Early Fetal Sex Determination using Cell-Free DNA in Micro-Volume of Maternal Plasma
Received: 08-Nov-2018 / Accepted Date: 25-Nov-2017 / Published Date: 02-Dec-2017 DOI: 10.4172/2376-127X.1000358
Abstract
Background: A qPCR-based assay, SneakPeek® Early Gender Test (Gateway Genomics), has been developed to determine fetal sex as early as 9 weeks gestation using a micro-volume of maternal plasma. The purpose of this study was to assess the clinical performance of SneakPeek for noninvasive prenatal testing (NIPT) of fetal sex.
Methods: A multicenter blinded study was conducted at fourteen ultrasound clinics with maternal blood samples collected from 241 pregnant women between 8.43 and 36.86 weeks of gestation. Plasma was separated from whole blood by double-centrifugation. Circulating cell-free DNA was isolated from a micro-volume of maternal plasma (100 μL) using a commercial DNA extraction kit (NucleoSpin® Plasma XS, Macherey-Nagel). Real-time quantitative PCR was performed to detect fetal DNA using a multi-copy sequence on the Y-chromosome. An autosomal control gene was used to measure total cell-free DNA (maternal and fetal cfDNA). Fifty-nine maternal plasma samples were tested twice, on different days, to assess the precision of SneakPeek. Cell-free DNA was detected in all maternal blood samples.
Results: Y-chromosome DNA was detected in all samples from women carrying a male fetus. Sneak Peek correctly identified fetal sex in 240 of the 241 samples. Fetal sex for all samples was unknown prior to genetic testing and was confirmed via sonographic evaluation at the conclusion of the study. SneakPeek accuracy, sensitivity and specificity were 99.6%, 100% and 99.1% for fetal sex identification, respectively.
Conclusion: This blinded multicenter study showed that SneakPeek Early Gender Test is highly accurate for fetal sex determination in early pregnancy. This micro volume noninvasive prenatal test for early fetal sex determination could simplify the collection of maternal blood and increase the accessibility of NIPT to a broad population.
Keywords: Fetal DNA; NIPT; Sneak peek; Early gender test; Pregnancy
Introduction
The potential of utilizing cell-free fetal DNA (cffDNA) in maternal plasma for noninvasive prenatal diagnosis was first described in 1997 by Lo et al. [1]. Lo et al. was the first investigator to demonstrate the presence of cell-free fetal DNA in maternal plasma [1]. cffDNA accounts for around 10% of the total maternal cell-free DNA, which helps increase the availability of noninvasive prenatal testing (NIPT) [2]. The maternal plasma samples in Lo’s study were obtained from pregnant women via venous blood draw. The process that Lo discovered was a ground breaking discovery for sex determination using noninvasive procedures [3].
Almost twenty years later, current non-invasive prenatal testing still requires a large volume of venous blood sampling, approximately 10-20 mL, for the detection and analysis of cffDNA [4]. The use of cffDNA decreases the necessity to utilize invasive procedures for genetic information about the fetus [5]. First trimester pregnancies have detectable sequences specific to the Y-chromosome in cffDNA, which also reduces the need for invasive procedures [6]. The ability to noninvasively obtain cffDNA for detailed information about the fetus in the first trimester, increases the accessibility of fetal testing to an extensive range of pregnant women [3].
Typically, early sex determination is completed using invasive techniques, such as chorionic villus sampling [7]. These invasive procedures carry an increased risk of miscarriage for the expectant mother [8]. An alternative method for sex determination is sonography. However, ultrasound evaluations for gender determination can only be conducted beginning in the second trimester [9]. First trimester sonography is often unreliable for sex determination due to the incomplete development of external genitalia [9].
cffDNA is derived from trophoblasts in the placenta and represents about 10% of the total cell-free DNA (cfDNA) that is in the maternal circulation at 11-13 week’s gestation [2]. Results from previous experiments have shown the possibility of fetal DNA in maternal plasma DNA for sex determination [10]. Sex determination is the most common clinical application in NIPT using cfDNA [7]. Different types Y-chromosome specific sequences have been used in previous studies, which vary in specificity and sensitivity [11]. For example, DYS14, a multicopy target and sex-determining region (SRY), a single copy gene, have been used in previous studies [7]. However, in these earlier studies a large volume of maternal blood had to be obtained [7]. There is a need for an improved method for noninvasive prenatal testing (NIPT) of fetal sex using cell-free fetal DNA obtained from a micro volume of maternal plasma.
In this multicenter blinded study, we recruited 241 pregnant women at fourteen ultrasound clinics to assess the performance of a qPCR-based NIPT, SneakPeek Early Gender Test (Gateway Genomics), for fetal sex using a micro-volume of maternal plasma. The accuracy, sensitivity, specificity and precision of SneakPeek fetal sex results in early pregnancy were calculated by comparing with ultrasound results.
