinetics are rather complex and a typical kinetic progress curve starts with a kinetic lag-phase, which has been related to peroxide- dependent enzyme activation [60]. At later time points the progress curve slows down and this is due to irreversible enzyme inactivation [61]. The molecular basis for this suicidal character of the ALOX15 reaction has not been clarified but covalent modification of the enzyme by reactive reaction intermediates has been suggested [62].
All naturally occurring omega-6 and omega-3 polyenoic fatty acids serve as suitable substrates for ALOX15 orthologs (low substrate specificity) but the reaction specificity of these enzymes is rather variable. ALOX15 orthologs of higher primates including men [49] and orangutans [63,64] exhibit dominant omega-6 oxygenase activity. In contrast, ALOX15 orthologs of lower mammals [mice [29,65], rats [66,67], pigs [68], cattles [69]] including lower primates [rhesus monkeys [63,64] exhibit major omega-9 oxygenase activity. Gibbons, which are flanked in evolution by rhesus monkeys on one side and orangutans on the other, express an ALOX15 ortholog that exhibits a pronounced dual positional specificity with almost equal distribution of omega-6 and omega-9 oxygenation (Adel et al., submitted). In other words, ALOX15 reaction specificity was systematically changed during late primate evolution from arachidonic acid 12-lipoxygenation in lower mammals to arachidonic 15-lipoxygenation in higher primates (Adel et al., submitted). The evolutionary driving force for this alteration remains to be explored but arachidonic acid 15- lipoxygenating ALOX15 orthologs have a higher biosynthetic capacity for anti-inflammatory lipoxins. Thus, this evolutionary switch in reaction specificity might be aimed at optimizing inflammatory resolution and thus may be considered a mechanism for fine-tuning the immune system of higher primates (Adel et al., submitted).
Mammalian ALOX15 genes and tissue specific gene expression: The gene encoding human ALOX15 is localized on chromosome 17 in a joint gene cluster, which also involves the genes for ALOX15B, ALOX12, ALOX12B and ALOXE3. The only human ALOX gene, which is not localized in this gene cluster, is that encoding for ALOX5 . The human ALOX15 gene consists of 14 exons and 13 introns. The mouse ALOX 15 gene has a very similar exon/intron structure and is located in a syntenic region on chromosome 11. The genes encoding for other mammalian ALOX15 orthologs look very similar. The promoter region of the human ALOX15 gene [70] involves a number of potential transcription factor binding sites but lacks classical TATAand CAAT boxes. However, it remains to be explored, which of these putative regulatory sequences are functional in vivo . When we evaluated the sequence data deposited in the database of the 1000 human genome project (www.1000genomes.org) we detected 78 single nucleotide polymorphisms in the ALOX15 gene (allelic frequency >1%). These data indicate an average genetic variability [71]. In addition, some 90 non-synonymous coding variations and eight nonsense mutations have been identified, which occur with lower genetic frequency [71]. Functionally important amino acids, such as the immediate iron ligands and the reaction specificity determinants are rarely affected by amino acid exchanges [71].
Human ALOX15 is high level expressed in reticulocytes, eosinophils and in airway epithelial cells [72]. Low level expression has been detected in polymorphonuclear leukocytes [73,74], alveolar macrophages [75], vascular cells [76,77], uterus [78], the male reproductive system [79], various parts of the brain [80,81] and in atherosclerotic lesions [82]. Resting human peripheral monocytes as well as B- and T-lymphocytes do not express significant amounts of ALOX15 . In some human cancer cells ALOX15 expression is silenced but can be reactivated by histone deacetylase inhibitors [83,84]. Mechanistic studies implicated the nucleosome remodeling and histone deacetylase repression complex (NuRD) in this transcriptional regulation of ALOX15 expression. In cancer cells NuRD appears to be recruited to the ALOX15 promoter, which silences ALOX15 expression [84].
In rabbits, a similar constitutive expression pattern of ALOX15 was observed but experimental anemia strongly upregulated ALOX15 expression in many cells and tissues including peripheral monocytes, lung, spleen, kidney and liver [85]. However, since the organs were not carefully saline- perfused to remove blood it might well be that the ALOX15 activity detected in these organs may originate from contaminating reticulocytes. When elicited rabbit peritoneal macrophages were incubated with arachidonic acid, 5S-HETE and leukotriene B4 were identified as major oxygenation products. In contrast, only small amounts of 15-HETE were found. When similar experiments were carried out with cell homogenates formation of LTB4 and 5S-HETE was strongly down and 15-HETE was the major arachidonic acid oxygenation product (Kuhn et al., unpublished data). Unfortunately, the mechanistic basis for this switch in reaction specificity has not been explored.
In mice, peripheral eosinophils are a rich source of ALOX15 expression but as in humans the biological relevance of high-level ALOX15 expression in these cells remains elusive. Similarly, high expression levels of ALOX15 have been reported in residential peritoneal macrophages [29] and immunohistochemistry indicated that more than 80% of these cells were ALOX15 positive. However, thioglycollate elicitation strongly decreased the share of ALOX15 positive cells to about 10% [86]. Mouse blood monocytes, mouse alveolar macrophages and mouse bone marrow derived macrophages express ALOX15 only at low levels [86], but the airway epithelium, which constitutes a major site of human ALOX15 expression in humans [72] and cattle [69], has not been tested. Taken together, these data indicate that mammalian ALOX15 orthologs exhibit speciesspecific expression patterns but the mechanistic details for the differences have not been explored in detail.
Biological implication of mammalian ALOX15 : Mammalian ALOX15 orthologs have been implicated in cell differentiation and maturation [red blood cells [87], epidermis [88,89], adipose tissue [42], sperms [90], neurons [91]] but they also play a role in the pathogenesis of various diseases [2]. A comprehensive review on the biological role of ALOX15 has recently been published [2] and thus, there is no need to discuss this topic extensively in this paper. In various diseases ALOX15 orthologs exhibit antagonizing activities depending on the disease model. For instance, in different types of animal inflammation models pro- [10,92,93] and anti- inflammatory [31,94] activities of ALOX15 have been reported. A similar situation was found in animal atherosclerosis models and in different types of cancer. Experiments with ALOX15 knockout mice suggested a pro-atherogenic activity of the enzyme [29,95-97] but overexpression studies revealed antiatherogenic effects [85,98,99]. ALOX15 overexpressing transgenics were protected from tumor growth and metastasis in two different mouse cancer models [100,101] and in different colorectal carcinoma cell lines ALOX15 also exhibited anti-tumor properties [32,101]. On the other hand, overexpression of ALOX15 in HCT116 colon carcinoma cells induced activation of ERK signaling, which upregulated the rate of cell proliferation. Treatment of these cells with a non-specific ALOX inhibitor blocked ERK activation, which is consistent with the pro-carcinogenic activity of the enzyme [102].
Cytokine-dependent Expression Regulation of Human ALOX15
IL4- and IL13-dependent expression regulation of ALOX15 in human monocytes
In human circulating blood monocytes ALOX15 is not expressed. However, when these cells were cultured in vitro in the presence of recombinant human IL4 (60-600 pM, which corresponds to 0.9-9 ng/ml) for 3 days ALOX15 expression was strongly upregulated as indicated by immunohistochemistry, Northern blotting and activity assays [16]. The induced enzyme reacted with endogenous substrates since specific ALOX15 products (15S-HETE) were detected in the membrane lipids. IL4-dependent ALOX15 expression was timedependent and maximal induction was reached after incubation periods longer than 48 h. These slow expression kinetics suggest that ALOX15 does not belong to the immediate early genes of the IL4 response [16]. Two years later [17] it was reported that IL13, another classical Th2 cytokine, does also upregulate ALOX15 expression. To explore the cell physiological context of IL4/13-induced ALOX15 expression microarray-based expression profiles were recorded with human peripheral monocytes [103]. After 3 days of continuous IL4/13 exposure the six most strongly upregulated gene products were the following: ALOX15 , fibronectin, monoamine oxidase-A, CD1c, CD23A and the coagulation factor XIII (transglutaminase). In fact, in IL4 treated monocytes ALOX15 mRNA was almost 300-fold higher than in IL4-deficient control incubations [103]. In contrast to upregulation of ALOX15 expression a number of classical proinflammatory gene products, such as tumor necrosis factor alpha, monocyte chemotactic protein-1, IL1, IL6, IL8, IL18, cyclooxygenase-2, as well as enzymes and receptors of the leukotriene signaling cascade (ALOX5, ALOX5-activating protein, leukotriene B4 receptors, cysteinyl leukotriene receptor 2) were significantly downregulated. These expression regulation profiles are consistent with the hypothesis that IL4 treatment forces peripheral monocytes to adopt a resolving phenotype [103].
