Functionalized Nanocontainers Operated as Controlled Release Systems and Bioactuators
Received: 12-Sep-2013 / Accepted Date: 14-Nov-2013 / Published Date: 04-Dec-2013 DOI: 10.4172/2090-4967.1000106
Abstract
Functional nanocontainers find applications in biosensors, bioelectronics, biochemical investigations, physico-chemical studies, pharmacology and medicine. In all possible applications of liposomes Smart systems responding to varying environmental conditions is desirable. For this purpose development of compound independent release systems that can be engineered for controlled compound release are reviewed in this article.
Introduction
The nanotechnology term is referred to the ability to engineer materials precisely at the nanometers scales. In fact, “engineered materials” is usually used to comprise design, characterization, production and application of materials and the scope has nowadays been widened to include systems and devices rather than just materials. Nanotechnology is thus defined as design and fabrication of materials, devices and systems with control at nanometer dimension.
Here we would like to approach the question why nanotechnology is often stated to be revolutionary. There are three distinct aspects: indirect, direct and conceptual. By indirect is meant the progressive miniaturization of existing technologies, which opens up new areas of application for those technologies. Direct refers to the application of novel, nano-engineered artifacts, either to enhance the performance of existing processes and materials or completely novel purposes. Finally there is the conceptual aspect of nanotechnology, in which all materials and processes are considered from a molecular or even atomic point of view, as living systems, in which complex molecules (like proteins) are partitioned into their constituent amino acids, which are then used for the template synthesis of new proteins.
Life is characterized by separation from the surroundings. Organisms are built out of cells, cells contain separate compartments. Compartmentalization is therefore an already existing principle in nature. Cells perform complex biosynthesis and transport within closed containers employing lipid membranes called vesicles. These natural systems served as a model system for encapsulation of pharmaceutical and cosmetic components. These naturally-derived lipid vesicles, so-called liposomes, showed significant success in their design and application in biotechnological applications due to their unique properties. Moreover liposomes are a very useful model, reagent and tool in various scientific disciplines. Liposomes are illustrating emerging trends at the interface of lipid/polymer chemistry and recombinant protein engineering. One of the best examples is the optimized solution of nature to overcome membrane permeability barrier: substances as nutrients, waste products as well as messenger molecules or ions have to betransported across membranes in the cells. The membrane permeability barrier is overcome by porins and transport proteins (channel proteins) present in the cell membrane.
Functionalized Nanocontainers
A current research field in material science is to formulate new types of responsive materials on a nanometer-sized scale. Although it is quite a challenge in the laboratory, this approach is largely inspired by a naturally occurring compartmentalization system: biological cells. For example, the outer cell wall of bacteria, such as E. coli, contains channel proteins that allow specific substrates to diffuse into the periplasmic space from where they are uptaken actively to inside [1]. Bacteria can open and close these channels upon specific triggers [2] and they have mechanisms to eject material outside. Nano-meter sized particles, micelles and vesicles [3] have already been used for, for instance, targeting cancer cells in medical applications. However a revolutionary new concept is the combination of these target-specific nanometer-sized nanocontainers with multifunctional biological components such as channel proteins. The goal is the creation of targetspecific and furthermore responsive nanosystems that can reach the desired cellular or subcellular target with the help of a defined targeting moiety and that exhibit a controlled and triggered diagnostic signal and therapeutic effect upon activation [4]. By combining sensing and effector functionality on the nanoscale, it will be possible to generate a defined response of nanostructure that depends on environmental factors such as pH, temperature, or substrate concentrations at the target site [4].
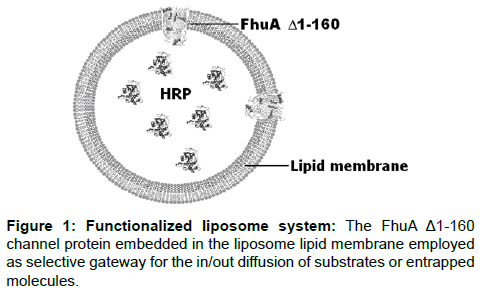
Functionalized nanocontainers are mechanically stable vesicles with a polymer and/or lipid membrane and a transmembrane channel for controlling the flux of compounds in and out of the lipid vesicle. Liposomes are nanometer-sized (50-1000 nm) and functionalized by loading them with reporter molecules such as enzymes or fluorophore molecules. A schematic representation of functionalized nanocontainer systems is given in Figure 1.
Horseradish peroxidase (HRP) is encapsulated in the vesicle and the outer membrane protein, FhuAΔ1-160, is reconstituted in the membrane and functions as a selective gate. A channel protein that is embedded in membranes offer the possibility to develop novel triggered drug release systems with potential applications in synthetic biology (pathway engineering), bioelectronics (biofuel cells) and medicine (drug release). So far only FhuA [5], OmpF [6-8] and Tsx [9] have been reconstituted functionally into synthetic block copolymer membranes. FhuA is a large monomeric transmembrane protein of 714 amino acids located in the E. coli outer membrane folded into 22 anti-parallel β-strands and two domains [10]. By removing the “cork” domain (deletion of amino acids 5-160 [11,12]) the resulting deletion variant functions as a large passive diffusion channel (FhuAΔ1-160) [5]. FhuA and engineered variants have a significantly wider channel than OmpF (elliptical cross section of OmpF is 7*11 Å [13] whereas FhuA is 39*46 Å [5]) allowing the translocation of even single stranded DNA [14].