Methods
Two hundred and forty-one participants from fourteen ultrasound clinics, who opted to have a sonographic evaluation for fetal sex, were recruited for this study. Written informed consent was obtained from all of the participants in accordance with the principles expressed in the Declaration of Helsinki guidelines for human subjects. At the time of sample collection, fetal gestational age of the participants ranged from 9-35 weeks. At the conclusion of the study, the clinics confirmed SneakPeek gender results via sonographic evaluation at 14 weeks’ gestation. At enrollment, 3-4 mL of maternal blood was collected by a phlebotomist via standard venipuncture. Plasma was separated from whole blood using a double-centrifugation protocol. First, samples were centrifuged at 1,200 g for 10 min. Next, the plasma layer was removed and subjected to a 16,000 g centrifugation for 10 min [12]. A microvolume of plasma, 100 uL, was processed with the NucleoSpin® Plasma XS kit (Macherey-Nagel) to extract and purify cfDNA according to the manufacturers’ recommended input ranges.
Following isolation of cell-free DNA, real-time quantitative PCR was performed to detect the presence of male cell-free fetal DNA using a multi-copy target sequence on the Y-chromosome. In addition to the Y-target, an autosomal control gene was used to ensure that a sufficient amount of cell-free DNA (maternal and fetal) was extracted from each plasma sample. PCR cycling conditions were 10 min at 95ºC for initial denaturation of the DNA and polymerase activation, followed by 45 cycles of 1 min at 60ºC and 15 s at 95ºC.
Fetal sex was determined using an algorithm that incorporates cycle threshold (CT) values of both the Y-target sequence and autosomal control gene PCR reactions.
To assess the precision and reproducibility of the CT values obtained during qPCR, we tested two separate 100 uL plasma aliquots from 59 maternal blood samples on different days. Assay precision was determined by the percent coefficient of variation (% CV) for the between-run replicates. For each plasma sample, we calculated the coefficient of variation (CV) using the CT values obtained for each target sequence (i.e., the Y-target sequence and autosomal control gene) as a ratio of standard deviation (SD) and mean. SneakPeek assay results for fetal sex were confirmed by subsequent ultrasound results obtained at each participating clinic.
Results
A total of 241 pregnant women from fourteen ultrasound clinics were recruited in this study. The gestational ages of the participants are shown in Table 1. The gestational age at the time of blood sampling ranged from 8.43 to 36.86 weeks with a median of 12.14 weeks (Table 1).
| Gestational Age | |
|---|---|
| Median (week) | 12.14 |
| Range (week) | 8.43-36.86 |
| 9-12 week (n, %) | 137 (56.85%) |
| 13-27 week (n, %) | 91 (37.76%) |
| =28 week (n, %) | 13 (5.39%) |
Table 1: Gestational age of the 241 participants.
The statistical parameters of the study are shown in Table 2. Fetal sex results were obtained from all pregnancies, 132 female bearing and 109 male bearing. Gestational age was similar between male and female pregnancies. Of the 109 participants carrying a male fetus, SneakPeek correctly identified all 109 cases, for a sensitivity of 100%. There was one false positive and no false negatives among the samples tested in this study. The positive predictive value was 99.1% for SneakPeek. SneakPeek also showed 99.2% specificity and correctly identified 131 of 132 female bearing pregnancies. The accuracy of the SneakPeek test for fetal sex determination was 99.6%. The precision data is shown in Table 2. The average CV values for the Y-target sequence (male fetal samples, n=26) and autosomal sequence (all samples n=59) averaged 0.77% and 1.20% respectively, demonstrating high reproducibility and low variability in laboratory results (Table 2).
| Samples Analyzed | 241 |
| Female Fetuses | 131 |
| Male Fetuses | 109 |
| False Positives | 1 |
| False Negatives | 0 |
| Sensitivity | 100% |
| Specificity | 99.2% |
| Accuracy | 99.6% |
| PPV | 99.1% |
| NPV | 100% |
| Average CV, Y-Target Sequence | 0.77% |
| Average CV, Autosomal Sequence | 1.20% |
Table 2: Statistical parameters of the study.