Specificity of cytokine-dependent expression regulation of LOXisoforms: To explore the specificity of cytokine-dependent expression regulation of LOXs, two aspects have been investigated: i) Cytokinespecificity of ALOX15 induction and ii) LOX-isoform specificity of IL4/13 induction.
i) ALOX15 induction in human peripheral monocytes was IL4/13- selective since other cytokines such as interleukins-1, -2, -3, -5 and -6 (IL1, IL2, IL3, IL5, IL6) as well as interferon-gamma (IFN-g), granulocyte monocyte colony stimulating factor (GM-CSF), monocyte colony stimulating factor (M- CSF), platelet derived growth factor (PDGF), tumor necrosis factor (TNF), transforming growth factor (TGF) and phorbol myristate acetate (PMA) did not induce ALOX15 expression [16]. Interestingly, incubation of the cells with IL4 in the presence of PMA or INFg did not lead to increased ALOX15 expression suggesting an antagonizing effect of these cytokines [16]. IL10 is another classical Th2 cytokine that exhibits anti-inflammatory properties [104]. It binds to several subtypes of a cell surface receptor and activates STAT3–dependent intracellular signaling cascades. In contrast to IL4/13 this Th2-cytokine does not induce ALOX15 expression in monocytes [17]. Erythropoietin is a hematopoietic cytokine, which regulates erythropoiesis on different levels [105]. It also exhibits activities outside the erythroid lineage [105] but does not induce expression of ALOX15 in cultured human peripheral monocytes nor in A549 cells (H. Kuhn, unpublished data).
ii) To explore whether IL4/13 do also regulate the expression of other LOX-isoforms in human monocytes ALOX12, ALOX15 and ALOX15B mRNA and protein was quantified during in vitro differentiation of isolated human monocytes [106]. Expression of ALOX15B was strongly upregulated (up to 10-fold) during the time course of in vitro incubation of the cells in the absence of any cytokines. This was not the case for ALOX15 and ALOX12. In the presence of 10 ng/ml IL4/13, expression of ALOX15 and ALOX15B was further upregulated whereas ALOX12 expression was hardly altered. Comparison of the extent of IL4/13-dependent expression regulation of ALOX15 and ALOX15B (fold change of mRNA concentrations in the absence and presence of cytokines) indicated that ALOX15 was more strongly induced. In fact, in the presence of IL4 ALOX15 expression was 300-fold upregulated whereas ALOX15B was only less than 3-fold up. A similar situation was observed for IL13 (80-fold upregulation of ALOX15 vs. 3-fold upregulation of ALOX15B ). These data are consistent with previous microarray and qRT-PCR data [103]. Interestingly however, if one quantifies the absolute copy numbers of ALOX15 and ALOX15B mRNA in IL4 treated monocytes, similar expression levels were observed. In the presence of IL13, expression of ALOX15B mRNA was even dominating. Here the copy number of ALOX15B mRNA was higher than that of ALOX15 . Taken together, these data indicate that IL4 and IL13 do not only induce expression of ALOX15 in cultured human peripheral monocytes but also of ALOX15B . However, the extent of induction is higher for ALOX15 . For the time being, the molecular mechanism of IL4-dependent ALOX15B expression has not been explored [106]. The different extents of induction, the different induction kinetics and the observation that expression of ALOX15B is already upregulated in the absence of any cytokine during in vitro cell culture (this is not the case for ALOX15 ) suggest mechanistic differences in the expression regulation of the two LOX-isoforms.
Cytokine-dependent ALOX15 expression in other cells: To explore the mechanistic basis for IL4-dependent upregulation of ALOX15 expression, a number of permanent cell lines were screened: HL60 (human myeloblastic), U937 (human promyelomonocyte), THP1 (human monocytic), MonoMac6 (human monocyte/macrophage), J774 and P388.D1 (mouse monocytic), A549, HTB56, HTB54 (all human lung carcinoma), HTB43 (human squamous head and neck carcinoma), HTB38 (human colon carcinoma), HMC1 cells (human mast cell). Unfortunately, most of them did not respond with upregulation of ALOX15 expression when they were cultured in the presence of IL4 [107]. However, when the lung carcinoma cell line A549 was maintained in the presence of IL4 or IL13 for 24 h or longer ALOX15 was expressed as indicated by RT-PCR, immunohistochemistry and activity assays. This effect was ALOX15 specific, since expression of ALOX5 and ALOX12 remained low. The IL4 mutant Y124D, which constitutes an IL4 receptor antagonist, counteracted the effect of the wild-type cytokine. These data indicate that IL4- dependent expression regulation is cell specific and involves in A549 cells binding of the cytokine to the IL4/13 cell surface receptor [107].
IL4 and IL13 have been implicated in transdifferentiation of dendritic cells from hematopoietic precursor cells [108]. In the absence of IL4, dendritic cells that had been generated from CD34- positive precursors in response to a mixture of stem cell factor, granulocytemacrophage colony stimulating factor and tumor necrosis factor alpha expressed high levels of ALOX5 and ALOX5 activating protein but hardly any ALOX15 . Addition of IL4 to the cytokine mixture led to selective downregulation of ALOX5 and strong upregulation of ALOX15 [108]. Transforming growth factor beta1 counteracted the IL4-dependent suppression of ALOX5 expression but did not alter ALOX15 expression. These findings were consistent with the results of metabolomic studies, which indicated that in the absence of IL4 5- HETE and leukotriene B4 were the major eicosanoids produced by dendritic cells derived from CD34-positive precursors. In contrast, 15- HETE and 5S,15S-diHETE were the major eicosanoids formed in the presence of IL4. These data indicate that dendritic cells, which were transdifferentiated from peripheral monocytes in vitro in the presence of IL4, express large amounts of ALOX15 [108]. However, it remains to be explored whether this effect is of any in vivo relevance. For the time being there is no convincing experimental evidence that in vivo differentiated dendritic cells express large amounts of ALOX15.
Orbital fibroblasts play a major role in tissue remodeling and have been implicated in the pathogenesis of Grave’s disease, an autoimmune inflammatory disorder, which affects the orbit around the eye. Upper eyelid retraction, lid lag, bulging eyes and conjunctivitis are characteristic symptoms of this disease [109]. Resting orbital fibroblasts do not express ALOX15 , but in vitro incubation of these cells in the presence of IL4/13 induce expression of the enzyme [110]. The intracellular signaling cascade involves Jak2 signaling since transient transfection of the cells with a dominant negative Jak2 mutant abolished ALOX15 expression. Interestingly, IL4/13-dependent ALOX15 induction was not detectable in dermal fibroblasts although these cells express the IL4 receptor and other elements of the intracellular IL4 signaling cascade [110].
Resting human umbilical vein endothelial cells (HUVEC) do not express ALOX15 but when cultured in the presence of IL4 transcription of the ALOX15 gene is induced [111]. However, activity assays and Western blotting did not provide any evidence for presence of the functional enzyme. Electrophoretic mobility shift assays indicated the activation of several transcription factors (STAT6, AP2, SP1, NF1), which have been implicated in expression regulation of ALOX15 and these data suggest a functional IL4 signaling cascade in these cells [111]. The most plausible explanation for the apparent contradiction (presence of ALOX15 mRNA vs. absence of functional ALOX15 protein) is that IL4 in HUVEC induces transcription of the ALOX15 gene, but that translation of the corresponding ALOX15 mRNA is silenced. In immature red blood cells translation of ALOX15 mRNA is inactivated by the binding of regulatory proteins to a repetitive sequence motif in the 3’-untranslated region of the mRNA [112,113]. If similar mechanisms of post-transcriptional regulation of ALOX15 expression also occur in vascular endothelial cells, the experimental data obtained in this cellular system become plausible. Unfortunately, the presence of downregulatory proteins under these experimental conditions has not been explored.
For adipocytes, IL4 has been implicated in energy homeostasis [114]. It inhibits adipogenesis by downregulating the expression of peroxisome proliferator-activated receptor-gamma and the CCAAT/ enhancer-binding protein-alpha [114]. In addition, IL4 upregulates the activity of hormone sensitive lipase (HSL) and thus, enhances lipolysis [114]. Unfortunately, it has not been explored yet whether ALOX15 expression is upregulated in this cellular system and whether the enzymes might be involved in any of these processes.
Molecular mechanism of IL4/13-dependent expression regulation of ALOX15 : IL4 and IL13 are two classical Th2 cytokines, which frequently exhibit anti-inflammatory activities. Both cytokines are capable of inducing ALOX15 expression in peripheral human monocytes but the intracellular signaling mechanisms in these cells are distinct [115]: i) IL4 induces Jak1 activation whereas the IL13- dependent signaling cascade involves Jak2 and Tyk2. ii) Tyk2 (IL13 response) controls STAT1 and STAT6 activation whereas Jak1 (IL4 response) upregulates STAT3 and STAT6. iii) IL13 utilizes both, the IL-4Ralpha/Jak2/Stat3 and IL-13Ralpha1/Tyk2/Stat1/Stat6 signaling pathways but IL4 can only employ the IL-4Ralpha/Jak1/Stat3/Stat6 cascade to upregulate the expression of ALOX15 .