Reduction Triggered Release System Based On Functionalized Polymersomes (Synthosomes)
Recently a nanocontainer based reduction triggered release system through an engineered transmembrane channel (FhuAΔ1-160) has been reported [15]. The aim of that study was the introduction of a triggering system by means of a reduction triggered “release-switch”, based on an engineered FhuA channel variant. Compound fluxes within the FhuAΔ1-160 channel protein are controlled sterically through labeled lysine residues. Quantifying the sterical contribution of each labeled lysine opened up an opportunity for designing compound specific release systems.
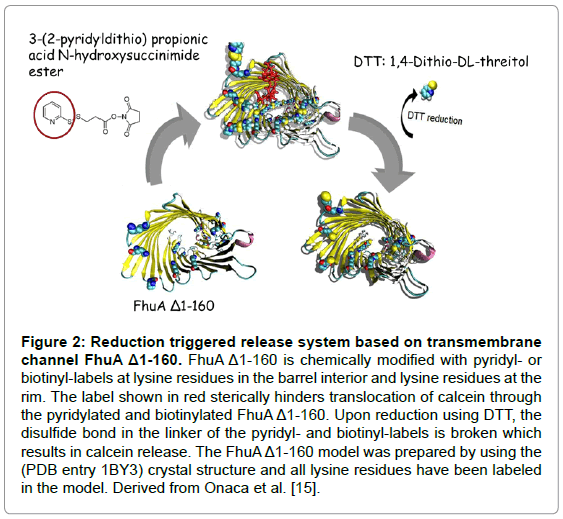
The Synthosome-calcein release system is a triggered release system in which the entrapped compound (calcein) is liberated through an engineered transmembrane channel (FhuAΔ1-160) upon addition of a reducing agent. These synthosomes were loaded with calcein at a selfquenching concentration (50 mM) and the fluorescence generation upon calcein release was used for recording release kinetics. In order to build a reduction triggered release system, the amino groups of lysines in the FhuAΔ1-160 were modified with a pyridyl- or biotin-label. Figure 2 shows the reaction schemes for eight chemically modified lysines (167, 226, 344, 364, 455, 537, 556, 586; FhuAΔ1-160) with a pyridyl- (left) and a biotinyl-label (right).
Figure 2: Reduction triggered release system based on transmembrane channel FhuA Δ1-160. FhuA Δ1-160 is chemically modified with pyridyl-or biotinyl-labels at lysine residues in the barrel interior and lysine residues at the rim. The label shown in red sterically hinders translocation of calcein through the pyridylated and biotinylated FhuA Δ1-160. Upon reduction using DTT, the disulfide bond in the linker of the pyridyl- and biotinyl-labels is broken which results in calcein release. The FhuA Δ1-160 model was prepared by using the (PDB entry 1BY3) crystal structure and all lysine residues have been labeled in the model. Derived from Onaca et al. [15].
As described above an exclusive translocation of calcein through an engineered transmembraneFhuAΔ1-160 which had been embedded in a tri-block copolymer membrane PMOXA-PDMS-PMOXA has been reported; where PMOXA = poly(2-methyl-2-oxazoline) and PDMS = poly(dimethyl siloxane); and could be opened up through a reduction triggered system [15]. The reported release kinetics of calcein was strongly modulated by the size of employed lysine-labeling reagents. Twenty nine lysines are present in the FhuAΔ1-160; 19 lysines located on the protein surface, 6 are inside the channel, and 4 are at the barrel rim [15]. The 19 lysines on the FhuA surface point into the outer membrane and are after purification covered by oPOE rendering pyridyl-labeling unlikely.
However channel proteins such as the engineered FhuAΔ1-160 often cannot insert into thick polymeric membranes due to a mismatch between the hydrophobic surface of the protein and the hydrophobic surface of the polymer membrane. To widen the range of applicable polymers, by a simple “semi-rational” approach the FhuAΔ1-160 hydrophobic transmembrane region was increased by 1 nm by doubling the last 5 amino acids of each of the 22 β-sheets, prior to the more regular periplasmatic β-turns (FhuAΔ1-159 Ext). This increase led to a predicted lower hydrophobic mismatch between the protein and polymer membrane, minimizing the insertion energy penalty. The already mentioned channel blocking by lysine group biotinylation in combination with the HRP assay system has been also employed to study the FhuA Δ1-159 Ext functionality [16].
Responsive Materials On Nanometer-sized Scale: Triggered Liposomal Systems
Reduction triggered liposomal nanocontainers
Typical and widespread model system for nanocontainers is liposomes [17]. After the discovery in 1965, liposomes became a promising tool for drug delivery [18]. Since then there have been many developments in liposomal composition, efficient drug encapsulation and retention, stability and targeting [19]. However in most cases the amount of drug passively released from such ideal long-circulating, sterically stable liposomes was not enough to show a therapeutic effect. To this end, more research efforts were directed towards triggered release in response to a specific stimulus at a target site [20].
