ESI-MS and Stavrox 3.6.0.1 Investigations of Crosslinking by an Aryl-Azido-NHS-Heterobifunctional Crosslinker
Received: 12-Mar-2018 / Accepted Date: 23-Mar-2018 / Published Date: 26-Mar-2018 DOI: 10.4172/2155-9872.1000402
Abstract
Chemical cross-linking-mass spectrometry (CX-MS) combined with bioinformatics tools is increasingly being used to analyze large-scale protein–protein interactions. It has gained importance in studies in proteomics, lipidomics, in systems and structural biology. Recently it has gained importance in preparation of homogeneous antibody-drug conjugates, which has been described as “a pinnacle of such targeting efforts.” What makes these approaches exciting is that using the “Click” and Bertozzi protocols in vivo studies can be carried out successfully. Using CX-MS combined with cryo-EM, structures of protein complexes can now be probed at almost molecular resolution (upto 3 Å). Chemical crosslinking is useful in materials science, as well. Major advances in both mass spectrometric techniques and bioinformatics tools today allow one to identify cross-linked peptides with highconfidence and with more user-friendly approaches. Crucial to this is the ability to capture intermolecular crosslinking reliably.
The use of a new small NHS-aryl azido heterobifunctional cross-linker based is described here, which picks intermolecular crosslinking better. Thus, Lysozyme has been crosslinked successfully as established by the ‘dimeric’ band observed in SDS-PAGE. its tryptic digestion, ‘zip tip’ enrichment, ESI-MS, MS/MS and the data generated analyzed using StavroX 3.6.0.1, a bioinformatics software, especially suited for determining intermolecular crosslinking.
Keywords: Chemical crosslinking; SDS-PAGE; ESI-MS; MS/MS; StavroX 3.6.0.1
Introduction
Chemical-crosslinking-mass spectrometry-bioinformatics has become an important technique for studying large scale protein-protein interactions, especially for capturing transient interactions [1-18]. This has become possible by the availability of a wide variety of crosslinkers, viz. both homo-and hetero-bifunctional crosslinkers. The former has identical displaceable groups on the two ends, e.g., the amine displaceable N-hydroxysuccinimide (NHS) group. One of more popular reagent of this type is BS2G (bis[sulfosuccinimydyl]glutarate), which has been used extensively.
For enhancing intermolecular crosslinking, more efficient crosslinkers are required. This limitation is overcome by using heterobifunctional crosslinkers, where two different groups are present on the two ends. One of these groups being thermally reactive (e.g., the NHS group) and the other one being photoreactive (e.g., the azide group), which also provides greater flexibility in experimental protocols. Small crosslinkers are known to be more effective for mapping interfaces, while the larger crosslinkers are more useful for identifying binding sites. X-ray diffraction and NMR continue to be the gold standards, but both have their own limitations. The former requires a single crystal, while the latter requires solubility in specific solvents and also demands larger amounts of the sample. Thus, both these techniques are not yet suitable for dynamic studies.
Despite the availability of a range of crosslinkers (including cleavable and isotope labeled crosslinkers) many laboratories continue to use conventional crosslinkers like formaldehyde, glutaraldehyde as crosslinkers, even though these lead to undesirable and extensive crosslinking. Even “zero crosslinkers” like DCC are still in use. This arises mainly due to the lack of awareness amongst many biochemists and others of the immense potential of this new technique of ‘chemical crosslinking mass spectrometry-bioinformatics’ particularly for studying dynamic interactions in living cells. It has been proposed that structures and functions of large protein complexes at the molecular or atomic level in dynamic situations can be studied by combining cryo-electron microscopy (cryo-EM) with crosslinking- mass spectrometry (CX-MS) [19].
Traditional crosslinking strategies generate an enormous amount of mass spectrometry data, which is extremely difficult to analyze with routine software tools. The situation has often been compared to “finding a needle in a haystack”. Tremendous advancement in mass spectrometry (MALDI-MS, MS/MS, ESI-MS) have provided much impetus in improving the quality of crosslinking studies. The use of different algorithms in each laboratory has led to better bioinformatics tools such as GPMAW, MSX3D, ProteinPilot, XQuest, ExPASy, StavroX, xTract. These have contributed greatly to making this technique a valuable tool for the identification of protein-protein interactions in dynamic systems.
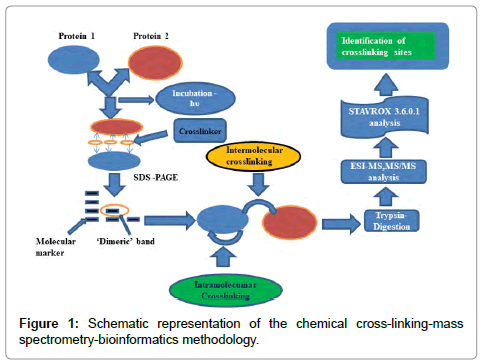
The protocol (Figure 1) commonly used for heterobifunctional crosslinkers involves, incubating the first protein with the crosslinker at room temperature and then in the second step the incubated sample is subjected to photolysis. In the second step, which could be done using either 254 nm or 366 nm UV lamp, a second and a different protein could be introduced. Shorter wavelengths could potentially damage proteins, while longer wavelength exposures are safer.