IL4-induced signalling: IL4 is typically liberated by activated Th2 cells (116). It binds to a cell surface receptor (IL4R), which consists of the common cytokine-receptor gamma subunit and the IL-4-binding chain (IL4Rα). IL4R is mainly expressed on naive T cells and signals via activation of various transcription factors such as STAT6 (signal transduction and activator of transcription 6), GATA3 (GATA binding transcription factor-3), NFAT (nuclear factor of activated T cells), activating protein-1 (AP1) and others. Activation of these transcription factors upregulates expression of the constituents of the IL4 gene cluster on chromosome 5, which involves the genes encoding for IL4, IL5, and IL13 genes [116]. Thus, stimulation of naive T-cells with IL4 induces IL4 expression by these cells (autocrine loop), which strongly accelerates Th2 cell differentiation. However, Th2 cells do not express ALOX15 and thus, the enzyme is unlikely to play a major role in T-cell differentiation.
The ALOX15 promoter involves putative STAT6 binding sites [117] and serial promoter deletion studies as well as STAT6 binding site mutations suggested their functionality [118]. When we studied the molecular mechanisms of IL4-dependent expression regulation in A459 cells, we found that genistein (tyrosine kinase inhibitor) reduced phopsphorylation of STAT6 and its recruitment to the ALOX15 promoter [119]. Moreover, IL4 activated the histone acetyltransferase activity of the CREB-binding protein (CBP)/p300, which catalyzes acetylation of nuclear histones and of STAT6. This protein acetylation appears to be essential for the IL4-induced signaling cascade leading to ALOX15 expression, since inhibition of the acetyltransferase activity of CBP/p300 by the E1A oncoprotein reduced histone and STAT6 acetylation and blocked transcriptional activation of the ALOX15 gene. We also observed that the inhibition of histone deacetylases (addition of sodium butyrate) enhanced the IL4 induced ALOX15 expression. Taken together, these data implicated STAT6 and the structure of nuclear histones in transcriptional activation of the ALOX15 gene [119]. The critical role of the histone structure for IL4-dependent expression regulation of ALOX15 was confirmed by recent studies on histone H3 trimethyl-lysine 27 (H3K27me3), which apparently interacts with the ALOX15 promoter [120]. When A549 cells were incubated with IL4 demethylation of H3K27me3 occurs. This demethylation is catalyzed by a H3K27me2/3-specific demethylase (UTX) and siRNA-induced expression knockdown of UTX significantly attenuated H3K27me3 demethylation and ALOX15 expression. The critical role of UTX in ALOX15 expression was confirmed in human peripheral monocytes but here the H3K27me3- demethylase activity does not play a major role [120].
To identify additional regulatory elements in the IL4-induced signaling cascade, which lead to upregulation of ALOX15 expression, differential protein binding studies were carried out [121]. For this purpose protein extracts of IL4-treated A549 cells and corresponding untreated controls were added to various promoter constructs and the binding proteins as well as their tryptic digest fragments were analyzed by MALDI-MS. The obtained proteinomic data identified the two major binding proteins as the Lupus KU autoantigens P86 and P70. Gel shift and supershift experiments employing monoclonal anti-Ku antibodies confirmed the physical interaction of the Ku antigens with the ALOX15 promoter and electroporation of neutralizing anti-Ku antibodies into A549 cells suppressed IL4-induced ALOX15 expression [121].
As indicated above, IL4 has been related to the energy metabolism of adipocytes [114]. AMP- activated protein kinase (AMPK) is a key enzyme in the energy metabolism of all cells [122] but until recently no connection between IL4-dependent induction of ALOX15 expression and AMPK has been described. Using primary human macrophages cultured in vivo in the presence of IL4 it has recently been shown, that activation of AMPK attenuated expression of ALOX15 mRNA and protein [123]. Activators of AMPK (phenformin, aminoimidazole-4- carboxamide-1-ß-D-ribofuranoside) also inhibited IL4-induced activation of STAT3, suggesting the involvement of this transcription factor in the IL4- dependent signaling cascade. Moreover, activation of AMPK (phenformin) prevented IL4-induced association of STAT6 and acetylation of histone H3 at the ALOX15 promoter (123). Taken together, these data suggest that activation of AMPK, which usually occurs under the conditions of energy deficiency, suppresses IL4- induced ALOX15 expression and thus, interconnects the energy metabolism of cellular systems with the ALOX15 pathway. Energy deficiency prevents IL4-dependent ALOX15 expression or, if one sees it the other way around, IL4-dependent ALOX15 expression might only occur in energy sufficient cells. Thus, the nutrition state of cells might impact their responsiveness to react with ALOX15 expression when stimulated with IL4.
IL13-induced signalling: IL13 induces ALOX15 expression in human peripheral monocytes [17] to a lesser extent than IL4 [106,124]. The IL13 induced intracellular signaling cascade, which leads to induction of ALOX15 expression, is rather diverse and involves Jak2 and Tyk2 kinases as well as the transcription factors STAT 1, 3, 5, and 6 [125]. When cells are stimulated with IL13, serine phosphorylation of both STAT1 and STAT3, as well as activation of p38 mitogen-activated protein kinase (MAPK) was detected. Pharmacological inhibition of the MAPK pathway inhibited IL13-induced STAT1 and STAT3 phosphorylation as well as the DNA binding capabilities of the transcription factors [125]. These data suggest that IL13 induces p38 MAPK activation that upregulates STAT1 and STAT3 phosphorylation, which in turn activates the ALOX15 gene.
In addition to MAPK another serine/threonine kinase, PKCdelta, has also been implicated in IL13-induced ALOX15 expression [126]. When cells are treated with IL13, PKCdelta was rapidly phosphorylated. Rottlerin, an isoform specific PKCdelta inhibitor, blocked IL13-induced ALOX15 expression but inhibitors of other PKC-isoforms were ineffective. Expression silencing of PKCdelta by siRNA oligonucleotides also inhibited ALOX15 expression [126]. Interestingly, IL13-mediated activation of PKCdelta on one hand and IL13-dependent activation of p38 MAPK on the other are independent pathways, since inhibition of one kinase activity had no effect on the other. These data suggest that the two pathways act in parallel and may variably contribute to upregulation of ALOX15 expression [126]. It remains to be worked out under which conditions the MAPK or the PKCdelta pathways prevail.
To make the situation even more complex, an ERK1/2-dependent signaling pathway has also been described [127]. When monocytes are exposed to IL13, rapid phosphorylation of ERK1/2 can be measured. Tyk2 kinase is required for ERK1/2 phosphorylation, which is independent of Jak2, p38MAPK and PKCdelta. To investigate the signaling mechanisms in more detail the possible involvement of various transcription factors was also explored. In these experiments IL13 induces nuclear accumulation of the erythroblast transformationspecific gene related protein-1 (EGR1) and phosphorylation of the cAMP response element binding protein (CREB). These events are markedly attenuated by pharmacological inhibition of ERK1/2 [127]. Most importantly, expression silencing of EGR1 and CREB inhibited IL13-induced ALOX15 expression. These results indicate an important regulatory role for ERK1/2 in mediating IL13-induced expression of ALOX15 via the transcription factors EGR1 and CREB. This additional pathway broadens the intracellular multiplicity of IL13- induced signaling, which leads to upregulation of ALOX15 expression.
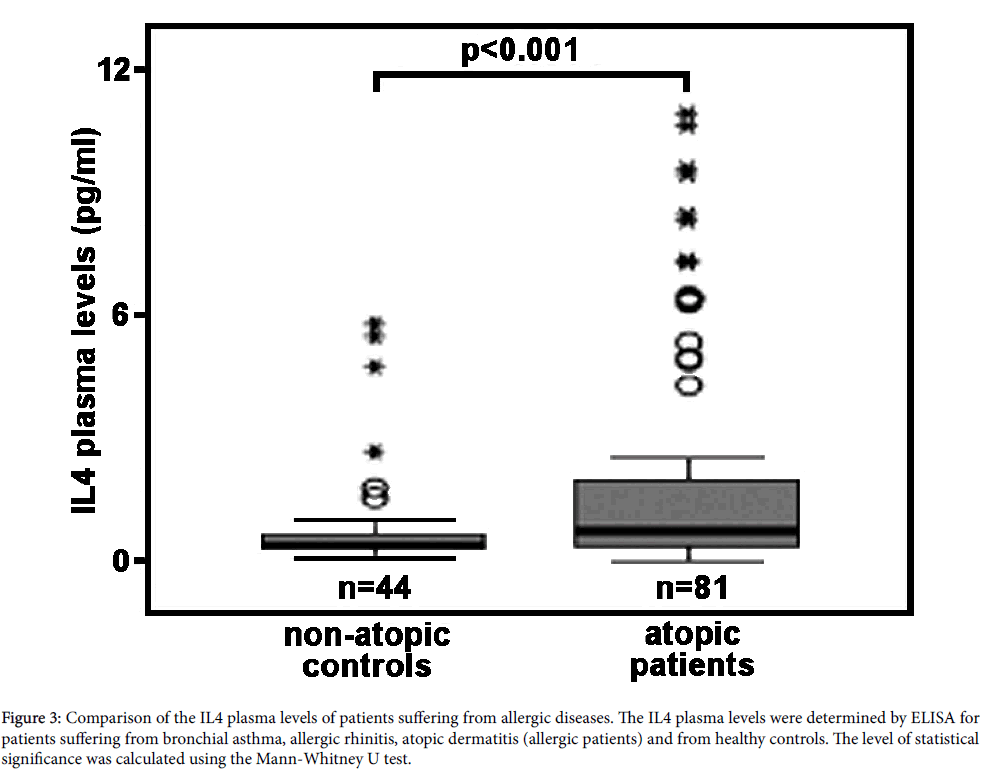
In vivo relevance of IL4-induced ALOX15 expression in humans: The majority of studies carried out so far on IL4/13-induced ALOX15 expression were performed in cellular in vitro systems, in which cultured human cells (primary cells or permanent cell lines) were incubated in the presence or absence of the cytokines. In most cases the observed effects were dramatic and robust but it still remains unclear whether they are of any in vivo relevance. IL4 is a classical Th2 cytokine, which is secreted in large amounts during the allergic reaction [128,129]. To answer the question of whether patients suffering from allergic diseases have elevated IL4 plasma levels and whether the IL4 concentrations reached under these conditions in vivo are sufficiently high to induce ALOX15 expression, we employed a dual research strategy. First, we compared the IL4 concentrations required for in vitro induction of ALOX15 in human monocytes with the plasma levels of IL4 measured in normal human beings and in patients with early rheumatoid arthritis. Here we found, that for in vitro induction of ALOX15 expression IL4 concentrations of 1-10 ng/ml are typically needed (Table 1). In contrast, measurements of the IL4 concentrations in healthy humans [130] and in patients suffering from early rheumatoid arthritis [131] revealed concentrations ranging from 5-10 pg/ml. When we determined the IL4 plasma levels (see below) in normal human beings and in patients suffering from allergic diseases (bronchial asthma, allergic rhinitis, atopic ekzema) we obtained values ranging between 0.1 to 11 pg/ml. Thus, the IL4 plasma concentrations determined in our (Figure 3) and in previous studies [130,131] are up to three orders of magnitude lower that the values required for in vitro induction of ALOX15.