There have been different strategies used for triggering liposomal release mostly based on destabilization of the liposomal bilayer. Formation of defects in the bilayer, lamellar-micellar or lamellar hexagonal phase transition, lipid phase separation and liposome fusion are some examples [21,22]. All these efforts have been focused on the lipid components of the liposome, and, for the most part, met with limited success [23]. As an alternative a bacterial channel protein could be used to enable controlled substance transport across otherwise impermeable membrane [24]. The channel protein, first of all, keeps its functionality when it is reconstituted into artificial lipid bilayers without requiring any other cellular components. Second, it is unique for its large and non-selective pore that allows the passage not only of ions but also small molecules. The attachment of synthetic modulators to biological channels is promising for designing universal drug release technology. However employing bacterial channel proteins has limitations, especially in the range of changes and responses that they can accomplish. In this regard, chemical modification provides a complementary approach. Variations can be introduced to the protein to confer features not achievable with the 20 amino acids that are genetically encoded. This is especially true when a combination of properties, such as reversibility, tenability, target specificity, sensitivity to external stimuli, and control over the timescale of the effect, are desired all at the same time [25].
Currently used systems (recently reviewed) [13] involved bacterial outer membrane protein F (OmpF) which was integrated into the synthetic bilayer [26,27] to build a pH responsive release and the mechanosensitive channel of large conductance (MscL) protein from E. coli [20,28] to work as a reversible diffusive channel in liposomes, modulating the open/closed states by a compound undergoing lightinduced or pH-induced charge separation. Latest progress was achieved in an exclusive translocation of calcein through a chemically triggered engineered transmembrane channel protein (FhuAΔ1-160 variant) embedded into a polymer membrane to develop a reduction triggered release system [15].
Following that, a mutational study onFhuAΔ1-160 has been reported [29]. In this study FhuA channel interior was modified through labeled lysines to gain better understanding on the sterically controlled diffusion of TMB through liposomal membranes. An average of four lysine residues per FhuAΔ1-160 was determined to be pyridyl labeled [15]. Based on the hypothesis that the 6 lysine inside the channel might mainly be responsible for restricting compound fluxes, two subsets of FhuAΔ1-160 variants were generated. In the six subset (A) variants only one of the six lysines in channel interior was substituted by alanine and in the six subset (B) variants only one lysine remained in the channel interior whereas all other five were substituted to alanines. For the in total 12 investigated FhuAΔ1-160 variants a HRP (Horseradish peroxidase) based colorimetric TMB (3,3',5,5'-tetramethylbenzidine) detection system was employed for quantifying the sterical hindrance of pyridyl-labeled lysines on the TMB substrateand the results led to the discovery of a flux controlling role of biotinyl- and pyridyl-labeled lysines inside the FhuA Δ1-160 pore [30]. The colorimetric HRP/TMB detection was preferred over the previously reported calcein detection system due to a superior reproducibility [5,15]. It’s important to note that the enzyme, HRP, entrapped in the liposomes is large enough to be kept locked in the liposomes however the substrate, TMB, for this enzyme and the product upon enzyme reaction are small to diffuse through channel protein. FhuAΔ1-160 lysine residues inside β-barrel structure were chemically labeled prior to reconstitution to block the channel from compound diffusion.Furthermore liposomes instead of a polymeric nanocontainer system were selected for characterizing the 12 FhuAΔ1-160 variants due to more simple and more rapid assay procedures [31], despite of drawbacks like leakiness, stability over time [31] and undesired biomolecule adsorption on the surface [32].
Six lysine residues were systematically analyzed in two subsets of engineered FhuAΔ1-160 channels. Analysis of 12 variants identified position K556 as a key substitution to sterically control compound fluxes through the inner channel of FhuAΔ1-160 embedded in liposome membrane. Additionally, b-factor analysis based on MD simulations identified position K556 as the least fluctuating lysine among the six investigated lysines suggesting a correlation between flexibility and steric control of TMB compound translocation through the inner FhuAΔ1-160 channel. The b-factor analysis indicates the dynamic mobility of an atom or group of atoms. The concept is derived from the X-ray scattering/crystallography theory, alternatively known as "temperature-factor" or "Debye-Waller factor" [33]. The subset B variant K556 of FhuAΔ1-160 represents therefore an excellent starting point to understand channel dynamics and to sterically control compound flux through engineered FhuAΔ1-160. Based on these results it seems promising that the reduction triggered release system can be advanced to a universal and compound independent release system which allows a size selective compound release through rationally reengineered FhuAΔ1-160 channels [29].
Furthermore a FhuAΔ1-160 variant with increased channel diameter has been constructed by the addition of a further β-sheet, again accomplished by a simple “copy-paste” approach using already present sequence information. The resulting protein FhuAΔ1-159 Exp was inserted into liposomes. Introduction of a reduction triggered release system was reached by biotinylation of lysine groups in the channel interior [34].
Light triggered liposomal nanocontainers
As already discussed above an important issue towards making the functional liposome systems useful for specific applications is their capability to respond to external stimuli such as temperature, pH and/or light. Approaches for advancing trigger systems are based on mainly chemically modifying the engineered channel protein to develop switchable channels which opens and closes in dependence on environmental conditions.
