Estimation of Genetic Variability, Heritability and Genetic Advance for Essential Oil Yield and Related Traits in Genus Ocimum
Received: 13-Mar-2018 / Accepted Date: 24-Mar-2018 / Published Date: 30-Mar-2018 DOI: 10.4172/2329-8863.1000350
Abstract
Assessing variability is fundamental to identify the most important traits in Ocimum improvement program. The objective of the present study was to estimate variability, heritability and genetic advance based on twelve morphological characters of Ocimum. The experiment was conducted in 2015/16 main cropping season at one location by using randomized complete block design with three replications. The results revealed highly significant differences (P<0.01) among genotypes for all characters considered. The phenotypic coefficient of variation (PCV) for all character was higher than genotypic coefficient of variation (GCV). The estimation of broad sense heritability ĥ2(BS)% was observed to be lower than those of broad sense heritability ĥ2(BS)% for all characters. Genetics advance was recorded as maximum for eugenol content (96.65). Highest heritability in broad sense ĥ2(BS)% recorded for days to maturity (99.46). On the basis of study accessions G-4, G-7, G-9, G-11, G-18 and G-25 were identified high oil of better quality. These accessions may be exploited for commercial cultivation.
Keywords: GCV; Genetic advance; Heritability; Ocimum; PCV; Variability
Introduction
Ocimum (family-Lamiaceae ) is a genus of about 200 species of annual and perennial aromatic herbs and shrubs. Most species are native to the tropical and warm temperate regions [1]. The dry herb (leaves), Ocimum leaf tea, essential oil and its chemical derivatives (eugenol, methyl–eugenol, linalool etc) are exported to European countries in sizable quantity every year [2,3] Basil has several medicinal properties. It is rich in carbohydrates, fibre, phosphorous, calcium, protein, iron, beta-carotene, vitamins B1 and B2 and in aromatic oils [4]. It is good for colds and coughs, indigestion, stomach pain, diarrhoea, nausea, ulcers, ringworm and asthma. It is said to lower blood sugar and increase lactation. The essential oil is also used as anti-perspirant and as fly and mosquito repellent [5]. Available genetic stocks at CSIR-CIMAP, 180 genetic stocks belonging to 5 Ocimum species-Ocimum sanctum-Krishna and Shyam tulsi, O. kilimandscharicum , O. africanum , O. gratissimum and O. basilicum including 8 varieties: CIM Ayu, CIM Angana, CIM Saumya, CIM Jyoti, CIM Sharada, CIM Kanchan, Kushmohak, Vikarsudha used in this investigation.
For any crop improvement program, germplasm collection and assessment of genetic variability is an important step. Hence, it is very essential to partition the observed variability into heritable and nonheritable components measured as genotypic coefficient of variation (GCV), phenotypic coefficient of variation (PCV), broad sense heritability ĥ2(BS)%, genetic advance (GA), and genetic advance expressed as percent mean (GAM%) [6]. Surveys of genetic variability with the help of suitable parameters such as GCV, heritability estimates, and GA are very necessary to start an efficient breeding program [7]. Heritability in associated with genetic advance over means (GAM%) is more effective and reliable in predicting the resultant effect of selection [8,9]. Thus, the present study was conducted in 60 genotypes of Ocimum to evaluate genetic variation among 12 quantitative characters.
Materials and Methods
The present investigation comprising different experiments was carried out at the Research Farm of CSIR-Central Institute of Medicinal and Aromatic Plants, Lucknow, Uttar Pradesh (INDIA) located 26.9°N latitude 80.5°E longitude with an elevation of 120 m above the mean sea level having the subtropical climatic conditions during Kharif season.