Much optimization of the duration of incubation, ratio of protein to the crosslinker ratio; exposure time, etc. are required before experimental success can be ensured. The sample at this stage is then subjected to SDS-PAGE. To establish intermolecular crosslinking, the ‘dimeric’ band is excised, trypsin digested and subjected to mass spectrometric analysis. The huge amount of data thus generated is then analyzed by suitable bioinformatics tools. As most of the protein (s) is not crosslinked, unlabeled peptide fragments usually dominate. Unlike the latter, labeled peptide fragments carry a positive charge (+1, +2, +3…) and can be thus enriched by strong cation exchange(SCX) (‘zip tip’) tips to separate them from the un-crosslinked fragments, which usually do not carry a charge. This has been referred to as the “double needle in a haystack” problem.
We employed the above protocol with the twin aims of confirming whether our new small heterobifunctional crosslinker does indeed bring about intermolecular crosslinking successfully and whether the same could be established using modern mass spectrometric methods (MALDI-MS, MS/ MS; ESI-MS) combined with StavroX 3.6.0.1. a bioinformatics software, especially suited for identifying intermolecular crosslinks reliably.
Our work arose out of Hagan Bayley’s [20] correct prediction based on the work of Banks et al. [21] on perfluorophenylazides, that reagents based on the latter could serve as efficient photo-affinity labelling agents, as such reagents involve ‘long-lived ’transients, which leads, in turn, to enhanced intermolecular reaction rates. This was confirmed by Platz et al. [22] who also showed that in this case, the singlet-triplet nitrene energy gap increases. The involvement of a “slippery potential energy surface” has also been demonstrated [23]. The concepts described above have found applications in diverse areas [24].
We have also published a series of papers in this area. Our reactions, however, do not involve any fluorination, which is a demanding step and yet we obtain comparable results. Further, in our cases, the initially formed singlet nitrene does not flip to the triplet state and instead forms the corresponding carbene (Nitrenecarbene conversion; Crow-Wentrup pathway) [25], which has also been substantiated by computational studies, yet our results parallel those based on perflourinated aryl azides. We first observed an unusual reaction involving a concomitant ring expansion and ring extrusion during thermolysis of ‘Dimethyl-Azido-m-hemipinate’ [26]. This was followed by establishing “nitrene insertion into an adjacent orthomethoxy group followed by the addition of the amine intermediate on to the corresponding heterocumulene intermediate” along with the isolation of carbene based products [27]. The life span of one such ‘long lived’ transient, in our case, has been shown to be 700 picoseconds [28]. Reaction of the somewhat more rigid azido-m-meconine led to intramolecular cyclisation via reaction with the adjacent o-methoxy group [29,30]. The reaction of the para analog, viz. ‘Dimethyl-azidosuccinylosuccinate’ led to similar results [31]. Recently we have published on crosslinking studies using a new heterobifunctional crosslinker based on an “introverted” carboxylic acid [32]. The present work is thus a part of our continued investigations in this field.
Results and Discussion
The new aryl azido-N-hydroxysuccinimide heterobifunctional crosslinker (I) was synthesized as shown in scheme I (SI-I). Thus, ‘dimethyl azido-m-hemipinate’ was subjected to selective alkaline hydrolysis, which was followed by reaction with N-hydroxysuccinimide (NHS) and dicyclohexylcarbodiimide (DCC) to yield (1), m.p. 165°C. (I) contains a photo-reactive aryl azido (-N3) group and an amine reactive N-hydroxysuccinimide (NHS) moiety, facilitating the two step protocol, viz. an initial incubation step followed by the photolysis step or vice-versa. The structure of (I) was established by detailed spectroscopic studies including, FT-IR, UV, MALDI-MS and MS/MS studies. NMR studies on (I) will be published separately.
Thus, FT-IR spectrum of (I) (in KBr) showed peaks at 3326, 2928, 2850, 2124, 1767, 1742, 1626, 1588, 1498, 1419,1351,1293, 1201, 1127, 1031, 968, 938, 913, 867, 641 cm-1. UV spectrum of (I) (λ max) in MeCN) showed 198 (2.17), 244(2.44), 2.87(0.49), 317(0.22) nm. MALDI-MS spectrum of the new cross-linker (I) (Figure 2) showed the [M+ H+] ion at m/z 379.0935 as the base peak yielding the molecular formula C15H14N4O8. In addition, other major peaks were observed at m/z 306.2756, 239.1829 and 225.1643. The MS/MS spectrum of the m/z 379 peak is given in SI-II.