The methods of chemical modification of proteins in solution generally involve the most reactive nucleophilic functions of proteins, namely the primary amino group of lysine residues, and the thiol group of cysteine residues [35]. Most importantly, protein modification reagents can be made highly selective of side chain groups. Acylation amidination and alkylation of lysine residues are well-known reactions in protein chemistry, whereas the thiol group of cysteine can be either selectively alkylated, carboxymethylated or even transformed to a disulphide with appropriate reagents [36]. Thus all these methods provide a good basis to design side-chain selective protein-labeling reagents. However, the use of many side-chain labeling reagents is severely restricted by their lack of complete specificity. Reactions with side chains of cysteine, histidine, tyrosine, serine, and threonine, occur frequently and have to be evaluated carefully. Remarkably specific reactions with amino groups have been obtained with a number of reagents under suitable conditions, such as acetic, succinic, maleic acid anhydrides [37], N-carboxyanhydrides of amino acids, cyanate (carbamylation), imidoesters, O-methylisourea (guanidination) [38].
It is often the case that the typical hydrolysis cleaving reaction is far too slow for investigating the initial rates of reaction for rapid biochemical processes. A desirable solution to these limitations would be a photo cleavable label that could be removed under neutral buffered aqueous conditions, thus avoiding any alterations to the substrate or to the natural biological environment [39]. Photolysis reactions present a remarkable and often ideal alternative to all other methods for introducing reagents or substrates into reactions or biological media. The ability to control the temporal and concentration variables by using light to photo chemically release a substrate provides the researcher with the ability to design more precisely the experimental applications in molecular biology. Covalent blocking of the functional groups of an enzyme essentially suspends its mode of action. Photo-release is sometimes termed a “traceless reagent process” because no reagents other than light are required. The advantage of a process that requires no further separation of spent reagents is attractive [39]. Photo cleavable labels should also have several other properties such as good aqueous solubility for biological studies, efficient photochemical release and formation of stable photoproducts [40]. Another important property that the photo cleavable agent should possess is an excitation wavelength longer than 300 nm because biological samples are frequently optically transparent in this wavelength range, and the potential photochemical damage to proteins and nucleotides that is anticipated from shorterwavelength irradiation is avoided [41].
In the earlier studies by Fodor and co-workers amino acids were protected with the photolabile, o-nitroveratryloxycarbonyl (NVOC) group, which was originally introduced by Patchornik et al., Houseman BT et al. and Vernet in 1970 [42-44]. Some improvement in the yield of photodeprotection was required since photolytic removal of NVOC was revealed to be very slow. Following that α-methylo- nitropiperonyloxycarbonyl (MeNPOC) was reported to be more efficiently photo-cleaved. However one very important drawback of using MeNPOC was that the photodegradation products of MeNPOC group include carbonyl compounds which can react with amino groups and reduce the synthetic yield [45,46]. More recently a new class of photosensitive 2-(2-nitrophenyl)ethoxycarbonyl group have been demonstrated that the efficiency of photolytic cleavage of 2-(2-nitrophenyl)propyloxycarbonyl (NPPOC) protected amino groups is significantly better than that for NVOC protected nucleotides [47]. One difference between the NPPOC group and the NVOC group is that the former is a derivative of 2-(2-nitrophenyl)ethyl alcohol, whereas the latter is derived from 2-nitrobenzyl alcohol. The additional methylene group in the NPPOC group leads to a different photocleavage mechanism [48].
The photocleavage of the o-nitrobenzyl group (e.g., NVOC) leads in an intramolecular redox reaction to o-nitrosobenzaldehyde whereby this process is initiated by a hydrogen abstraction from the benzylside- chain, followed by the formation of an intermediary carbo-cation, quenching by a nucleophile and finally dissociation of the intermediate [49]. On the other hand, the photocleavage of the 2-(2-nitrophenyl) ethoxycarbonyl groups can be explained with an entirely new mechanism consisting of a light-induced β-elimination process [49]. In the first step of photo-elimination a hydrogen abstraction from the β-position in the side-chain is most likely, then intersystem crossing of the diradical takes place followed by β-elimination forming CO2. Since this cleavage does not involve a chemical step like in the case of o-nitrobenzyl cleavage an accelerated breakdown of the molecule can be expected explaining the experimental findings in a rational manner [49].
Light activation is a promising method for triggering vesicle contents release since it provides a broad range of adjustable parameters (such as pulse duration, intensity, pulse cycle, and wavelength) that can be optimized for biological compliance [50-53]. Actual compound release systems based on light triggering implies mostly a compound diffusion controlled through a physical change of the membrane surface involving lipid bilayer disruption [49] permanently opened channel pores in either liposomes [54] or polymersomes [4] or temporary temperature-triggered lipid-packing defects in liposomes [52,55]. Alternatively, diffusion control can be accomplished by using specific light triggered channels/pores embedded into the liposome or polymersome membrane. In these studies, the above described classical photochemical approach was aimed to synthesize an inert “caged” precursor of the active compound and use flash photolysis to rapidly release the compound in a spatially controlled manner.
Considering the importance of different triggering stimuli to which the channel must react, the ideal situation would be the development and use of a universal protein scaffold, easily obtained introducing specific modifications by protein engineering techniques [56].