180 collections were assembled from different state of India including few exotic from 6 countries (Tanzania, Thailand, Singapore, Slovak Republic, USA and South Africa). Removing duplicates, sixty genetic stocks were examined for high herb and essential oil. These sixty genetic stocks of Ocimum are belonging to five species: 46- Ocimum basilicum (French basil, Sweet basil, Zanzibar basil, Indian basil, and Thai basil); 5-Ocimum sanctum (Krishna/holy basil, Shyam tulsi and Scare basil); 2-Ocimum kilimandscharium (Champhor tulsi); 5-Ocimum africanum (Hoary basil); 2-Ocimum gratissimum L. (African basil/Van tulsi/Tree basil/clove basil) (Table 1).
| S.No | Accessions/Genotypes/cultivar | Botanical name | Origin |
| 1 | CIM-Ayu | Ocimum sanctum | CSIR-CIMAP, Lucknow U.P. (India) |
| 2 | Vikarsudha | Ocimum basilicum | CSIR-CIMAP, Lucknow U.P. (India) |
| 3 | CIM-Angana | Ocimum sanctum | CSIR-CIMAP, Lucknow U.P. (India) |
| 4 | French basil | Ocimum basilicum | Bangalore, Karnataka (India) |
| 5 | French basil | Ocimum basilicum | Mangalore, Karnataka (India) |
| 6 | CIM-Jyoti | Ocimum africanum | CSIR-CIMAP, Lucknow U.P. (India) |
| 7 | Sweet basil | Ocimum sanctum | Gandhi Nagar Gujarat (India) |
| 8 | Selection I | Ocimum africanum | Chennai, A.P. (India) |
| 9 | Sweet basil | Ocimum basilicum | Singapore |
| 10 | Sweet basil (Kushmohak) | Ocimum basilicum | CSIR-CIMAP, Lucknow U.P. (India) |
| 11 | CIM-Kanchan | Ocimum santum | CSIR-CIMAP, Lucknow U.P. (India) |
| 12 | Indian basil | Ocimum basilicum | CSIR-CIMAP, Lucknow U.P. (India) |
| 13 | French basil | Ocimum basilicum | Mangalore, Karnataka (India) |
| 14 | Indian basil | Ocimum basilicum | Muzaffarpur, Bihar (India) |
| 15 | Indian basil (CIM Saumya) | Ocimum basilicum | CSIR-CIMAP, Lucknow U.P. (India) |
| 16 | Sweet basil | Ocimum basilicum | Udaipur, Rajasthan (India) |
| 17 | Zanzibar basil | Ocimum basilicum | Tanzania |
| 18 | Indian basil | Ocimum basilicum | Bareilly, Uttaranchal, (India) |
| 19 | French basil | Ocimum basilicum | CSIR-CIMAP, Lucknow U.P. (India) |
| 20 | Indian basil | Ocimum basilicum | Lucknow, U.P. (India) |
| 21 | Indian basil | Ocimum basilicum | Lakhimpur (Kheri), U.P. (India) |
| 22 | Kapoor/camphor tulsi | Ocimum kilimandscharicum | CSIR-CIMAP, Lucknow U.P. (India) |
| 23 | French basil | Ocimum basilicum | Nasik, Maharashtra (India) |
| 24 | French basil | Ocimum basilicum | Lucknow, U.P. (India) |
| 25 | Indian basil (sel-2) | Ocimum basilcum | CSIR-CIMAP, Lucknow U.P. (India) |
| 26 | Sweet basil (Selection-1) | Ocimum basilicum | CSIR-CIMAP, Lucknow U.P. (India) |
| 27 | Sweet basil | Ocimum basilicum | Trivandrum, Kerala (India) |
| 28 | French basil | Ocimum basilicum | Lucknow, U.P. (India) |
| 29 | CIM- Surabhi | Ocimum basilicum | CSIR-CIMAP, Lucknow U.P. (India) |
| 30 | French basil | Ocimum basilicum | Haridwar, Uttaranchal (India) |
| 31 | Sweet basil | Ocimum basilicum | CSIR-CIMAP, Lucknow U.P. (India) |
| 32 | Thai basil | Ocimum basilicum var. thyrsiflora | Thailand |
| 33 | Shaym tulsi | Ocimum sanctum | Puralia, W.B. (India) |
| 34 | Camphor basil | O. Kilimandscharicum | CSIR-CIMAP, Lucknow U.P. (India) |
| 35 | Sweet basil | Ocimum basilicum | Jammu (J. and K. (India) |
| 36 | Lemon basil | Ocimum africanum | Phagwara, Punjab (India) |