The new heterobifunctional crosslinker (I) was incubated with Lysozyme. This was followed by photolysis at 366 nm (using a 6W TLC visualization UV lamp) and SDS-PAGE (SI-III) with standard protocol (SI-IV,) gave a ‘dimeric’ band at 28 kDa. This confirmed successful intermolecular cross-linking. The ‘dimeric’ band was excised, trypsin digested (SI-V) and enrichment by using zip tip C-18 with standard protocol (SI-VI). In early work on this project, we had no access ‘in house’ mass spectrometric facilities and we received only MALDIMS data with no MS/MS data. Even the bioinformatics software had limitations. In the current centre we had access to all modern MS facilities under one roof. Even here, too, we initially carried out only MALDI-MS (SI-VII and SI- VIII) investigations. We had to switch to ESI-MS, MS/ MS as ESI-MS data ((SI-IX) which works better for the bioinformatics software, StavroX 3.6.0.1, (used by us in the current study) works on ESI-MS data alone. The ESI-MS chromatogram for the crosslinked Lysozyme is shown in (SI-X). The ESI-MS data thus obtained was stored in the form of .mgf file and then fed into the StavroX 3.6.0.1 software (SI-XI).
StavroX 3.6.0.1 software
For analysis, the original FASTA sequence of Lysozyme along with the ESI-MS data as .mgf file was uploaded into StavroX 3. 6. 0. 1 software to identify the intermolecularly cross-linked peptides. StavroX 3.6.0.1 is a recent version of this software and it enables quick and efficient identification of the intermolecularly cross-linked peptides. This software calculates the theoretical cross-links and estimates them to the precursors of the ESI-MS data stored in the form of .mgf file [33]. This further leads to the identification of the hits and scores which are then tabulated. The software provides options to select the desired crosslinker along with the scope to add new cross-linkers. The software itself calculated the actual mass of our new crosslinker as 236.2803 amu, based on the loss of one N-hydroxysuccinimide (NHS) unit and one molecule of N2 (loss of nitrogen from the azide), which are lost during the incubation and the photolysis steps, respectively. No changes were made in the amino acid sequence section. An unspecific digest option was selected along with minimum peptide length as 1 and maximum peptide length as 10. The precursor precision was selected to be 250.0 ppm, fragment ion precision as 1.0 Da with the lower mass limit as 200.0 Da and upper mass limit as 6000.0 Da. The S/N ratio was selected to be 2. Only ‘b’ and ‘y’ ions were selected with the score cut-off of -1 and pre-score intensity greater than 10%.
After the data was uploaded into the StavroX 3.6.0.1 software along with the FASTA sequence of Lysozyme, appropriate settings were selected as stated above and the software was run to allow the data to be processed. As a result, 123 spectral peaks were compared to 1024687 theoretical candidates out of which, 3791 possible cross-links were identified within 1 minute and 17 seconds of the run. Out of these, the top nine crosslinked peptide fragments with their m/ z values and scores for intermolecular crosslinking are shown in Table 1. Out of all the possible cross-linked candidates, the highest score obtained was 98 (for the m/z 1383.523 fragment).
| NR | SCORE | M/Z | Z | M+H+ | Calc. | Dev | Peptide (1) | Protein (1) |
from | to | Peptide (2) |
Protein (2) |
from | to | Site (1) |
Site (1) |
rank | scan | RT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 98 | 461.846 | +3 | 1383.523 | 1383.725 | -146 | [KIVS] | >5K70:A | 97 | 101 | [AWRNR] | >5K70:A | 110 | 115 | S4 | R5 | 1 | Locus:1.1 | 683 |
| 2 | 97 | 460.264 | +3 | 1378.778 | 1378.643 | 97.61 | [KVF] | >5K70:A | 0 | 4 | [CKGTDVQ] | >5K70:A | 115 | 122 | K1 | C1 | 1 | Locus:1.1 | 1250 |
| 3 | 95 | 461.948 | +3 | 1382.83 | 1383.83 | 75.79 | [KIVS] | >5K70:A | 97 | 101 | [AWRNR] | >5K70:A | 110 | 115 | S4 | R5 | 1 | Locus:1.1 | 919 |
| 4 | 95 | 606.885 | +2 | 1212.762 | 1212.636 | 104.6 | [KK] | >5K70:A | 96 | 98 | [AWRNR] | >5K70:A | 110 | 115 | K2 | A1 | 1 | Locus:1.1 | 1129 |
| 5 | 90 | 771.053 | +3 | 2311.145 | 2311.109 | 15.67 | [TDVQAWIR] | >5K70:A | 118 | 126 | [RHGLDNYRG | >5K70:A | 14 | 23 | T1 | G3 | 1 | Locus:1.1 | 488 |
| 6 | 88 | 478.792 | +4 | 1912.146 | 1911.893 | 132 | [QATNR] | >5K70:A | 41 | 46 | [RHGLDNYRG] | >5K70:A | 14 | 23 | T3 | H2 | 1 | Locus:1.1 | 1055 |
| 7 | 86 | 403.862 | +5 | 2015.28 | 2014.957 | 160 | [NWVCAAK] | >5K70:A | 27 | 34 | [TDVQAWIR] | >5K70:A | 118 | 126 | K7 | I7 | 1 | Locus:1.1 | 623 |
| 8 | 85 | 714.923 | +2 | 1428.84 | 1428.714 | 88.08 | [AAMKR] | >5K70:A | 10 | 15 | [QINSR] | >5K70:A | 57 | 62 | K4 | I2 | 1 | Locus:1.1 | 548 |
| 9 | 85 | 403.862 | +5 | 2015.28 | 2014.999 | 139 | [ILQINSR] | >5K70:A | 55 | 62 | [KIVSDGNGm] | >5K70:A | 97 | 106 | R7 | K1 | 2 | Locus:1.1 | 623 |
| 10 | 84 | 48.792 | +4 | 1912.146 | 1911.824 | 168.3 | [FESNF] | >5K70:A | 34 | 39 | [MNAWVAWR] | >5K70:A | 105 | 113 | S3 | W4 | 2 | Locus:1.1 | 1055 |
Table 1: Major peaks of the crosslinked peptide fragments.