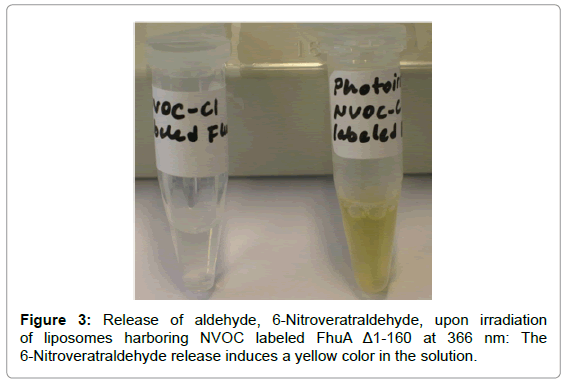
Moving from a chemical to a light triggered system, the same engineered set of FhuAΔ1-160 variants have been used and tested [57]. The FhuAΔ 1-160 variants were chemically modified by the aminogroup labeling agent 6-nitroveratryloxycarbonyl chloride (NVOCCl) and 2-(2-nitrophenyl)propyloxycarbonyl chloride (NPPOC-Cl) embedded into liposomes membranes encapsulating horseradish peroxidase (HRP). HRP substrate 3,3',5,5'-tetramethylbenzidine (TMB) conversion was determined by diffusion limited translocation through the used FhuAΔ 1-160 variant channels using a well-established previously reported colorimetric HRP/TMB detection system [5,29,30]. Upon external light stimulus at 366 nm, NVOC or NPPOC is cleaved off, releasing an aldehyde detected for further proof and quantitative measurements of the lysine chemical modifications [58]. Authors presented the successful equipping of a liposome platform with light triggered functionality responsive to external conditions by integrating bacterial channel protein, FhuAΔ1-160, into lipid membrane. This system demonstrates the feasibility of a nanometer-sized bioreactor with size-selective substance diffusion and a defined trigger mechanism that allows external control of a defined functionality inside the enzyme-protecting, aqueous pore containing synthetic liposome [57].
In addition, a colorimetric aldehyde [58] determination test was performed for further proof of the lysine chemical modifications. Upon irradiation of liposomes harboring NVOC labeled FhuAΔ1-160 at 366 nm the release of aldehyde, 6-Nitroveratraldehyde, induced a yellow color in the solution (Figure 3).
Labeling all lysine amino acids inside the β-barrel showed that the product formation was hindered which was related to the hindered substrate diffusion and upon light triggering the product formation was regained. Beer-Lambert law was used to calculate average TMB conversion. The previous findings demonstrated 65 nM/s TMB diffusion through FhuAΔ1-160 upon external stimuli (light) [29,57] whereas 29 μM calcein release was observed through FhuAΔ1-160 embedded in nanocontainers upon external stimuli (1,4-dithio-d,l-threitol (DTT) reduction) [15]. Another study demonstrated 43% calcein release of liposomal content in 50 min in the case of nanocontainers harboring photo-induced MscL pore forming protein [28]. These examples show that different sizes of molecules and shape and characteristics of channels greatly effect molecule diffusion rates. Most of the studies to our knowledge including our previous work did not elaborate many important properties of pore forming proteins. For example channel selectivity, the distribution of the pore charges and their discreteness, thermodynamic equilibrium of molecules in the pore with solution, the shape of the channel and the profile of the dielectric constant, water structuring in pores and pore and membrane deformability/ adjustability [59] are crucial effects that have to be considered in order to discuss compound diffusion through transmembrane channel proteins embedded in lipid bilayers. For example, Singh et al. [60] showed the relevant kinetic and electric parameters of antibiotic enrofloxacin interaction with the channelsurface in the presence of magnesium. Ion current fluctuation analysis has often been used to characterize permeation of water-soluble antibiotics through a membrane channel. Favorable interaction increases the concentration of the drug inside the channel and enhances the number of translocation events. Although most of the antibiotic shows a very low affinity for the OmpF channel, enrofloxacin is an exception. The presence of magnesium chloride alters the affinity of the antibiotic for the channel wall. Binding of Mg2+ produce a local charge inversion that favors the antibiotic to cross the constriction region and further translocate with a faster kinetics [60]. The penetration of antibiotic molecules into the channels and subsequentinteraction of the drug with the channel may interrupt the ion current [61]. The analysis of ion current fluctuation allows us to obtain permeation rates as previously shown for sugars and ampicillin [60,62]. In another example, Bajaj etal. [63] investigated the relation between pore properties, the structure of the antibiotic, and the correlation with the uptake of these molecules. In their study a single OmpPst1 was reconstituted into an artificial planar lipid bilayer and time-resolved ion current fluctuations were characterized in the presence of antibiotics. Single-channel analysis of ion currents through a porin in the presence of antibiotics revealed effective binding constants and subsequently the transport parameters at a single-molecule level [63].
Functionalized nanocontainers controlled by enzyme logic systems
Recent advances in signal processing with cascades of enzymatic reactions realizing logic gates, as well as progress in networking these gates and coupling of the resulting systems to signal-responsive systems for output readout, have opened new biosensing opportunities. Digitally operating biosensors/bioactuators logically processing multiple biochemical signals through Boolean logic networks composed of biomolecular systems, yielding the final output signal as YES/NO responses. Biochemical signal processing systems are being developed for novel biosensor concepts, with multiple input signals being processed via enzymatic or antibody affinity recognition processes, in combination with electrochemical or optical output signal. Extensive research in unconventional chemical computing resulted in the formulation of various logic systems processing chemical signals according to Boolean logic using chemical means [64-68]. The use of biomolecular/biological tools for logic operations [69-71], such as proteins [72-75], DNA [76] and whole-cells [77], broadened the research area of signal processing benefiting from the specificity of biomolecular interactions. Recently pioneered enzymatic logic gates allowed signal processing mimicking various Boolean logic operations: AND, OR, XOR, NAND, NOR, etc [78].