| 37 | Indian basil | Ocimum basilicum | Barabanki U.P., (India) |
| 38 | Clove basil. | Ocimum gratissimum L. | Shillong, Meghalaya (India) |
| 39 | Indian basil | Ocimum basilicum | Razaganj, U.P. (India) |
| 40 | Indian basil | Ocimum basilicum | Rishikesh, Uttaranchal (India) |
| 41 | French basil | Ocimum basilicum | Bangalore, Karnataka (India) |
| 42 | French basil | Ocimum basilicum | Mangalore, Karnataka (India) |
| 43 | French basil | Ocimum basilicum | Chandigarh |
| 44 | Sweet basil | Ocimum basilicum | CSIR-CIMAP, Lucknow U.P. |
| 45 | Sweet basil | Ocimum basilicum | Singapore |
| 46 | Sweet basil | Ocimum basilicum | Singapore |
| 47 | Sweet basil | Ocimum basilicum | Singapore |
| 48 | Sweet basil | Ocimum basilicum | CSIR-CIMAP, Lucknow U.P. (India) |
| 49 | Sweet basil | Ocimum basilicum | Košice, Slovak Republic |
| 50 | Lemon basil | Ocimum africanum | Gandhi Nagar Gujarat (India) |
| 51 | CIM-Sharada | Ocimum basilicum | CSIR-CIMAP, Lucknow U.P. (India) |
| 52 | Lemon basil | Ocimum africanum | Mangalore, Karnataka (India) |
| 53 | Sweet basil | Ocimum basilicum | Košice, Slovak Republic |
| 54 | Sweet basil | Ocimum basilicum | Gandhi Nagar Gujarat (India) |
| 55 | Sweet basil | Ocimum basilicum | Singapore |
| 56 | French basil | Ocimum basilicum | Chennai, A.P., (India) |
| 57 | Clove basil | Ocimum gratisimum | CSIR-CIMAP, Lucknow U.P. (India) |
| 58 | Sweet basil | Ocimum basilicum | Gandhi Nagar Gujarat (India) |
| 59 | Sweet basil | Ocimum basilicum | Gandhi Nagar Gujarat (India) |
| 60 | Sweet basil | Ocimum basilicum | Singapore |
Table 1: Accessions of Genus Ocimum their botanical name and geographic origin.
Observations
Twelve morph-metric observations were recorded on the following economic traits as below:
Days to flowering (50%), Plant height (cm), Number of branches/ plant, Fresh herb yield/ plant (g), Oil content (%), Oil yield/plant (g), Inflorescence length (cm), Days to maturity, Methyl chavicol content (%), Linalool content (%), Citral content (%), Eugenol content (%).
Essential oil analysis
The fresh 100 gm aerial part of Ocimum spices were collected from plants of the field of CSIR-CIMAP, were processed by hydrodistillation for 3 hrs in a Clevenger apparatus to obtain the essential oil [10]. Identification of the essential oil composition was analyzed by gas chromatography (GC) and (MS).
Statistical Analysis
Data were tabulated and mean data were subjected to statistical analysis using computer program package developed at the Data Processing Division of the CIMAP, Lucknow, which based on Singh and Choudhary [11].
Analysis of variance (ANOVA): The analysis of variance was based on the model; Xijk=m+gi+rj+eijk
Where, Xijk=effect of kth observation of ith genotype in jth replication, m=general mean, gi=effect of ith genotype, rj=effect of jth replication, eijk=environmental effect associated with ijkth observation.
The ANOVA based on this model led to the following break up of variance components (Table 2).
| Sources of Variation |
df | Mean sum of product (MSS) | Expectation |
|---|---|---|---|
| Replications | r-1 | Rms | |
| Genotypes | g-1 | Gms | 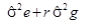 |
| Error | (r-1) (g-1) | Ems | 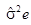 |
| Total | (rg-1) |
Table 2: Break up of variance components.