After the analysis was done, a window opened on top, showing a bar chart, where the number of candidates identified in a certain score range (number of hits) to the score hit is plotted. The Decoy analysis figure helped to estimate the quality of the score in our experiment. The blue bars represent the number of candidates from our data set and the red bars represent the number of false positive candidates from a decoy data set, which is obtained from the inverted sequence of the FASTA file (SI-XII). More enriched real data set candidates in the score region indicates towards better crosslinking. The decoy analysis data for fragment at m/z 1404.900 is shown in Figure 3.
Out of the candidates with high scores, the detailed spectrum for the peak value m/z 1404. 900, the peptide fragments involved in the process of intermolecular cross-linking are shown in Figure 4. The spectrum panel shows the MS2 spectrum for the identified peaks. This one example with its annotation is shown here as a representative. In the deviation diagram (printed below the spectrum panel, deviation of the identified signals is plotted against the m/z values) less deviation in the annotation, points towards better results in the crosslinking experiment. Similar detailed spectra of the other intermolecularly crosslinked fragments have also been obtained via StavroX 3. 6. 0.1, from our experimental data, but these have not been exhibited here (Tables 2 and 3).
| b | b-H2O | b-NH3 | AA | y | y-H2O | y-NH3 |
|---|---|---|---|---|---|---|
| Peptide: ɑ | ||||||
| Charge: +1 | ||||||
| 742.39 | 724.379 | 725.363 | L | |||
| 813.427 | 795.416 | 796.4 | A | 663.361 | 645.35 | 646.334 |
| 884.464 | 866.453 | 867.437 | A | 592.324 | 574.313 | 575.297 |
| 955.501 | 937.49 | 938.474 | A | 521.285 | 503.276 | 504.26 |
| 1102.536 | 1084.526 | 1085.51 | M | 450.249 | 432.239 | 433.223 |
| 1230.631 | 1212.621 | 1213.605 | K | 303.214 | 285.203 | 286.187 |
| R | 175.119 | 157.108 | 158.092 | |||
| Charge: +2 | ||||||
| 371.698 | 362.693 | 363.185 | L | |||
| 407.217 | 398.212 | 398.704 | A | 332.184 | 323.179 | 323.671 |
| 442.736 | 433.73 | 434.222 | A | 296.665 | 287.660 | 288.152 |
| 478.254 | 469.249 | 469.741 | A | 261.147 | 252.142 | 252.634 |
| 551.772 | 542.767 | 543.259 | M | 225.628 | 216.623 | 217.115 |
| 615.819 | 606.814 | 607.306 | K | 152.111 | 143.105 | 143.597 |
| R | 88.063 | 79.058 | 79.55 | |||
| Charge: +3 | ||||||
| 248.135 | 242.131 | 242.459 | L | |||
| 271.814 | 265.81 | 266.138 | A | 221.792 | 215.788 | 216.116 |
| 295.493 | 289.489 | 289.817 | A | 198.113 | 192.109 | 192.437 |
| 319.172 | 313.168 | 313.496 | A | 174.434 | 168.43 | 168.758 |
| 368.184 | 362.18 | 362.508 | M | 150.755 | 144.751 | 145.079 |
| 410.882 | 404.878 | 405.206 | K | 101.743 | 95.739 | 96.067 |
| R | 59.045 | 53.041 | 53.369 | |||
| Charge: +4 | ||||||
| 186.353 | 181.85 | 182.096 | L | |||
| 204.112 | 199.609 | 199.855 | A | 166.596 | 162.093 | 162.339 |
| 221.871 | 217.369 | 217.615 | A | 148.836 | 144.334 | 144.580 |
| 239.631 | 235.128 | 235.374 | A | 131.077 | 126.574 | 126.82 |
| 276.39 | 271.887 | 272.133 | M | 113.318 | 108.815 | 109.061 |
| 308.413 | 303.911 | 304.157 | K | 76.559 | 72.056 | 72.302 |
| R | 44.535 | 40.033 | 40.279 | |||
| Peptide: β | ||||||
| Charge: +1 | ||||||
| 1140.596 | 1122.585 | 1123.569 | K | |||
| 1239.664 | 1221.653 | 1222.637 | V | 265.155 | 247.144 | 248.128 |
| F | 166.086 | 148.076 | 149.06 | |||
Table 2: Modified fragment ions obtained from the StavroX 3.6.0.1 analysis. Where, ‘b’ and ‘y’ ions for m/z 1404.900 identified by StavroX 3.6.0.1 analysis are shown in Table 3.