| Reference | Cell type | IL4/13 (ng/ml) |
| Conrad et al. [16] | peripheral monocytes | 1-10 |
| Nassar et al. [17] | peripheral monocytes | 2 |
| Brinckmann et al. [107] | alveolar epithelial cells (A549) | 10 |
| Heydeck et al. [132] | peripheral monocytes, macrophages | 15 |
| Kelavkar et al. [121] | alveolar epithelial cells (A549) | 0.2 |
| Lee et al. [111] | human umbilical vein endothelial cells | 10-50 |
| Shankaranarayanan et al. [119] | alveolar epithelial cells (A549) | 10 |
| Spanbroek et al. [108] | dendritic cells (monocyte derived) | 50 |
| Chaitidis et al. [103] | peripheral monocytes | 10 |
| Chen et al. [110] | orbital fibroblasts | 10 |
| Wüst et al. [106] | peripheral monocytes | 10 |
| Bhattacharjee et al. [115] | periphera lmonocytes | 10 |
| Han et al. [120] | alveolar epithelial cells (A549) | 50 |
| Namgaladze et al. [123] | peripheral monocytes | 20 |
Table 1: IL4 concentrations used for in vitro induction of ALOX15 expression.
Figure 3: Comparison of the IL4 plasma levels of patients suffering from allergic diseases. The IL4 plasma levels were determined by ELISA for patients suffering from bronchial asthma, allergic rhinitis, atopic dermatitis (allergic patients) and from healthy controls. The level of statistical significance was calculated using the Mann-Whitney U test.
It should, however, been stressed that local IL4 concentrations, for instance in the bone marrow or in certain environments of peripheral tissue, might be much higher than equilibrium concentrations in the plasma. Moreover, unknown factors in the plasma might sensitize monocytes towards IL4. To address this problem we next quantified the arachidonic acid oxygenase activity of monocytes prepared from patients suffering from allergic diseases and from healthy controls. For this purpose we collected blood from patients suffering from different entities of allergic disorders (bronchial asthma, allergic rhinitis, atopic ekzema), prepared the peripheral monocytes and carried out in vitro activity assays measuring the formation of 15S-HETE from exogenous arachidonic acid. In parallel, we quantified the corresponding plasma levels of both IgE (readout parameter for the severity of the allergic disease) and IL4. As indicated in Table 2 the IgE levels of the patients suffering from the allergic diseases were significantly (p<0.001) elevated indicating the activity state of the disease. Next, we compared the IL4 plasma concentrations of the patients with those of healthy controls and found elevated IL4 levels in allergic patients (Figure 3). Although the extent of elevation was not dramatic, the difference was highly significant (p<0.001). Quantification of the arachidonic acid oxygenase activity of the monocytes revealed a higher activity of the “allergic monocytes” when compared with “healthy control” cells (Table 3). However, more detailed analysis of the chemical structure of the oxygenation products (SP-HPLC) indicated that these products did not originate from the ALOX15 pathway. In fact, a mixture of all major HETE isomers was detected and the 15-HETE identified in this mixture was racemic. These results are inconsistent with an ALOX15 origin of the oxygenation products. We also attempted to detect the ALOX15 protein in some of the monocyte preparations using our anti- ALOX15 antibody (immunoblotting) but the amounts of ALOX15 in the cells were below detection limits. Finally, we correlated the arachidonic acid oxygenase activity of the monocytes with the individual IL4 plasma concentrations but did not observe a significant correlation.
| IgE level (kU/ml) | n | median | 25% percentile | 75% percentile | Significance |
| Healthy controls | 44 | 12 | 6 | 30 | - |
| Allergic rhinitis | 46 | 1028 | 165 | 1265 | 0.001 |
| Bronchial asthma | 20 | 473 | 156 | 1060 | 0.001 |
| Atopic ekzema | 15 | 564 | 347 | 1412 | 0.001 |
Table 2: Plasma IgE concentrations of patients suffering from allergic diseases and of healthy controls. EDTA blood was drawn from the patients, the plasma was prepared and aliquots (5 μl) were used to quantify the IgE levels by solid phase enzyme immunoassay of DPC Biermann. The experimental raw data were evaluated with the Mann-Whitney U test and the significance values p were calculated in comparison to the healthy controls.
| IgE level (kU/ml) | n | median | 25% percentile | 75% percentile | Significance |
| Healthy conrols | 42 | 2.5 | 1.1 | 8.8 | - |
| Allergic rhinitis | 45 | 12.9 | 6.6 | 34.9 | <0.001 |
| Bronchial asthma | 19 | 6.9 | 2.4 | 13.6 | <0.001 |
| Atopic ekzema | 13 | 6.0 | 3.1 | 7.8 | <0.001 |
Table 3: Arachidonic acid oxygenation capacity of peripheral monocytes prepared from allergic patients and healthy controls. 50 ml of EDTA blood was drawn from the patients and the mononuclear cells were prepared by density gradient centrifugation (Ficoll gradient). The cells were plated in Petri-dishes overnight and the non-adhering cells were discarded. The adhering monocytes were scraped off, spun down and reconstituted in 1 ml of PBS. Arachidonic acid (100 μM final concentration) was added and the cells were incubated for 20 min at room temperature. 1 ml of methanol was added, protein precipitate was spun down and aliquots of the clear supernatant were injected to RP-HPLC for quantification of the arachidonic acid oxygenation products. A solvent system of methanol/water/acetic acid (85:15:0.1, by vol) was used to develop the chromatograms and the absorbance at 235 nm was recorded. The chromatographic scale was calibrated with known amounts of 15-HETE. The experimental raw data were evaluated with the Mann-Whitney U test and the significance values p were calculated in comparison to the healthy controls.
Consequently, one has to conclude that for the time being there is no experimental evidence for an increased expression of ALOX15 in the monocytes of allergic patients. Thus, it still remains to be explored of whether or not the IL4/13- dependent induction of ALOX15 expression is of any biological relevance in humans.
Regulation of ALOX15 in mice
ALOX15 expression in mouse monocytes and macrophages: In contrast to the human ortholog, mouse ALOX15 is a 12-lipoxygenating enzyme [65]. To explore whether human and mouse ALOX15 orthologs exhibit a similar sensitivity for cytokine dependent expression regulation, the impact of IL4/13 on ALOX15 expression was studied in mouse peritoneal macrophages and peripheral monocytes. Activity assays indicated that IL4 and IL13, but not IL10 upregulated ALOX15 activity in a dose-dependent manner [132]. Interferon gamma completely prevented the induction of ALOX15 [16,133]. In contrast, basal expression of ALOX15 in mouse peritoneal macrophages was not suppressed by interferon gamma [133]. Moreover, the time course of IL4-dependent expression regulation was dramatically different between men and mice. In human monocytederived macrophages ALOX15 expression is up for a time period of up to 72 h but then declines at longer incubation periods. In contrast, residential mouse peritoneal macrophages constitutively express ALOX15 but their activity declines by more than 90% when the cells are cultured in the absence of IL4. Interestingly, addition of IL4 prevented this decline [133].