Systems specifically designed for analysis of biomedical conditions use medically relevant biomarkers as input signals at their physiological concentrations. This poses additional challenge, compared to biomolecular logic systems which process external chemical or/and physical signals without medical relevance and use the physical zero and a conveniently high concentrations of chemical signals to mimic logic 0 and 1 values, respectively [79]. Enzyme logic systems were applied to control states of signal-responsive materials in the form of a switchable membrane [80], an emulsion [81] and a nanoparticle assembly [82], which operated as nanostructured actuators reversibly changing the structure and material properties upon receiving signals from the biomolecular systems [79]. Signal-responsive materials, in the form of polymer brushes, were bound to electrode surfaces allowing reversible switching of electrochemical activities ON/OFF by complex biochemical signals processed through enzyme biocatalytic cascades [83,84]. Privman et al. [79] presented an example of a switchable electrode interface logically controlled by the enzyme cascade processing biomarkers characteristic of an injury. A liver injury, using the alanine transaminase (ALT) and lactate dehydrogenase (LDH) as the biomarker inputs for the AND logic gate [85], was selected to illustrate this concept. The biochemically controlled actuation achieved in that study present integrated “smart” ‘Sense/Act’ (biosensor-bioactuator) systems processing biomedical signals, making programmed decision, and then performing an actuation according to the biomedical needs [79].
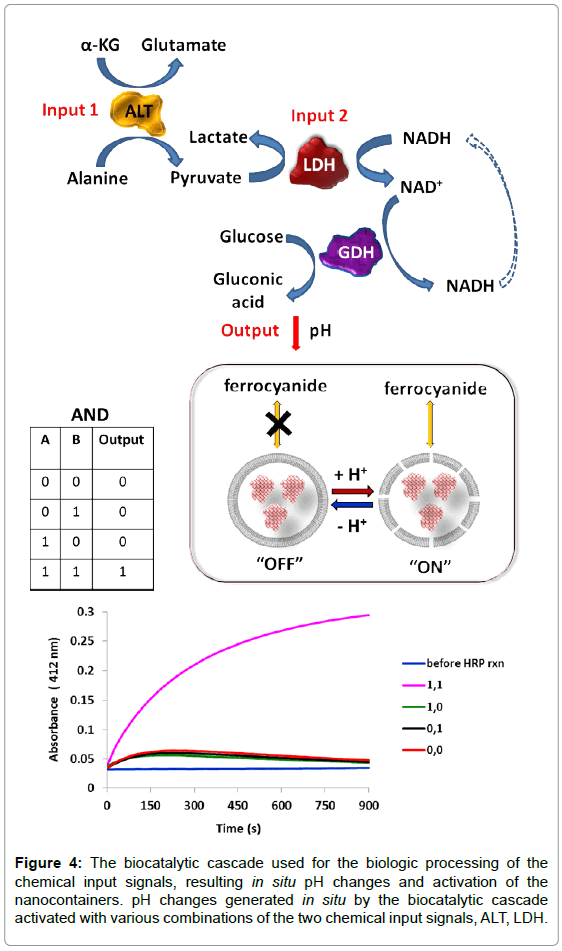
An enzymatic cascade which includes cooperative action of both biomarkers mimicking the AND Boolean logic gate was designed to signal the presence of these biomarkers [86]. Figure 4 shows the biocatalytic cascade activated by the biomarker inputs, ALT and LDH, primary resulting in the conversion of NADH to its oxidized form NAD+. In addition to modified electrodes [84], in order to convert the NAD+ signal to pH changes affecting the state of the pH sensitive liposomal nanocontainers (without the channel proteins in this case), glucose dehydrogenase (GDH) and glucose were added to the system to operate as the last step of the enzyme cascade. As it is expected, in situ generation of NAD+ resulted in the oxidation of glucose catalyzed by the dehydrogenase enzyme, causing an acidification of the solution, and producing ΔpH as the final output signal. The biocatalytic cascade lowering the pHamount can only be completed in the presence of both the input signals (ALT and LDH), thus mimicking the AND logic operation. The challenge of this work was to achieve the bioactivation only with the signal combination 1, 1 (Figure 4), while all other combinations preserve the OFF (i.e., closed vesicle) state of the nanocontainer. In order to realize this specific switchable behavior of the nanocontainer, its pH-controlled transition from the OFF to ON statemust be coordinated with the pH output signals produced by the biocatalytic system of liver injury.
Figure 4: The biocatalytic cascade used for the biologic processing of the chemical input signals, resulting in situ pH changes and activation of the nanocontainers. pH changes generated in situ by the biocatalytic cascade activated with various combinations of the two chemical input signals, ALT, LDH.