| Intens. | Rel Intens. | m/z | calc. | Dev(Da) | Type | Z | Peptide | Loss |
|---|---|---|---|---|---|---|---|---|
| 4999.0 | 100.0 | 202.107 | 198.113 | 3.994 | y5 | +3 | α | 0 |
| 4999.0 | 100.0 | 202.107 | 204.112 | -2.005 | b2 | +4 | α | 0 |
| 2568.0 | 51.4 | 123.115 | 113.318 | 9.798 | y3 | +4 | α | 0 |
| 2568.0 | 51.4 | 123.115 | 131.077 | -7.962 | y4 | +4 | α | 0 |
| 2568.0 | 51.4 | 123.115 | 133.081 | -9.966 | y2 | +2 | β | 0 |
| 2226.6 | 44.5 | 216.122 | 221.792 | -5.67 | y6 | +3 | α | 0 |
| 2226.6 | 44.5 | 216.122 | 221.871 | -5.749 | b3 | +4 | α | 0 |
| 2226.6 | 44.5 | 216.122 | 225.628 | -9.506 | y3 | +2 | α | 0 |
| 1611.0 | 32.2 | 567.361 | 570.801 | -3.441 | b1 | +2 | β | 0 |
| 1358.7 | 27.2 | 258.168 | 261.147 | -2.978 | y4 | +2 | α | 0 |
| 1358.7 | 27.2 | 258.168 | 265.155 | -6.986 | y2 | +1 | β | 0 |
| 1285.8 | 25.7 | 485.285 | 478.254 | 7.03 | b4 | +2 | α | 0 |
| 1050.0 | 21.0 | 341.242 | 332.184 | 9.058 | y6 | +2 | α | 0 |
| 1050.0 | 21.0 | 341.242 | 347.439 | -6.196 | P0 | +4 | 18 | |
| 1050.0 | 21.0 | 341.242 | 347.685 | -6.442 | P0 | +4 | 17 | |
| 958.0 | 19.2 | 285.181 | 276.39 | 8.792 | b5 | +4 | α | 0 |
| 958.0 | 19.2 | 285.181 | 285.904 | -0.723 | b1 | +4 | β | 0 |
| 906.9 | 18.1 | 145.048 | 148.836 | -3.788 | y5 | +4 | α | 0 |
| 906.9 | 18.1 | 145.048 | 150.755 | -5.706 | y3 | +3 | α | 0 |
| 906.9 | 18.1 | 145.048 | 152.111 | -7.062 | y2 | +2 | α | 0 |
| 827.8 | 16.6 | 324.217 | 319.172 | 5.045 | b4 | +3 | α | 0 |
| 824.2 | 16.5 | 624.421 | 615.819 | 8.601 | b6 | +2 | α | 0 |
| 824.2 | 16.5 | 624.421 | 620.336 | 4.085 | b2 | +2 | β | 0 |
| 541.0 | 10.8 | 245.149 | 239.631 | 5.519 | b4 | +4 | α | 0 |
| 541.0 | 10.8 | 245.149 | 248.135 | -2.986 | b1 | +3 | α | 0 |
| 476.2 | 9.5 | 416.227 | 407.217 | 9.01 | b2 | +2 | α | 0 |
| 476.2 | 9.5 | 416.227 | 410.882 | 5.345 | b6 | +3 | α | 0 |
| 476.2 | 9.5 | 416.227 | 413.893 | 2.334 | b2 | +3 | β | 0 |
| 452.5 | 9.1 | 367.259 | 368.184 | -0.925 | b5 | +3 | α | 0 |
| 452.5 | 9.1 | 367.259 | 371.698 | -4.439 | b1 | +2 | α | 0 |
| 428.0 | 8.6 | 528.326 | 521.285 | 7.039 | y4 | +1 | α | 0 |
| 408.0 | 8.2 | 159.063 | 166.086 | -7.023 | y1 | +1 | β | 0 |
| 408.0 | 8.2 | 159.063 | 166.596 | -7.532 | y6 | +4 | α | 0 |
| 386.0 | 7.7 | 301.212 | 295.493 | 5.719 | b3 | +3 | α | 0 |
| 386.0 | 7.7 | 301.212 | 296.665 | 4.547 | y5 | +2 | α | 0 |
| 386.0 | 7.7 | 301.212 | 303.214 | -2.001 | y2 | +1 | α | 0 |
| 386.0 | 7.7 | 301.212 | 308.413 | -7.201 | b6 | +4 | α | 0 |
| 386.0 | 7.7 | 301.212 | 310.671 | -9.459 | b2 | +4 | β | 0 |
| 335.2 | 6.7 | 459.271 | 450.249 | 9.022 | y3 | +1 | α | 0 |
| 335.2 | 6.7 | 459.271 | 462.916 | -3.645 | P0 | +3 | 18 | |
| 335.2 | 6.7 | 459.271 | 463.244 | -3.973 | P0 | +3 | 17 | |
| 335.2 | 6.7 | 459.271 | 468.919 | -9.648 | P0 | +3 | 0 | |
| 335.0 | 6.7 | 685.383 | 693.87 | -8.487 | P0 | +2 | 18 | |
| 335.0 | 6.7 | 685.383 | 694.362 | -8.979 | P0 | +2 | 17 | |
| 263.0 | 5.3 | 385.24 | 380.87 | 4.37 | b1 | +3 | β | 0 |
| 263.0 | 5.3 | 595.408 | 592.324 | 3.085 | y5 | +1 | α | 0 |
| 238.4 | 4.8 | 699.397 | 702.875 | -3.478 | P0 | +2 | 0 | |
| 180.0 | 3.6 | 811.551 | 813.427 | -1.876 | b2 | +1 | α | 0 |
Table 3: The ‘b’ and ‘y’ ions for m/z 1404.900 identified by StavroX 3.6.0.1 analysis.