In vivo relevance of the inducing effect IL4 on ALOX15 expression: Murine ALOX15 is constitutively expressed at high levels in peritoneal macrophages [65,132,134] and addition of IL4 to in vitro cell cultures augmented the expression level in peritoneal macrophages [132]. Interestingly, no upregulation of ALOX15 activity was observed when macrophages of STAT6-deficient mice were employed suggesting the involvement of STAT6 in the signaling cascade. In contrast, macrophages prepared from transgenic mice, which systemically overexpress IL4 exhibited a 3-4-fold higher ALOX15 activity when compared with cells prepared from control mice [132]. A similar upregulation was detected in other organs (heart, spleen, lung). Unfortunately, it has not been tested whether this gain in activity might be related to contaminating monocytes in the organs. These data suggest that as for human ALOX15 , expression of the mouse ortholog can be upregulated in vitro by the addition of IL4.
However, this experimental setup did not answer the question of whether or not constitutive expression of ALOX15 in mouse peritoneal macrophages involves IL4 as decisive signaling molecule. This question was answered by testing the ALOX15 activity of peritoneal macrophages prepared from IL4+/+- and IL4-/--mice [133]. Interestingly, there was no difference in the ALOX15 activity of these two cell preparations and these data indicate that IL4 is not required for constitutive expression of this enzyme in mouse macrophages.
Open questions and perspectives
The classical Th2 cytokines IL4 and IL13 are powerful inducers of ALOX15 expression in human [16,17] and mouse peripheral monocytes [132], although in mice constitutive expression of this enzyme in peritoneal macrophages is not IL4-dependent [133]. A number of other cytokines including IL1, IL2, IL3, IL4, IL5, IL6, IFNg, GM-CSF, M-CSF, PDGF, TNF and TGF did not induce expression of this enzyme. However, most other cytokines have not been tested. The multiplicity of the cytokine family is constantly growing and it is about time to initiate a systematic screen of untested cytokines for their ALOX15 inducing capacity. The assay systems (qRT-PCR, immunoblotting, activity assays) for such as screen have all been worked out and the required tools (specific amplification primers, ALOX15 specific antibody, activity assay protocols) are available. The major problem for such experiments is the relatively high cost of the recombinant cytokines.
The recent finding that IL4-induced ALOX15 expression is prevented by upregulation of AMPK interconnects the energy metabolism with eicosanoid biosynthesis, in particular with the ALOX15 pathway. If such coupling can also be observed in other cellular systems, ALOX15 might constitute part of cellular fuel sensors, which adapt the energy metabolism to fuel supply. If this is the case ALOX15 inhibitors might interfere with the energy metabolism, which could be of biological relevance for lipid storage diseases, such as obesity or on the other hand, lipodystrophy. The recent finding that IL4 in adipocytes inhibits triglyceride synthesis and upregulates the activity of hormone sensitive lipase suggests the relevance of IL4 for storage lipid homeostasis [114]. However, a possible role of ALOX15 in this metabolic scenario remains to be explored.
The molecular mechanisms for IL4/13-induced ALOX15 expression have extensively been studied and the current mechanistic picture is already very complex. IL4/13 cell surface receptors are clearly involved but the downstream signaling cascades are quite diverse. A number of different protein kinases have been implicated, which lead to activation of several transcription factors. The current picture suggests that there is not a single but several distinct signaling cascades, which include modification of the histone structure. In complex regulatory networks like this there is always room for alternative or supplemental signaling events. One specific question that needs to be addressed is the molecular basis for AMPK-dependent suppression of IL4-induced expression regulation. It has been suggested that AMPK activation may directly interfere with STAT3 and/or STAT6 activation or histone acetylation [123] but for the time being there are no experimental data supporting this hypothesis.
The biological relevance of IL4/13-induced upregulation of ALOX15 expression remains to be explored in more detail. In mice, constitutive expression of ALOX15 in peritoneal macrophages does not depend on IL4 expression [133]. In humans preliminary data suggest that the plasma concentrations of IL4 are simply not high enough to induce expression of the enzyme in peripheral monocytes and it remains questionable whether IL4 concentrations in the range of 1-10 ng/ml, which are required for the in vitro effect, can be reached in vivo . One major problem in this respect is, that to the best of our knowledge, there is no major human disease, which is consistently associated with strongly elevated plasma IL4 levels. In different allergic diseases (see 3.1.4.) we did not find sufficient elevations of the IL4 plasma levels. IL4 has been used in clinical trials for the treatment of chronic lymphatic leukemia [135]. In this study IL4 was well tolerated but the anti-tumor activity observed in previous in vitro studies could not be confirmed. Unfortunately, the impact of the administered IL4 on ALOX15 expression in peripheral monocytes has not been explored in this study. Although the systemic IL4 concentrations in the treated patients have not been quantified, a rough estimate suggested that plasma concentrations in the lower ng/ml range should have been reached. IL4 was administered subcutaneously at doses varying between 2-6 μg/kg day. If one neglects drug elimination and assumes homogenous distribution of the administered IL4 in the body water (which is probably not the case), IL4 concentrations in the one digit ng/ml range should have been reached. Since there are no reliable data characterizing distribution, compartimentation and elimination of exogenous IL4 in humans such model calculations neglecting these parameters may not be very informative.
References
- Haeggström JZ, Funk CD (2011) Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem Rev 111: 5866-5898.
- Kuhn H, Banthiya, van Leyen K (2015) Mammalian lipoxygenases and their biological relevance. BiochimBiophysActa 1851: 308-330.
- Horn T, Adel S, Schumann R, Sur S, Kakularam KR, et al. (2015) Evolutionary aspects of lipoxygenases and genetic diversity of human leukotriene signaling. Prog Lipid Res 57: 13-39.
- Nugteren DH (1975) Arachidonatelipoxygenase in blood platelets. BiochimBiophysActa 380: 299-307.
- Hamberg M, Samuelsson B (1974) Prostaglandin endoperoxides. Novel transformations of arachidonic acid in human platelets. ProcNatlAcadSci U S A 71: 3400-3404.
- Schewe T, Halangk W, Hiebsch C, Rapoport SM (1975) A lipoxygenase in rabbit reticulocytes which attacks phospholipids and intact mitochondria. FEBS Lett 60: 149-152.
- Funk CD, Chen XS, Johnson EN, Zhao L (2002) Lipoxygenase genes and their targeted disruption. Prostaglandins & other lipid mediators 68-69: 303-312.
- Fierro IM, Colgan SP, Bernasconi G, Petasis NA, Clish CB, et al. (2003) Lipoxin A4 and aspirin-triggered 15-epi-lipoxin A4 inhibit human neutrophil migration: comparisons between synthetic 15 epimers in chemotaxis and transmigration with microvessel endothelial cells and epithelial cells. J Immunol 170: 2688-2694.
- Pace-Asciak CR (2015) Pathophysiology of the hepoxilins. BiochimBiophysActa 1851: 383-396.
- Sachs-Olsen C, Sanak M, Lang AM, Gielicz A, Mowinckel P, et al. (2010) Eoxins: a new inflammatory pathway in childhood asthma. J Allergy ClinImmunol 126: 859-867.
- Serhan CN, Petasis NA (2011) Resolvins and protectins in inflammation resolution. Chem Rev 111: 5922-5943.
- Serhan CN, Dalli J, Colas RA, Winkler JW, Chiang N (2015) Protectins and maresins: New pro-resolving families of mediators in acute inflammation and resolution bioactive metabolome. Biochimica et biophysicaacta 1851: 397-413
- Kuhn H, Belkner J, Wiesner R, Brash AR (1990) Oxygenation of biological membranes by the pure reticulocyte lipoxygenase. J BiolChem 265: 18351-18361.
- Belkner J, Wiesner R, Kühn H, Lankin VZ (1991) The oxygenation of cholesterol esters by the reticulocyte lipoxygenase. FEBS Lett 279: 110-114.
- Kühn H, Heydeck D, Brinckman R, Trebus F (1999) Regulation of cellular 15-lipoxygenase activity on pretranslational, translational, and posttranslational levels. Lipids 34 Suppl: S273-279.
- Conrad DJ, Kuhn H, Mulkins M, Highland E, Sigal E (1992) Specific inflammatory cytokines regulate the expression of human monocyte 15-lipoxygenase. ProcNatlAcadSci U S A 89: 217-221.
- Nassar GM, Morrow JD, Roberts LJ 2nd, Lakkis FG, Badr KF (1994) Induction of 15-lipoxygenase by interleukin-13 in human blood monocytes. J BiolChem 269: 27631-27634.
- Ivanov I, Kuhn H, Heydeck D (2015) Structural and functional biology of arachidonic acid 15-lipoxygenase-1 (ALOX15). Gene 573: 1-32.
- Rådmark O, Werz O, Steinhilber D, Samuelsson B (2015) 5-Lipoxygenase, a key enzyme for leukotriene biosynthesis in health and disease. BiochimBiophysActa 1851: 331-339.
- Funk CD, Furci L, FitzGerald GA (1990) Molecular cloning, primary structure, and expression of the human platelet/erythroleukemia cell 12-lipoxygenase. Proceedings of the National Academy of Sciences of the United States of America 87: 5638-5642
- Andreou A, Feussner I (2009) Lipoxygenases - Structure and reaction mechanism. Phytochemistry 70: 1504-1510.