Parameters of genetic variability: In order to evaluate and quantify the genetic variability among the genotypes for the characters and study the following parameters were estimated (Table 3).
| Genetic parameters | Days to flowering (50%) | Plant height (cm) | Number of branches | Inflorescence length (cm) | Fresh herb yield/plant (g) | Oil content (%) | Oil yiel/plant (g) | Days to maturity | Methyl chavicol content (%) | Linalool content (%) | Citral content (%) | Eugenol content (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotypic variance (σ2g) |
74.05 | 371.97 | 6.15 | 72.33 | 98350.78 | 0.048 | 11.597 | 764.39 | 243.16 | 135.69 | 59.71 | 6.58 |
| Phenotypic variance (σ2g) | 79.799 | 523.31 | 15.74 | 76.23 | 216420.4 | 0.0594 | 19.63 | 768.47 | 247.86 | 137.14 | 60.45 | 12.27 |
| Genotypic coefficient of variation (G.C.V.) % | 5.47 | 18.05 | 22.05 | 13.16 | 43.57 | 34.64 | 23.93 | 74.65 | 94.41 | 263.73 | 258.71 | 55.47 |
| Phenotypic coefficient of variation (P.C.V.) % | 5.68 | 21.41 | 35.28 | 13.51 | 64.63 | 38.37 | 31.14 | 74.85 | 95.32 | 265.12 | 260.32 | 75.74 |
| Heritability (broad sense) ĥ2(BS)% | 92.78 | 71.08 | 39.05 | 94.87 | 45.44 | 81.49 | 59.07 | 99.46 | 98.11 | 98.94 | 98.77 | 53.63 |
| Genetic advance over mean (GAM %) | 25.63 | 26.42 | 17.72 | 86.43 | 40.79 | 57.81 | 89.89 | 36.01 | 85.21 | 51.03 | 63.59 | 94.65 |
Table 3: Genotypic (σ2g) and phenotypic (σ2p) variance, Genotypic (GCV) and phenotypic (PCV) coefficient of variation, heritability ĥ2(BS) and genetic advance (GA as % of mean) of twelve quantitative traits of 60 genotypes of Genus Ocimum.
Estimation of variance components:
Genotypic and phenotypic coefficient of variability: Genotypic and phenotypic coefficients of variability were computed according to Burton and Devane [12].
Genotypic coefficient of variability 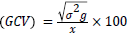
Phenotypic coefficient of variability 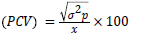
Environmental coefficient of variability 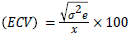
Where, σ2g=Genotypic variance, σ2p=Phenotypic variance and σ2e=Environmental variance, 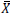 =General mean of character
=General mean of character
The PCV and GCV values are ranked as low, medium and high [13] as follows: 0-10%-Low; 10-20%-Moderate; >20%-High.
Heritability: Broad sense heritability was estimated based on the ratio of genotypic variance to the phenotypic variance and was expressed in percentage [14].
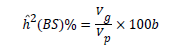
Where ĥ2 (BS)%=heritability in broad sense; Vg=Genotypic variance; Vp=Phenotypic variance. Heritability values are categorized as low, moderate and high [15] as follows: 0-30%: Low; 30-60%: Moderate; 60% and above: High.
Genetic advance: The extent of genetic advance is expected by selecting certain proportion of the superior progeny was calculated by using the following formula [15] given by Robinson et al.
Genetic advance (GA)=k. σp.h2
Where: k=selection intensity at 5% (k=2.06); σp=Phenotypic standard deviation; ĥ2(BS)%=Heritability in broad sense.
Genetic advance expressed as percentage over Mean (GAM%):
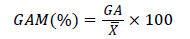
Where: GAM (%)=Genetic advance over mean; 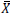 =General mean of the character
=General mean of the character
Results and Discussion
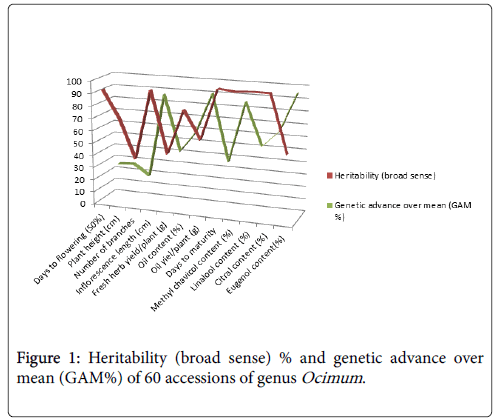
The present investigation was carried out to assess the genetic variability, path analysis between traits as related by genotypic and phenotypic relationship (correlations) and character contribution in 180 genetic stocks of Ocimum [16]. All the twelve traits were highly significant (p<0.01%). The phenotypic variances are always greater than the genotypic variances. The maximum amount of genotypic coefficient of variation (GCV) in percent recorded for the traits was linalool content (263.73%) followed by citral content (258.71%), methyl chavicol content (94.41%), date of maturity (74.65%), eugenol content (55.47%) and fresh herb yield (43.57%) was found suitable for favourable selection for further genetic improvement. Other traits like, oil content (34.64%), oil yield/plant (23.93%), number of branches (22.05%), plant height (18.05%), inflorescence length (13.16%) were expressed medium GCV. The days of flowering (50%) was recorded least (5.47%) genotypic coefficient variance. The phenotypic coefficient of variation (PCV) values were higher than their corresponding GCV for all traits under study, which reflects that the apparent variations were not only due to genotypes alone but environment also. The maximum amount of PCV was displayed by these characters namely, linalool content (265.12), chiral content (260.32%), methyl chavicol content yield (95.32%), eugenol content (75.74%), days to maturity (74.85%), fresh herb yield (64.63%), oil content (38.37%), number of branches (35.28%), oil yield/ plant (31.14%), plant height (21.41%) and inflorescence length (13.51%). The lowest estimate of PCV (5.68%) was observed for days of 50% flowering respectively [17].