An intermolecularly crosslinked peptide fragment m/z 1404.900 is shown in Figure 5. It contains Peptide 1 (α-peptide) “KVF” crosslinked to peptide 2 (β-peptide) “LAAAmKR”, which shows that intermolecular cross-linking has occurred between K1 (Lysine-1 in the primary structure of Lysozyme) of the α-peptide with L8 (Leucine-8 in the primary structure of Lysozyme) of the β-peptide of another moiety of Lysozyme. This is shown as a representative example from many such intermolecular crosslinks established by StavroX 3.6.0.1.
PyMol software
Using the software PyMol, 3D (SI-XIII) representation of the intermolecular cross-link between the two Lysozyme molecules as obtained by using our new heterobifunctional cross-linker and StavroX 3.6.0.1 analysis is depicted in Figure 6.
Crosslinking studies on Lysozyme using homobifunctional crosslinkers was discussed in a seminal and highly cited paper by A. Sinz’s group [3]. Cross-linking reactions with sulfo-DST and sulfo- EGS yielded no cross-linking products, while the cross-linking reaction with BS3 gave two cross-linking products. The details of the nature of this cross-linking (mostly intramolecular cross-linking) are included in Table 4, reproduced from this earlier work.
| Cross-linking reagent | Cross-linking product | Observed mass | Sequence (amino acid) |
|---|---|---|---|
| BS3 | N-Terminal-K1 | 744.442 | 1-5+XL |
| K96-K97 | 1643.868 | 87-100+Xl |
Table 4:Earlier reported Identification of crosslinking of Lysozyme with the homo bifunctional cross-linker, the di-NHS ester, BS3.
In addition, our experiments also identified many intermolecular cross-linking in MS and ESI-MS based crosslinked peptide not detected by the earlier workers. In comparison, our experiments have led to enhanced intermolecular cross-linking (Table 5).
| Cross-linking reagent | Cross-linking product | Observed mass | Sequence (amino acid) |
|---|---|---|---|
| CXL-379.0935 | N-Terminal of S-R S 100, R 114 | 1383.523 | 100-114, 90-120 + XL |
| K1-C115 | 1378.778 | 1-120 + XL | |
| K96-R114 | 1383.830 | 90-120 + XL | |
| K96-A110 | 1212.763 | 95-120 + XL | |
| T118-G16 | 2311.145 | 110-30 + XL | |
| Q50-Y20 | 1912.146 | 40+30 + XL |
Table 5: Intermolecular cross-links identified by StavroX 3.6.0.1 with our new heterobifunctional cross-linker.
Our results thus justify the hypothesis originally put up by Hagan Bayley [loc.cit.] based on perflourophenylazides and extended by us to our aryl azides, which do not require ortho-flanking fluorines, as in the case of perflourophenylazides. Aryl azides that lead to ‘long-lived’ transients bring about more efficient intermolecular cross-linking, which is the case with our new hetero-bi-functional cross-linker. As stated earlier, this observation can have an impact on diverse areas of science.
Conclusions
A new small aryl-azido-NHS-hetero-bifunctional cross-linker has been synthesized and characterized spectroscopically. It has been successfully used to crosslink Lysozyme as a ‘proof of concept’. This is done in two steps, i.e., via an initial incubation step, which is then followed by the second step of photolysis (366 nm) using a 6 W TLC visualization UV lamp. It was then subjected to SDS-PAGE, excision of the ‘dimeric’ band, trypsin digestion, desalting using ziptip and analysis by ESI-MS. The data thus obtained was fed into the StavroX 3.6.0.1, a bioinformatics tool, especially suited for studies on intermolecular crosslinking, which established the crosslinking sites.