- Hawkins DJ, Brash AR (1987) Eggs of the sea urchin, Strongylocentrotuspurpuratus, contain a prominent (11R) and (12R) lipoxygenase activity. The Journal of biological chemistry 262: 7629-7634.
- Mortimer M, Järving R, Brash AR, Samel N, Järving I (2006) Identification and characterization of an arachidonate 11R-lipoxygenase. Arch BiochemBiophys 445: 147-155.
- Brash AR, Boeglin WE, Chang MS, Shieh BH (1996) Purification and molecular cloning of an 8R-lipoxygenase from the coral Plexaurahomomalla reveal the related primary structures of R- and S-lipoxygenases. J BiolChem 271: 20949-20957.
- Eek P, Järving R, Järving I, Gilbert NC, Newcomer ME, et al. (2012) Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca2+ regulation. J BiolChem 287: 22377-22386.
- Jansen C, Hofheinz K, Vogel R, Roffeis J, Anton M, et al. (2011) Stereocontrol of arachidonic acid oxygenation by vertebrate lipoxygenases: newly cloned zebrafishlipoxygenase 1 does not follow the Ala-versus-Gly concept. The Journal of biological chemistry 286: 37804-37812
- van Leyen K, Duvoisin RM, Engelhardt H, Wiedmann M (1998) A function for lipoxygenase in programmed organelle degradation. Nature 395: 392-395.
- Rapoport SM, Schewe T (1986) The maturational breakdown of mitochondria in reticulocytes. BiochimBiophysActa 864: 471-495.
- Cyrus T, Witztum JL, Rader DJ, Tangirala R, Fazio S, et al. (1999) Disruption of the 12/15-lipoxygenase gene diminishes atherosclerosis in apo E-deficient mice. J Clin Invest 103: 1597-1604.
- Kriska T, Cepura C, Magier D, Siangjong L, Gauthier KM, et al. (2012) Mice lacking macrophage 12/15-lipoxygenase are resistant to experimental hypertension. American journal of physiology. Heart and circulatory physiology 302: 2428-2438
- Kronke G, Katzenbeisser J, Uderhardt S, Zaiss MM, Scholtysek C, et al.(2009) 12/15-lipoxygenase counteracts inflammation and tissue damage in arthritis. Journal of immunology 183: 3383-3389
- Cimen I, Astarci E, Banerjee S (2011) 15-lipoxygenase-1 exerts its tumor suppressive role by inhibiting nuclear factor-kappa B via activation of PPAR gamma. J Cell Biochem 112: 2490-2501.
- Jin G, Arai K, Murata Y, Wang S, Stins MF, et al. (2008) Protecting against cerebrovascular injury: contributions of 12/15-lipoxygenase to edema formation after transient focal ischemia. Stroke; a journal of cerebral circulation 39: 2538-2543
- Johnson EN, Brass LF, Funk CD (1998) Increased platelet sensitivity to ADP in mice lacking platelet-type 12-lipoxygenase. Proceedings of the National Academy of Sciences of the United States of America 95: 3100-3105
- Johnson EN, Nanney LB, Virmani J, Lawson JA, Funk CD (1999) Basal transepidermal water loss is increased in platelet-type 12-lipoxygenase deficient mice. J Invest Dermatol 112: 861-865.
- Zhao L, Moos MP, Gräbner R, Pédrono F, Fan J, et al. (2004) The 5-lipoxygenase pathway promotes pathogenesis of hyperlipidemia-dependent aortic aneurysm. Nat Med 10: 966-973.
- CobanoÄŸlu B, Toskala E, Ural A, Cingi C (2013) Role of leukotriene antagonists and antihistamines in the treatment of allergic rhinitis. Curr Allergy Asthma Rep 13: 203-208.
- Chen XS, Sheller JR, Johnson EN, Funk CD (1994) Role of leukotrienes revealed by targeted disruption of the 5-lipoxygenase gene. Nature 372: 179-182.
- Epp N, Fürstenberger G, Müller K, de Juanes S, Leitges M, et al. (2007) 12R-lipoxygenase deficiency disrupts epidermal barrier function. J Cell Biol 177: 173-182.
- Krieg P, Rosenberger S, de Juanes S, Latzko S, Hou J, et al. (2013) Aloxe3 knockout mice reveal a function of epidermal lipoxygenase-3 as hepoxilin synthase and its pivotal role in barrier formation. J Invest Dermatol 133: 172-180.
- Eckl KM, de Juanes S, Kurtenbach J, Nätebus M, Lugassy J, et al. (2009) Molecular analysis of 250 patients with autosomal recessive congenital ichthyosis: evidence for mutation hotspots in ALOXE3 and allelic heterogeneity in ALOX12B. J Invest Dermatol 129: 1421-1428.
- Hallenborg P, Jorgensen C, Petersen RK, Feddersen S, Araujo P, et al. (2010) Epidermis-type lipoxygenase 3 regulates adipocyte differentiation and peroxisome proliferator-activated receptor gamma activity. Molecular and cellular biology 30: 4077-4091
- Ivanov I, Heydeck D, Hofheinz K, Roffeis J, O'Donnell VB, et al. (2010) Molecular enzymology of lipoxygenases. Arch BiochemBiophys 503: 161-174.
- Hammel M, Walther M, Prassl R, Kuhn H (2004) Structural flexibility of the N- terminal beta-barrel domain of 15-lipoxygenase-1 probed by small angle X-ray scattering. Functional consequences for activity regulation and membrane binding. Journal of molecular biology 343: 917-929.
- Shang W, Ivanov I, Svergun DI, Borbulevych OY, Aleem AM, et al. (2011) Probing dimerization and structural flexibility of mammalian lipoxygenases by small-angle X-ray scattering. J MolBiol 409: 654-668.
- Walther M, Anton M, Wiedmann M, Fletterick R, Kuhn H (2002) The N-terminal domain of the reticulocyte-type 15-lipoxygenase is not essential for enzymatic activity but contains determinants for membrane binding. The Journal of biological chemistry 277: 27360- 27366.
- Toledo L, Masgrau L, Maréchal JD, Lluch JM, González-Lafont A (2010) Insights into the mechanism of binding of arachidonic acid to mammalian 15-lipoxygenases. J PhysChem B 114: 7037-7046.
- Kuban RJ, Wiesner R, Rathman J, Veldink G, Nolting H, et al. (1998) The iron ligand sphere geometry of mammalian 15-lipoxygenases. Biochem J 332 : 237-242.
- Bryant RW, Bailey JM, Schewe T, Rapoport SM (1982) Positional specificity of a reticulocyte lipoxygenase. Conversion of arachidonic acid to 15-S-hydroperoxy- eicosatetraenoic acid. The Journal of biological chemistry 257: 6050-6055.
- Bryant RW, Schewe T, Rapoport SM, Bailey JM (1985) Leukotriene formation by a purified reticulocyte lipoxygenase enzyme. Conversion of arachidonic acid and 15-hydroperoxyeicosatetraenoic acid to 14, 15-leukotriene A4. J BiolChem 260: 3548-3555.
- Kühn H, Wiesner R, Alder L, Fitzsimmons BJ, Rokach J, et al. (1987) Formation of lipoxin B by the pure reticulocyte lipoxygenase via sequential oxygenation of the substrate. Eur J Biochem 169: 593-601.
- Romano M (2010) Lipoxin and aspirin-triggered lipoxins. ScientificWorldJournal 10: 1048-1064.
- Lee HN, Surh YJ (2012) Therapeutic potential of resolvins in the prevention and treatment of inflammatory disorders. BiochemPharmacol 84: 1340-1350.
- Belkner J, Wiesner R, Rathman J, Barnett J, Sigal E, et al. (1993) Oxygenation of lipoproteins by mammalian lipoxygenases. Eur J Biochem 213: 251-261.
- Kuhn H, Chaitidis P, Roffeis J, Walther M (2007) Arachidonic Acid metabolites in the cardiovascular system: the role of lipoxygenase isoforms in atherogenesis with particular emphasis on vascular remodeling. Journal of cardiovascular pharmacology 50: 609-620.
- Garssen GJ, Vliegenthart JF, Boldingh J (1971) An anaerobic reaction between lipoxygenase, linoleic acid and its hydroperoxides. Biochem J 122: 327-332.
- de Groot JJ, Garssen GJ, Vliegenthart JF, Boldingh J (1973) The detection of linoleic acid radicals in the anaerobic reaction of lipoxygenase. BiochimBiophysActa 326: 279-284.
- Kuhn H, Salzmann-Reinhardt U, Ludwig P, Ponicke K, Schewe T, et al. (1986) The stoichiometry of oxygen uptake and conjugated diene formation during the dioxygenation of linoleic acid by the pure reticulocyte lipoxygenase. Evidence for aerobic hydroperoxidase activity. Biochimica et biophysicaacta 876: 187-193.