Conclusion
In order to reduce the toxicity and increase the efficacy of drugs, there is a need for smart drug delivery systems. Liposomes are one of the promising tools for this purpose. For drug delivery to specific tissues and organs, a localized, triggerable and controlled compound release by chemical or physical stimuli is an important attribute for efficient therapies. An ideal liposomal delivery system should be stable, long-circulating, accumulating at the target site and releasing its drug in a “time and dose-dependent” manner. Even though there have been many developments to this end, the dilemma of having a stable liposome during circulation but converting it into a leaky structure at the target site is still a major challenge. So far, most attempts have focused on destabilizing the liposome in response to a specific stimulus at a target site, but with limited success. Passive release of the encapsulated agent is often too slow to achieve an optimal therapeutic effect. This realization has stimulated new efforts directed toward the development of triggered release mechanisms that accelerate drug release rates and promote maximum efficacy. Our approach enables us to design novel triggered release mechanisms employing channel plugs with tunable release kinetics which can rapidly be resized to the compound of interest by keeping the stable liposome and building in a remotecontrolled valve. The valve is a pore-forming bacterial membrane protein. It has been engineered such that, after being reconstituted into the liposomes, its opening and closing can be controlled on command by the ambient pH or light.
Recent development of triggered release mechanisms was described. Stable polymer and liposome structuresand reconstituted pore-forming bacterial membrane proteinsare used as the release mechanism. Channel proteins like the engineered, bacterial channel protein FhuAΔ1-160 can be an alternative to artificial chemically synthesized nanopores. In order to be competitive, channel proteins must be flexible enough to be modified in their geometry, i.e. length and diameter. Such tuned geometry can give rise to a new set of synthetic nanochannels with desired lengths and diameters broadening the FhuAΔ1-160 applications.
The FhuAΔ1-160 and its variants, have been used as an externally controlled gate in sterically stable liposomes for a triggered release of the liposomal content on command. The channel protein has been engineered and chemically modified to respond to specific signals present in the environment. This strategy provides flexibility for fine-tuning the liposome’s response to its environment. The release mechanism has been investigated in reduction and light triggered systems. In this approach, a universal molecule-scale compound release system based on an engineered biological unit acting as an intelligent triggered compound release device has been developed. What makes our technology superior is its ability to allow us to design channel plugs with tunable release kinetics which can rapidly be resized to the compound of interest and modulated to be responsive for different type of external stimuli. However, one very important requirement yet has to be fulfilled which is a reversible system for the development of triggered and time-controlled (at a time or long-lasting) release depending on the application.
Liposomal nanocontainers are applied for the chemical actuation triggered by appropriate combinations of medically relevant biomarkers. Nanocontainer systems were selectively activated by the input signals combination, which was processed by the enzyme logic systems. Functionalized nanocontainer systems triggered upon receiving output systems can be used for biosensor and bioactuator integrated systems.
Acknowledgements
The authors thank to Penn State Hershey Medical Center for financial support; Jacobs University Bremen (Bremen, Germany) and Clarkson University (Potsdam, NY) for the resources offered.
References
- Schirmer T (1998) General and specific porins from bacterial outer membranes. J Struct Biol 121: 101-109.
- Delcour AH (1997) Function and modulation of bacterial porins: insights from electrophysiology. FEMS Microbiol Lett 151: 115-123.
- Duncan R (2003) The dawning era of polymer therapeutics. Nat Rev Drug Discov 2: 347-360.
- Broz P, Driamov S, Ziegler J, Ben-Haim N, Marsch S, et al. (2006) Toward intelligent nanosize bioreactors: a pH-switchable, channel-equipped, functional polymer nanocontainer. Nano Lett 6: 2349-2353.
- Nallani M, Benito S, Onaca O, Graff A, Lindemann M, et al. (2006) A nanocompartment system (Synthosome) designed for biotechnological applications. J Biotechnol 123: 50-59.
- Nardin C,WidmerJ, Winterhalter M, Meier W(2001) Amphiphilic block copolymer nanocontainers as bioreactors. European Physical Journal E 4: 403-410
- Ranquin A, Versées W, Meier W, Steyaert J, Van Gelder P (2005) Therapeutic nanoreactors: combining chemistry and biology in a novel triblock copolymer drug delivery system. Nano Lett 5: 2220-2224.
- Meier W, Nardin C, Winterhalter M (2000) Reconstitution of Channel Proteins in (Polymerized) ABA Triblock Copolymer Membranes This work was supported by the Swiss National Science Foundation. We thank Dr. T. Hirt and Dr. J. Leukel for the synthesis of the triblock copolymer, Dr. P. Van Gelder and Dr. F. Dumas for bright and helpful discussions, and T. Haefele for his contribution to the experimental part. Angew Chem Int Ed Engl 39: 4599-4602.
- Ye J, van den Berg B (2004) Crystal structure of the bacterial nucleoside transporter Tsx. EMBO J 23: 3187-3195.
- Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W (1998) Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282: 2215-2220.
- Braun M, Killmann H, Braun V (1999) The beta-barrel domain of FhuADelta5-160 is sufficient for TonB-dependent FhuA activities of Escherichia coli. Mol Microbiol 33: 1037-1049.
- Braun M, Killmann H, Maier E, Benz R, Braun V (2002) Diffusion through channel derivatives of the Escherichia coli FhuA transport protein. Eur J Biochem 269: 4948-4959.
- Koebnik R, Locher KP, Van Gelder P (2000) Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol 37: 239-253.
- Nallani M, Onaca O, Gera N, Hildenbrand K, Hoheisel W, et al. (2006) A nanophosphor-based method for selective DNA recovery in Synthosomes. Biotechnol J 1: 828-834.