The above study confirms that the new crosslinker successfully brings about intermolecular crosslinking and that the use of ESI-MS/ MS/MS along with StavroX 3.6.0.1, the bioinformatics software, greatly facilitates the analysis of intermolecular crosslinking of the two protein molecules.
The above technique has implications in diverse fields, e.g., for studies on protein-protein interactions, for proteomics/lipidomics and in systems and structural biology. It is expected to help in preparing monoclonal antibody-drug conjugates, which specifically target tumor cells representing “the pinnacle of such targeting efforts.” Recently it has been shown that combining cryo-electron microscopy (cryo-EM) with chemical crosslinking will pave the way for highly efficient in vivo studies. The technique is also important in many areas of materials science.
Supplementary Information (SI)
A new Heterobifunctional cross-linker has been characterized spectroscopically its molecular formula has been established based on its observed [M+ H+] m/z 379.0935 calculated value for [M+ H+] m/z 378.299 The new cross-linker has amine reactive N-Hydroxysuccinimide and a photo-reactive aryl azide group. It has been used to cross-linked two lysozyme molecules which was proved by the appearance of ‘Dimeric band’ in the SDS-PAGE. This band was subjected to Trypsin digestion, ESI-MS and StravroX 3.6.0.1 the bio-informatics software helped establish the crosslinking sites. The, study has implication in proteinprotein interaction and diverse areas of science.
Acknowledgements
Authors thank Dr Sudhanshu Vrati, Executive Director, Regional Centre for Biotechnology (RCB) Faridabad, Haryana for research facilities and AcSIR for Emeritus Professoship (Hony.) (to SVE). We thank the Advanced Technology Platform Center (ATPC, RCB) and Dr Nirpender Singh (ESI-MS, MS/MS studies) for his advice and foir supervising the MS studies. We are grateful to Dr David Cane, Brown University, Providence, Rhode Island, USA (FAB- MS); Prof Vani Bramhachari, Ambedkar Centre for Biomedical Research, University of Delhi, Delhi (initial SDS-PAGE work); Dr Andrea Sinz, University of Halle-Salle, Germany (for helpful discussions) and Dr Michael Goetze from the same group (for StavroX 3. 6.0.1 software and its use).
References
- Rappsilber J (2011) The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. Struct Biol 173: 530-540.
- Sinz A (2014) The advancement of chemical-cross linking and mass spectrometry for structural proteomics. Expert Review of Proteomics, pp: 733-743.
- Stengel F, Abersold R, Carol VR (2012) Joining Forced: Integrating Proteomics and Cross Linking with Mass Spectrometry of intact complexes. Mol Cell Proteomics 11: R111.01402.
- Hermanson GT (2008) Bioconjugate Techniques. 2nd edn. Academic Press, New York, USA.
- Ewens CA, Panico S, Kloppsteck P, McKeown C, Ebong IO, et al. (2014) The p97-FAF1 Protein Complex Reveals a Common Mode of p97 Adapter Binding. J Biol Chem 114: 559591.
- Kannan R, Santoshkumar P, Mooney BP, Sharma KK (2013) Identification of subunit-subunit Interaction Sites in a A-WT Crystallin and mutant a A-G98R Crystallin using Isotope-Labeled Cross-Linker and Mass Spectrometry. PLoS ONE 8: 6561.
- Leitner A, Fain M, Stengel F, Aebersold R (2016) Crosslinking and Mass Spectrometry:An Integrated Technology to Understand the Structure and Function of Molecular Machines. Trends in Biochem Sci 41: 20-32.
- Fischer R, Chen ZA (2008) Quantitative cross-linking/mass spectrometry using isotope-labelled cross linkers. J Proteomics 88: 120-128.
- Walzthoeni T, Joachimaik LA, Rosenberger G, Röst HL, Malmströ L, et al. (2015) xTract: Software for characterizing conformational changes of protein complexes by quantitative cross-linking mass spectrometry. Nat Methods 12: 1185-1190.
- Russel D, Lasker K, Webb B, Velázquez-Muriel J, Tjioe E, et al. (2012) Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol 1001244.
- Dihazi GH, Sinz A (2003) Mapping low-resolution three-dimensional protein structures using chemical cross-linking and Fourier transform ion-cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom 17: 2005-2014.
- Back JW, Sanz MA, De Jong L, De Koning LJ, Nijtmansw LGJ, et al. (2012) A structure for the yeast prohibitin complex: Structure prediction and evidence from chemical crosslinking and mass spectrometry. Protein Science 11: 247.
- Andrew N (2015) XL-MS: Protein cross-linking coupled with mass spectrometry. Methods 89: 54-63.