- Feltenmark S, Gautam N, Brunnstrom A, Griffiths W, Backman L, et al. (2008) Eoxins are proinflammatoryarachidonic acid metabolites produced via the 15-lipoxygenase-1 pathway in human eosinophils and mast cells. Proceedings of the National Academy of Sciences of the United States of America 105: 680-685.
- Ludwig P, Holzhütter HG, Colosimo A, Silvestrini MC, Schewe T, et al. (1987) A kinetic model for lipoxygenases based on experimental data with the lipoxygenase of reticulocytes. Eur J Biochem 168: 325-337.
- Härtel B, Ludwig P, Schewe T, Rapoport SM (1982) Self-inactivation by 13-hydroperoxylinoleic acid and lipohydroperoxidase activity of the reticulocyte lipoxygenase. Eur J Biochem 126: 353-357.
- Wiesner R, Suzuki H, Walther M, Yamamoto S, Kuhn H (2003) Suicidal inactivation of the rabbit 15-lipoxygenase by 15S-HpETE is paralleled by covalent modification of active site peptides. Free RadicBiol Med 34: 304-315.
- Vogel R, Jansen C, Roffeis J, Reddanna P, Forsell P, et al. (2010) Applicability of the triad concept for the positional specificity of mammalian lipoxygenases. The Journal of biological chemistry 285: 5369-5376.
- Johannesson M, Backman L, Claesson HE, Forsell PK (2010) Cloning, purification and characterization of non-human primate 12/15-lipoxygenases. Prostaglandins, leukotrienes, and essential fatty acids 82: 121-129.
- Freire-Moar J, Alavi-Nassab A, Ng M, Mulkins M, Sigal E (1995) Cloning and characterization of a murine macrophage lipoxygenase. BiochimBiophysActa 1254: 112-116.
- Pekárová M, Kuhn H, Bezáková L, Ufer C, Heydeck D (2015) Mutagenesis of triad determinants of rat Alox15 alters the specificity of fatty acid and phospholipid oxygenation. Arch BiochemBiophys 571: 50-57.
- Watanabe T, Medina JF, Haeggström JZ, Rådmark O, Samuelsson B (1993) Molecular cloning of a 12-lipoxygenase cDNA from rat brain. Eur J Biochem 212: 605-612.
- Yokoyama C, Shinjo F, Yoshimoto T, Yamamoto S, Oates JA, et al. (1986) Arachidonate 12-lipoxygenase purified from porcine leukocytes by immunoaffinity chromatography and its reactivity with hydroperoxyeicosatetraenoic acids. The Journal of biological chemistry 261: 16714-16721.
- De Marzo N, Sloane DL, Dicharry S, Highland E, Sigal E (1992) Cloning and expression of an airway epithelial 12-lipoxygenase. The American journal of physiology 262: 198-207.
- Kelavkar U, Wang S, Montero A, Murtagh J, Shah K, et al. (1998) Human 15-lipoxygenase gene promoter: analysis and identification of DNA binding sites for IL-13-induced regulatory factors in monocytes. MolBiol Rep 25: 173-182.
- Horn T, Reddy Kakularam K, Anton M, Richter C, Reddanna P, et al. (2013) Functional characterization of genetic enzyme variations in human lipoxygenases. Redox Biol 1: 566-577.
- Nadel JA, Conrad DJ, Ueki IF, Schuster A, Sigal E (1991) Immunocytochemical localization of arachidonate 15-lipoxygenase in erythrocytes, leukocytes, and airway cells. J Clin Invest 87: 1139-1145.
- Narumiya S, Salmon JA, Flower RJ, Vane JR. (1982) Purification and properties of arachidonate-15-lipoxygenase from rabbit peritoneal polymorphonuclear leukocytes. Advances in prostaglandin, thromboxane, and leukotriene research 9: 77-82.
- Vanderhoek JY, Bailey JM (1984) Activation of a 15-lipoxygenase/leukotriene pathway in human polymorphonuclear leukocytes by the anti-inflammatory agent ibuprofen. The Journal of biological chemistry 259: 6752-6756.
- Levy BD, Romano M, Chapman HA, Reilly JJ, Drazen J, et al. (1993) Human alveolar macrophages have 15-lipoxygenase and generate 15(S)-hydroxy-5,8,11-cis- 13-trans-eicosatetraenoic acid and lipoxins. The Journal of clinical investigation 92: 1572-1579.
- Takayama H, Gimbrone MA, Schafer AI (1987) Vascular lipoxygenase activity: synthesis of 15-hydroxyeicosatetraenoic acid from arachidonic acid by blood vessels and cultured vascular endothelial cells. Thrombosis research 45: 803-816.
- Kuhn H, Ponicke K, Halle W, Wiesner R, Schewe T, et al. (1985) Metabolism of [1-14C]-arachidonic acid by cultured calf aortic endothelial cells: evidence for the presence of a lipoxygenase pathway. Prostaglandins, leukotrienes, and medicine 17: 291-303.
- Lei ZM, Rao CV (1992) The expression of 15-lipoxygenase gene and the presence of functional enzyme in cytoplasm and nuclei of pregnancy human myometria. Endocrinology 130: 861-870.
- Fischer KA, Van Leyen K, Lovercamp KW, Manandhar G, Sutovsky M, et al. (2005) 15-Lipoxygenase is a component of the mammalian sperm cytoplasmic droplet. Reproduction 130: 213-222.
- van Leyen K, Kim HY, Lee SR, Jin G, Arai K, et al. (2006) Baicalein and 12/15-lipoxygenase in the ischemic brain. Stroke 37: 3014-3018.
- Han J, Sun L, Xu Y, Liang H, Cheng Y (2015) Activation of PPARgamma by 12/15- lipoxygenase during cerebral ischemia-reperfusion injury. International journal of molecular medicine 35: 195-201.
- Abecasis GR, Auton A, Brooks LD, DePristo MA, et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56-65.
- Zuo X, Shen L, Issa JP, Moy O, Morris JS, et al. (2008) 15-Lipoxygenase-1 transcriptional silencing by DNA methyltransferase-1 independently of DNA methylation. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 22: 1981-1992.
- Zuo X, Morris JS, Broaddus R, Shureiqi I (2009) 15-LOX-1 transcription suppression through the NuRD complex in colon cancer cells. Oncogene 28: 1496-1505.
- Trebus F, Heydeck D, Schimke I, Gerth C, Kuhn H (2002) Transient experimental anemia in cholesterol-fed rabbits induces systemic overexpression of the reticulocyte-type 15- lipoxygenase and protects from aortic lipid deposition. Prostaglandins, leukotrienes, and essential fatty acids 67: 419-428.
- Kühn H, O'Donnell VB (2006) Inflammation and immune regulation by 12/15-lipoxygenases. Prog Lipid Res 45: 334-356.
- Schewe T, Rapoport SM, Kuhn H (1986) Enzymology and physiology of reticulocyte lipoxygenase: comparison with other lipoxygenases. Advances in enzymology and related areas of molecular biology 58: 191-272.
- Krieg P, Fürstenberger G (2014) The role of lipoxygenases in epidermis. BiochimBiophysActa 1841: 390-400.
- Munoz-Garcia A, Thomas CP, Keeney DS, Zheng Y, Brash AR (2014) The importance of the lipoxygenase-hepoxilin pathway in the mammalian epidermal barrier. Biochimica et biophysicaacta 1841: 401-408.
- Moore K, Lovercamp K, Feng D, Antelman J, Sutovsky M, et al. (2010) Altered epididymal sperm maturation and cytoplasmic droplet migration in subfertile male Alox15 mice. Cell Tissue Res 340: 569-581.
- DeCostanzo AJ, Voloshyna I, Rosen ZB, Feinmark SJ, Siegelbaum SA (2010) 12-Lipoxygenase regulates hippocampal long-term potentiation by modulating L-type Ca2+ channels. J Neurosci 30: 1822-1831.
- Claesson HE (2009) On the biosynthesis and biological role of eoxins and 15-lipoxygenase-1 in airway inflammation and Hodgkin lymphoma. Prostaglandins Other Lipid Mediat 89: 120-125.
- Martinez-Clemente M, Ferre N, Titos E, Horrillo R, Gonzalez-Periz A, et al. (2010) Disruption of the 12/15-lipoxygenase gene (Alox15) protects hyperlipidemic mice from nonalcoholic fatty liver disease. Hepatology 52: 1980-1991.
- Munger KA, Montero A, Fukunaga M, Uda S, Yura T et al. (1999) Transfection of rat kidney with human 15-lipoxygenase suppresses inflammation and preserves function in experimental glomerulonephritis. Proceedings of the National Academy of Sciences of the United States of America 96: 13375- 13380.
- George J, Afek A, Shaish A, Levkovitz H, Bloom N, et al. (2001) 12/15-Lipoxygenase gene disruption attenuates atherogenesis in LDL receptor-deficient mice. Circulation 104: 1646-1650.
- Cyrus T, Praticò D, Zhao L, Witztum JL, Rader DJ, et al. (2001) Absence of 12/15-lipoxygenase expression decreases lipid peroxidation and atherogenesis in apolipoprotein e-deficient mice. Circulation 103: 2277-2282.