Estimates of heritability ĥ2 (BS)% in broad sense
Information on heritability in broad sense ĥ2(BS) and genetic advance of yield attributing traits and their association helps plant breeder to identify characters for effective selection [18]. The concept of heritability explains whether differences observed among individuals rose as a result of differences in genetic makeup or due to environmental forces. According to Robinson et al. [15], heritability values are categorized as low from 0-30%, moderate from 30-60% and 60% and above are high. Considering this benchmark, heritability estimate of this study are described as follows.
Heritability in broad sense ĥ2(BS)% parameter also played a major role in unison with character association and path analysis in genetic improvement program of Ocimum . The high heritability in broad sense ĥ2(BS)% was observed for date of maturity (99.46%) followed by linalool content (98.94%), citral content (98.77%), methyl chavicol content (98.11%), inflorescence length (94.87%), days to flowering 50% (92.78%) and oil content (81.49%) in order. Hence, the selections of above mentioned traits are easy in this crop. Moderate heritability was observed for plant height (71.08%) followed by oil yield/plant (59.07%), eugenol content (53.63%) and fresh herb yield (45.44%) respectively. Low heritability was observed in branches/ plant (39.05%). Therefore, selections of these traits are difficult due to high environmental influences [19].
Genetic advance over mean (GAM%)
The expected genetic advance (GA) values for 12 characters of the genotypes evaluated are presented in Table 2. High genetic advance over mean were observed for the traits eugenol content (94.65) followed by oil yield/plant (89.89), inflorescence length (86.43) and methyl chavicol content (85.21). The medium genetic advance was noticed in citral content (63.59), oil content (57.81), linalool content (51.03), fresh herb yield/plant (40.79) and days to maturity (36.01). Remaining three characters exhibited low genetic advance for example plant height (26.42), days of flowering (50%) (25.45) and number of branches (17.72) respectively (Figure 1).
The estimates of genetic advance help in understanding the type of gene action involved in the expression of various polygenic characters. High values of genetic advance are indicative of additive gene action whereas low values are indicative of non-additive gene action [20]. Accordingly, Heritability and genetic advance are important selection parameters. The estimate of genetic advance is more useful as a selection tool when considered jointly with heritability estimates [21].
Conclusion
Analysis of variance disclosed highly significant difference among the accessions for all the parameters studied. In this research the phenotypic and genotypic coefficient of variation was found to be high for citral content and methyl chavicol content while, moderate GCV and PCV was noticed for days to maturity and eugenol content. Days to flowering (50%) recorded least phenotypic and genotypic coefficient of variation. The heritability estimates was noticed to be high for all characters studied. The characters viz. Days to maturity, linalool content, citral content, methyl chavicol content, inflorescence length and days to flowering (50%) exhibited high heritability coupled with a high genetic advance representing that simple selection scheme would be good enough for these traits to enhance genetic improvement in desired direction. On the basis of study accessions G-4, G-7, G-9, G-11, G-18 and G-25 were identified high oil of better quality. These accessions may be exploited for further crop improvement for the development of high essential oil yielding Ocimum cultivars of better quality.