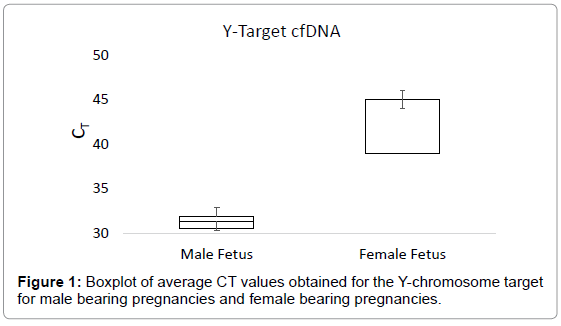
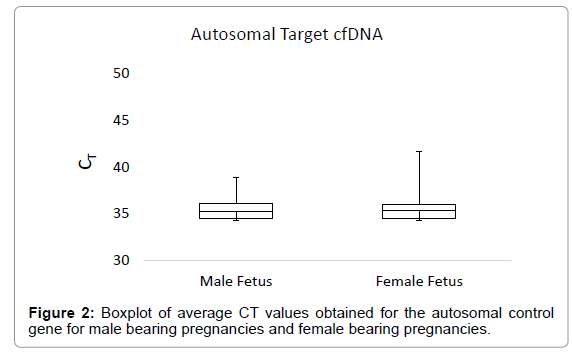
The boxplot in Figure 1 shows the average CT values obtained for the Y-chromosome target in plasma samples from male bearing pregnancies and female bearing pregnancies. The average CT value for plasma from male bearing pregnancies was 31.12, with a range of 26.88-33.66. For plasma from female bearing pregnancies, the average CT value was 42.23, with a range of 34.41-45 (Figure 1). The boxplot in Figure 2 shows the CT values obtained for the autosomal control gene from male bearing pregnancies and female bearing pregnancies (a measure of total cfDNA present in the plasma).
Total cfDNA levels did not differ between male and female bearing pregnancies. The average CT value for total DNA in male bearing pregnancies was 35.37, with a range of 31.76-45.00. For female bearing pregnancies, the average CT value for total DNA was 35.24, with a range of 28.77-45.00.
Discussion and Conclusion
In this blinded multicenter study, we found that SneakPeek had 99.6% accuracy for the determination of fetal sex as early as 9 weeks gestation in a prenatal population. The findings of this study demonstrate that using a micro-volume of maternal plasma as early as 9 weeks into pregnancy can be used to accurately determine fetal sex. We used a multi-copy Y-chromosome target sequence to detect the presence of male fetal DNA in a micro-volume of maternal plasma and an autosomal control gene to confirm adequate DNA was recovered from plasma samples.
Significant differences in the average CT values obtained from qPCR for the Y-chromosome target sequence in plasma samples from male and female-bearing pregnancies were observed. Total cfDNA levels did not differ between male and female bearing pregnancies indicating good reproducibility of cfDNA yield from the DNA extraction and purification process (Figure 2).
Fetal sex was accurately determined using an algorithm that incorporates cycle threshold (CT) values of both the Y-target sequence and autosomal control gene PCR reactions.
Laboratory assay precision was determined by testing two separate 100 μL plasma aliquots from each of 59 maternal blood samples on different days. The CV values for the Y-target sequence and autosomal sequence averaged 0.77% and 1.20%, respectively, demonstrating high reproducibility and low variability in laboratory test results.
All gender results were confirmed via ultrasound by a licensed sonographer. One false-positive and no false-negative results were observed in this study. False-positive detection of Y-chromosome sequences in this case is unlikely to be due to contamination or persistence of fetal DNA from a previous male pregnancy, but more likely to be due to a vanishing twin, whose lysis might have led to large numbers of fetal cells and subsequent release of fetal DNA into the maternal circulation (Table 2) [13].
To our knowledge, this is the first study using a micro-volume (100 μL) of maternal plasma for fetal sex determination during the first trimester of pregnancy. A volume of 200 μL of whole blood would be sufficient to yield 100 μL of plasma. This smaller volume of whole blood needed for SneakPeek is about 1/50th of the amount of whole blood required for current NIPTs [14-16].
The ability to determine fetal sex early in pregnancy is not without bioethical concerns [14]. However, significant benefits from such prenatal technology have been reported. For example, Hyett et al. [14] showed a reduction in diagnostic and therapeutic interventions following non-invasive determination of fetal sex in early pregnancy. Knowledge of fetal g ender in early pregnancy is clinically indicated if sex-linked genetic disorders are present. Lewis et al. [16] reported significant benefits of early sex determination for expectant mothers including several psychological advantages (perceived control, early-reengagement, normalization of pregnancy and peace of mind).
In conclusion, the SneakPeek Early Gender Test was shown to be highly accurate for fetal sex determination as early as 9 weeks gestation. This new method of using maternal blood for prenatal diagnosis of fetal gender is simple, accurate and reliable. This study shows that genetic testing with only a micro-volume of maternal plasma is feasible for gender determination and has potential for various applications for noninvasive prenatal testing. The ability to use a micro-volume of maternal plasma has the potential to increase the accessibility of noninvasive prenatal testing for broad population screenings. For example, such a small volume of maternal blood could be obtained by finger stick. The option to collect patient samples by finger stick could facilitate a more convenient collection process especially in regions where phlebotomy services are not readily available [17].