- Ehresmann C, Moine H, Mouge M, Dondon J, Grunberg M, et al. (1986) Translational Control of Ribosomal protein S15. Nucleic Acids Research 14: 4803-4821.
- Sutherland BW, Toews J, Kast J (2008) Utility of formaldehyde cross-linking and mass spectrometry in the study of protein protein interactions. J Mass Spectrometry 43: 699-715.
- Fritzsche R, Ihling CH, Götze M, Sinz A (2012) Optimizing the enrichment of cross-linked products for mass spectrometric protein analysis Rapid. Commun Mass Spectrom 26: 653-658.
- Kao A, Chiu CL, Vellucci D, Yang D, Patel VR, et al. (2010) Design of CID-Cleavable Protein Cross-Linkers: Identical Mass Modifications for Simpler Sequence Analysis. Mol Cell Proteomicsmcp-M110.
- Cravatt BF, Gabriel MS, Johny R, Iii Jr (2007) The biological impact of mass-spectrometry based proteomics. Nature 450: 991-1000.
- Schmidt C, Urlaub H (2017) Combining cry-electron microscopy (cryo EM) and (CX-MS) for structural elucidation of large protein assemblies. Curr Opin Struct Biol 46: 157-168.
- Bayley H (1983) Photogenerated Reagents in Biochemistry and Molecular Biology. Elsevier Science, p: 186.
- Banks RE, Venayak ND, Hamor TA (1980) Decisive evidence for occurrence of ring-expansion during pyrolysis of azidopentafluorobenzene: X-ray crystallographic analysis of pentafluorophenylnitrene dimer. J Chem Soc Chem Commun 19: 900-901.
- Platz MS (1995) Comparison of phenylcarbene and phenylnitrene. Acc Chem Res 12: 487-492.
- Tomioka H, Ichikawa N, Kamatsu K (1993) Photochemistry of 2-(methoxycarbonyl) phenyl azide studied by matrix-isolation spectroscopy. A new slippery energy surface for phenylnitrene. J Am Chem Soc 115: 8621-8626.
- Liu LH, Yan M (2011) Nitrogen-doped graphene nanosheets with excellent lithium storage properties. J Mater Chem 10: 3273-3276.
- Crow WD, Wentrup C (1968) Decomposition and Isomerization of organic compounds. Tetrahedron Letters 9: 3115-3118.
- Eswaran SV, Neela HY, Ramkumar S, Viswamitra MA (1996) Soluble functionalized fullerenes for Photovoltains. J Heterocyclic Chem 33: 1333-1337.
- Kaur D, Luk HL, Coldren W, Srinivas MP, Sridhar L, et al. (2014) Concomitant nitrene and carbene insertion accompanying ring expansion: spectroscopic, X-ray, and computational studies. J Org Chem 79: 1199-1205.
- Xue J, Luk LH, Eswaran SV, Platz MS (2012) Ultrafast Infrared and UV-vis Studies of the Photochemistry of 2-Methoxy-6-Methoxycarbonylphenyl Azide in Solution. J Phys Chem A 116: 5325-5334.
- Eswaran SV, Kaur D, Khamaru K, Prabhakar S, Sony T, et al. (2016) Tuning the strain effect to induce selectivity through intramolecular nitrene insertion into an adjacent methoxy C single bond H bond leading to form a new benzoxazole: experimental and computational studies. Tetrahedron Letters 57: 1899-1902.
- Sharma M, Naik AA, Gaur M, Raghunathan P, Eswaran SV (2012) Azido-m-meconine-'high ortho'Novolak resin-based negative photoresists for deep UV lithography. J Chem Sci 124: 395-401.
- Eswaran SV, Kaur D, Jana K, Khamaru K, Prabhakar S, et al. (2017) Nitrene insertion into an adjacent o-methoxy group followed by nucleophilic addition to the heterocumulene intermediate: Experimental and computational studies. Tetrahedron 73: 5280-5288.
- Thakur KS, Eswaran SV (2017) A New Heterobifunctional Cross-linker Based on an “Introverted’ Acid: Mass Spectrometric and Bioinformatics Studies, Analysis of Intermolecular Crosslinking of Protein. J Anal Bioanal Tech 8.
- Götze M, Pettelkau J, Fritzsche R, Ihling CH, Schäfer M, et al. (2015) Automated assignment of MS/MS cleavable cross-links in protein 3D-structure analysis. J Am Soc Mass Spectrom 26: 83-97.
Citation: Thakur KS, Eswaran SV (2018) ESI-MS and Stavrox 3.6.0.1 Investigations of Crosslinking by an Aryl-Azido-NHS-Heterobifunctional Crosslinker. J Anal Bioanal Tech 9: 402. DOI: 10.4172/2155-9872.1000402
Copyright: © 2018 Thakur KS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 5466
- [From(publication date): 0-2018 - Nov 09, 2025]
- Breakdown by view type
- HTML page views: 4541
- PDF downloads: 925