Acknowledgements
The work was carried out under the CSIR funded project BSC0203. I am Thankful to the Director CSIR-CIMAP for providing the facility. I am also thankful to my PI and Central Facility of CSIR-CIMAP.
References
- Verma RS, Bisht PS, Padalia RC, Saikia D, Chauhan A (2011) Chemical composition and antibacterial activity of essential oil from two Ocimum spp. grown in subtropical India during spring-summer cropping season. J of Traditional Medicine 6: 211-217.
- Lal RK, Khanuja SPS, Agnihotri AK, Mishra HO, Shasany AK, et al. (2003) High yielding eugenol rich oil producing variety of Ocimum sanctum. Cim-Ayu 25: 746-747.
- Lal RK (2014) Breeding for new chemotypes with stable high essential oil yield in Ocimum. Industrial Crops and Products 59: 41-49.
- Ismaile M (2006) Central properties and chemical composition of Ocimum basilicum essential oil. Pharmacuitical Biology 8: 619-626.
- Ojo OD, Adebayo OS, Olaleye O, Orkpeh U (2012) Basil (Ocimum basilicum) genetic variability and viral disease assessment in Nigeria. Asian Journal of Agricultural Sciences 4: 1-4.
- More AD, Borkar AT (2016) Analysis of genetic variability, heritability and genetic advance in Phaseolus vulgaris L. Int J Curr Microbiol App Sci 5: 494-503.
- Atta BM, Haq MA, Shah TM (2008) Variation and inter relationships of quantitative traits in chickpea (Cicer arietinum L.). Pak J Bot 40: 637–647.
- Patil YB, Madalageri BB, Biradar BD, Hoshmani RM (1996) Variability studies in okra (Abelmoschus esculentus L. Moench.). Karnataka. J Agr Sci 9: 289–293.
- Ramanjinappa V, Arunkumar KH, Hugar A, Shashibhaskar MS (2011) Genetic variability in okra (Abelmoschus esculentus L. Moench.). Plant Arch 11: 435–437.
- Clevenger JF (1928) Apparatus for determination of essential oil. J Am Pharma Assoc 17: 346-349.
- Singh RK, Chaudhury BD (1979) Biometrical methods of quantitative genetic analysis. Kalyani Pub. Ludhiana, New Delhi, p: 318.
- Burton GW, Devane EM (1953) Estimating heritability in tall Festuca (Restucaarundinaceae) from donar material. Agron J 45: 1476-1481.
- Shivasubramanian S, Menon M (1973) Heterosis and inbreeding depression in rice. Madras Agric J 60: 1139.
- Falconer DS, Mackay TFC (1996) Introduction to quantitative genetics, Longman, London, UK.
- Robinson HF, Comstock RE, Harvey PH (1949) Estimates of heritability and degree of dominance in corn. Agron J 41: 353-359.
- Arora PP (1991) Genetic variability and its relevance in chickpea improvement. International Chickpea Newsletter 25: 9-10.
- Gutu B (2015) Genetic variability and path coefficient analysis for yield and yield related traits in common bean (Phaseolus vulgaris L.). MSc thesis, Haramaya University, Hararge, Ethiopia, p: 62.
- Misra RC (1991) Stability of heritability, genetic advance and character association estimates in chickpea. International Chickpea News letter 25: 10-11.
- Dixit M, Shukla RS (2016) Estimation of parameters of genetic variability, heritability and genetic advance in elite lines of wheat collected from different parts of Madhya pradesh (India). International Journal of Agriculture Sciences 8: 1723-1726.
- Singh P, Narayanan SS (1993) Biometrical techniques in plant breeding. Kalayani Publishers, New Delhi, India.
- Johanson RW, Robinson HF, Comstock RE (1995) Estimates of genetic and environmental variability in soybeans. Agron J 47: 314-318.
Citation: Smita S and Kishori RL (2018) Estimation of Genetic Variability, Heritability and Genetic Advance for Essential Oil Yield and Related Traits in Genus Ocimum . Adv Crop Sci Tech 6: 350. DOI: 10.4172/2329-8863.1000350
Copyright: © 2018 Smita S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 8581
- [From(publication date): 0-2018 - Dec 02, 2025]
- Breakdown by view type
- HTML page views: 7476
- PDF downloads: 1105