Acknowledgement
SneakPeek is a registered trademark of Gateway Genomics, LLC. Primacio and Jacob conceived and designed the study. Primacio, Milot and Jacob participated in the data collection and analysis and drafted the manuscript. Primacio and Milot have received consultancy payments from Gateway Genomics regarding their role as laboratory scientists for this study. Jacob has shareholdings in Gateway Genomics.
References
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, et al. (1997) Presence of fetal DNA in maternal plasma and serum. Lancet 350: 485-487.
- Ashoor G, Syngelaki A, Poon LCY, Rezende JC, Nicolaides KH (2012) Fetal fraction in maternal plasma cell-free DNA at 11-13 weeks' gestation: Relation to maternal and fetal characteristics. Ultrasound Obstet Gynecol 41: 26-32.
- Marina SP, Arago AB, Horcajada L, Tiebas, MT, Sanchez IL, et al. (2013) Overview of five-years of experience performing non-invasive fetal sex assessment in maternal blood. Diagnostics 3: 283-290.
- Rahman H (2014) Non-invasive prenatal diagnosis using cell free fetal DNA in maternal blood: Current efforts and future perspectives. J Obstet Gynaecol Barpeta 1: 70-77.
- Khorshid H, Zargari M, Sadeghi M, Edallatkhah H, Shahhosseiny M, et al. (2013) Early fetal gender detemination using real-time PCR analysis of cell-free fetal DNA during 6th-10th weeks of gestation. Acta Med Iran 51: 209-214.
- Ordoñez E, Rueda L, Canadas M, Fuster C (2013) Development and validation of multiplex real-time PCR assay for noninvasive prenatal assessment of fetal RhD status and fetal sex in maternal plasma. Fetal Diagn Ther 34: 13-18.
- Lim J, Park S, Kim S, Kim D, Choi J, et al. (2012) Effective detection of fetal sex using circulating fetal DNA in first-trimester maternal plasma. FASEB J 26: 250-258.
- Tabor A, Philip J, Madsen M, Bang J, Obel EB, et al. (1986) Randomised controlled trial of genetic amniocentesis in 4606 low-risk women. Lancet 1: 1287-1293.
- Odeh M, Granin V, Kais M, Ophir E, Bornstein J (2009) Sonographic fetal sex determination. Obstet Gynecol Surv 64: 50-57.
- Aragones AB, Gonzalez C, De Alba MR, Ainse E, Ramos C (2010) Non-invasive prenatal diagnosis using ccffDNA in maternal blood: State of the art. Expert Rev Mol Diagn 10: 197-205.
- Johnson KL, Dukes KA, Vidaver J, LeShane ES, Ramirez I, et al. (2004) Interlaboratory comparison of fetal male DNA detection from common maternal plasma samples by real-time PCR. Clin Chem 50: 516-521
- Zimmermann B, EL-Sheikhah A, Nicolaides K, Holzgreve W, Hahn S (2005) Optimized real-time quantitative PCR measurement of male fetal DNA in maternal plasma. Clin Chem 51: 1598-1604.
- Dayre McNally J, Matheson LA, Sankaran K, Rosenberg AM (2008) Capillary blood sampling as an alternative to venipuncture in the assessment of serum 25 hydroxyvitamin D levels. J Steroid Biochem Mol Biol 112: 164-168.
- Hyett JA, Gardener G, Mikic TS, Finning KM, Martin PG, et al. (2005) Reduction in diagnostic and therapeutic interventions by non-invasive determination of fetal sex in early pregnancy. Prenat Diagn 25: 1111-1116.
- Puri S, Nachtigall RD (2010) The ethics of sex selection: A comparison of the attitudes and experiences of primary care physicians and physician providers of clinical sex selection services. Fertil Steril 93: 2107-2114.
- Lewis C, Hill M, Skirton H, Chitty LS (2012) Non-invasive prenatal diagnosis for fetal sex determination: benefits and disadvantages from the service users’ perspective. Eur J Hum Genet 20: 1127-1133.
- Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, et al. (1998) Quantitative analysis of fetal DNA inmaternal plasma and serum: Implications for noninvasive prenatal diagnosis. Am J Hum Genet 62: 768-775.
Citation: Primacio R, Milot H, Jacob C (2017) Early Fetal Sex Determination using Cell-Free DNA in Micro-Volume of Maternal Plasma. J Preg Child Health 4: 358. DOI: 10.4172/2376-127X.1000358
Copyright: ©2017 Primacio R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 31645
- [From(publication date): 0-2017 - May 01, 2025]
- Breakdown by view type
- HTML page views: 30413
- PDF downloads: 1232