- Poeckel D, Zemski Berry KA, Murphy RC, Funk CD (2009) Dual 12/15- and 5- lipoxygenase deficiency in macrophages alters arachidonic acid metabolism and attenuates peritonitis and atherosclerosis in ApoE knock-out mice. The Journal of biological chemistry 284: 21077-21089.
- Shen J, Herderick E, Cornhill JF, Zsigmond E, Kim HS, et al. (1996) Macrophage-mediated 15-lipoxygenase expression protects against atherosclerosis development. J Clin Invest 98: 2201-2208.
- Merched AJ, Ko K, Gotlinger KH, Serhan CN, Chan L (2008) Atherosclerosis: evidence for impairment of resolution of vascular inflammation governed by specific lipid mediators. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 22: 3595-3606.
- Harats D, Ben-Shushan D, Cohen H, Gonen A, Barshack I, et al. (2005) Inhibition of carcinogenesis in transgenic mouse models over- expressing 15-lipoxygenase in the vascular wall under the control of murine preproendothelin- 1 promoter. Cancer letters 229: 127-134.
- Cimen I, Tunçay S, Banerjee S (2009) 15-Lipoxygenase-1 expression suppresses the invasive properties of colorectal carcinoma cell lines HCT-116 and HT-29. Cancer Sci 100: 2283-2291.
- Yoshinaga M, Buchanan FG, DuBois RN (2004) 15-LOX-1 inhibits p21 (Cip/WAF 1) expression by enhancing MEK-ERK 1/2 signaling in colon carcinoma cells. Prostaglandins & other lipid mediators 73: 111-122.
- Chaitidis P, O'Donnell V, Kuban RJ, Bermudez-Fajardo A, Ungethuem U, et al. (2005) Gene expression alterations of human peripheral blood monocytes induced by medium-term treatment with the TH2-cytokines interleukin-4 and -13. Cytokine 30: 366-377.
- Mosser DM, Zhang X (2008) Interleukin-10: new perspectives on an old cytokine. Immunol Rev 226: 205-218.
- Cernaro V, Lacquaniti A, Buemi A, Lupica R, Buemi M (2014) Does erythropoietin always win? Curr Med Chem 21: 849-854.
- Wuest SJ, Crucet M, Gemperle C, Loretz C, Hersberger M (2012) Expression and regulation of 12/15-lipoxygenases in human primary macrophages. Atherosclerosis 225: 121-127.
- Brinckmann R, Topp MS, Zalán I, Heydeck D, Ludwig P, et al. (1996) Regulation of 15-lipoxygenase expression in lung epithelial cells by interleukin-4. Biochem J 318 : 305-312.
- Spanbroek R, Grabner R, Lotzer K, Hildner M, Urbach A, et al. (2003) Expanding expression of the 5- lipoxygenase pathway within the arterial wall during human atherogenesis. Proceedings of the National Academy of Sciences of the United States of America 100: 1238-1243.
- Chen B, Tsui S, Boeglin WE, Douglas RS, Brash AR, et al. (2006) Interleukin-4 induces 15-lipoxygenase-1 expression in human orbital fibroblasts from patients with Graves disease. Evidence for anatomic site-selective actions of Th2 cytokines. The Journal of biological chemistry 281: 18296-18306.
- Lee YW, Kühn H, Kaiser S, Hennig B, Daugherty A, et al. (2001) Interleukin 4 induces transcription of the 15-lipoxygenase I gene in human endothelial cells. J Lipid Res 42: 783-791.
- Ostareck-Lederer A, Ostareck DH, Standart N, Thiele BJ (1994) Translation of 15- lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3' untranslated region. The EMBO journal 13: 1476-1481.
- Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, et al. (1997) mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3' end. Cell 89: 597-606.
- Tsao CH, Shiau MY, Chuang PH, Chang YH, Hwang J (2014) Interleukin-4 regulates lipid metabolism by inhibiting adipogenesis and promoting lipolysis. Journal of lipid research 55: 385-397.
- Bhattacharjee A, Shukla M, Yakubenko VP, Mulya A, Kundu S, et al. (2013) IL-4 and IL-13 employ discrete signaling pathways for target gene expression in alternatively activated monocytes/macrophages. Free radical biology & medicine 54: 1-16.
- Ansel KM, Djuretic I, Tanasa B, Rao A (2006) Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol 24: 607-656.
- Conrad DJ, Lu M (2000) Regulation of human 12/15-lipoxygenase by Stat6-dependent transcription. Am J Respir Cell MolBiol 22: 226-234.
- Liu C, Schain F, Han H, Xu D, Andersson-Sand H, et al. (2012) Epigenetic and transcriptional control of the 15-lipoxygenase-1 gene in a Hodgkin lymphoma cell line. Exp Cell Res 318: 169-176.
- Shankaranarayanan P, Chaitidis P, Kuhn H, Nigam S (2001) Acetylation by histone acetyltransferase CREB-binding protein/p300 of STAT6 is required for transcriptional activation of the 15-lipoxygenase-1 gene. The Journal of biological chemistry 276: 42753- 42760.
- Han H, Xu D, Liu C, Claesson HE1, Björkholm M, et al. (2014) Interleukin-4-mediated 15-lipoxygenase-1 trans-activation requires UTX recruitment and H3K27me3 demethylation at the promoter in A549 cells. PLoS One 9: e85085.
- Kelavkar UP, Wang S, Badr KF (2000) Ku autoantigen (DNA helicase) is required for interleukins-13/-4-induction of 15-lipoxygenase-1 gene expression in human epithelial cells. Genes Immun 1: 237-250.
- Hardie DG, Schaffer BE, Brunet A (2016) AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends Cell Biol 26: 190-201.
- Namgaladze D, Snodgrass RG, Angioni C, Grossmann N, Dehne N, et al. (2015) AMP-activated protein kinase suppresses arachidonate 15-lipoxygenase expression in interleukin 4-polarized human macrophages. The Journal of biological chemistry 290: 24484-24494.
- Hersberger M (2010) Potential role of the lipoxygenase derived lipid mediators in atherosclerosis: leukotrienes, lipoxins and resolvins. Clinical chemistry and laboratory medicine : CCLM / FESCC 48: 1063-1073.
- Xu B, Bhattacharjee A, Roy B, Xu HM, Anthony D, et al. (2003) Interleukin-13 induction of 15-lipoxygenase gene expression requires p38 mitogen-activated protein kinase-mediated serine 727 phosphorylation of Stat1 and Stat3. Molecular and cellular biology 23: 3918-3928.
- Xu B, Bhattacharjee A, Roy B, Feldman GM, Cathcart MK (2004) Role of protein kinase C isoforms in the regulation of interleukin-13-induced 15-lipoxygenase gene expression in human monocytes. J BiolChem 279: 15954-15960.
- Bhattacharjee A, Mulya A, Pal S, Roy B, Feldman GM, et al. (2010) Monocyte 15-lipoxygenase gene expression requires ERK1/2 MAPK activity. J Immunol 185: 5211-5224.
- Wynn TA (2015) Type 2 cytokines: mechanisms and therapeutic strategies. Nat Rev Immunol 15: 271-282.
- Kleiner G, Marcuzzi A, Zanin V, Monasta L, Zauli G (2013) Cytokine levels in the serum of healthy subjects. Mediators Inflamm 2013: 434010.
- Pavlovic V, Dimic A, Milenkovic S, Krtinic D (2014) Serum levels of IL-17, IL-4, and INFγ in Serbian patients with early rheumatoid arthritis. J Res Med Sci 19: 18-22.
- Heydeck D, Thomas L, Schnurr K, Trebus F, Thierfelder WE, et al. (1998) Interleukin-4 and -13 induce upregulation of the murine macrophage 12/15- lipoxygenase activity: evidence for the involvement of transcription factor STAT6. Blood 92: 2503-2510.
- Cornicelli JA, Welch K, Auerbach B, Feinmark SJ, Daugherty A (1996) Mouse peritoneal macrophages contain abundant omega-6 lipoxygenase activity that is independent of interleukin-4. Arteriosclerosis, thrombosis, and vascular biology 16: 1488-1494.
- Sun D, Funk CD (1996) Disruption of 12/15-lipoxygenase expression in peritoneal macrophages. Enhanced utilization of the 5-lipoxygenase pathway and diminished oxidation of low density lipoprotein. The Journal of biological chemistry 271: 24055-24062.
- Lundin J, Kimby E, Bergmann L, Karakas T, Mellstedt H, et al. (2001) Interleukin 4 therapy for patients with chronic lymphocytic leukaemia: a phase I/II study. Br J Haematol 112: 155-160.
Citation: Kuhn H, Gehring T, Schröter A, Heydeck D (2016) Cytokine-Dependent Expression Regulation of ALOX15. J Cytokine Biol 1: 106. DOI: 10.4172/2576-3881.1000106
Copyright: © 2016 Kuhn H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 18354
- [From(publication date): 8-2016 - Sep 04, 2025]
- Breakdown by view type
- HTML page views: 17102
- PDF downloads: 1252