- Onaca O, Sarkar P, Roccatano D, Friedrich T and Hauer, B.et al. (2008) Functionalized nanocompartments (synthosomes) with a reduction-triggered release system. Angewandte Chemie-International Edition 47:7029-7031.
- Muhammad N, Dworeck T, Fioroni M, Schwaneberg U (2011) Engineering of the E. coli outer membrane protein FhuA to overcome the hydrophobic mismatch in thick polymeric membranes. J Nanobiotechnology 9: 8.
- Bangham AD(1994) Review of Lasic, liposomes: from physics to applications. Biophysical Journal 67: 1358-1359
- Bangham AD, Standish MM, Watkins JC (1965) Diffusion of univalent ions across the lamellae of swollen phospholipids. J Mol Biol 13: 238-252.
- Woodle MC, Lasic DD (1992) Sterically stabilized liposomes. Biochim Biophys Acta 1113: 171-199.
- Koçer A (2007) A remote controlled valve in liposomes for triggered liposomal release. J Liposome Res 17: 219-225.
- Thomas JL, Tirrell DA (1992) Polyelectrolyte-sensitized phospholipid vesicles. Accounts of Chemical Research 25:336-342
- Meers P (2001) Enzyme-activated targeting of liposomes. Adv Drug Deliv Rev 53: 265-272.
- Guo X, Szoka FC Jr (2003) Chemical approaches to triggerable lipid vesicles for drug and gene delivery. Acc Chem Res 36: 335-341.
- Drummond DC, Meyer O, Hong K, Kirpotin DB, Papahadjopoulos D (1999) Optimizing liposomes for delivery of chemotherapeutic agents to solid tumors. Pharmacol Rev 51: 691-743.
- Graff A, Winterhalter M, Meier W (2001) Nanoreactors from polymer-stabilized liposomes. Langmuir 17: 919-923
- Koçer A, Walko M, Bulten E, Halza E, Feringa BL, et al. (2006) Rationally designed chemical modulators convert a bacterial channel protein into a pH-sensory valve. Angew Chem Int Ed Engl 45: 3126-3130.
- Sauer M, Streich D, Meier W (2001) pH-sensitive nanocontainers. Advanced Materials 13: 1649-1651.
- Koçer A, Walko M, Meijberg W, Feringa BL (2005) A light-actuated nanovalve derived from a channel protein. Science 309: 755-758.
- Güven A, Fioroni M, Hauer B, Schwaneberg U (2010) Molecular understanding of sterically controlled compound release through an engineered channel protein (FhuA). J Nanobiotechnology 8: 14.
- JosephyPD, Eling T, Mason R.P (1982) The horseradish peroxidase catalyzed oxidation of 3,5,3',5'-tetramethylbenzidine free radical and charge transfer complex intermediates. Journal of Biological Chemistry 257: 3669-3675.
- Ruysschaert T, Germain M, Gomes JF, Fournier D, Sukhorukov GB, et al. (2004) Liposome-based nanocapsules. IEEE Trans Nanobioscience 3: 49-55.
- SabÃn J, Prieto G, Ruso JM, Hidalgo-Alvarez R, Sarmiento F (2006) Size and stability of liposomes: a possible role of hydration and osmotic forces. Eur Phys J E Soft Matter 20: 401-408.
- Schlessinger A, Rost B (2005) Protein flexibility and rigidity predicted from sequence. Proteins 61: 115-126.
- Krewinkel M, Dworeck T, Fioroni M (2011) Engineering of an E. coli outer membrane protein FhuA with increased channel diameter. J Nanobiotechnology 9: 33.
- Salmain M, Jaouen G (2003) Side-chain selective and covalent labelling of proteins with transition organometallic complexes. Perspectives in biology. ComptesRendusChimie 6 : 249-258
- Means GE, Feeney RE (1990) Chemical modifications of proteins: history and applications. Bioconjug Chem 1: 2-12.
- Butler PJ, Harris JI, Hartley BS, Leberman R (1967) Reversible blocking of peptide amino groups by maleic anhydride. Biochem J 103: 78P-79P.
- Glazer AN (1970) Specific chemical modification of proteins. Annu Rev Biochem 39: 101-130.
- Givens RS, Timberlake GT, Conrad PG 2nd, Yousef AL, Weber JF, et al. (2003) A photoactivated diazopyruvoyl cross-linking agent for bonding tissue containing type-I collagen. Photochem Photobiol 78: 23-29.
- Sheehan JC, Umezawa K (1973) Phenacyl photosensitive blocking groups. Journal of Organic Chemistry 38: 3771-3774.
- McCray JA, Trentham DR (1989) Properties and uses of photoreactive caged compounds. Annu Rev Biophys Biophys Chem 18: 239-270.
- Patchornick A, Amit B, Woodward RB (1970) Photosensitive Protecting Groups. Journal of the American Chemical Society 92: 6333-6337
- Houseman BT, Huh JH, Kron SJ, Mrksich M (2002) Peptide chips for the quantitative evaluation of protein kinase activity. Nat Biotechnol 20: 270-274.
- Vernet G (2002) DNA-chip technology and infectious diseases. Virus Res 82: 65-71.
- McGall GH, Barone AD, Diggelmann M, Fodor SPA and Gentalen E et al. (1997) The efficiency of light-directed synthesis of DNA arrays on glass substrates. Journal of the American Chemical Society119: 5081-5090
- Bochet CG (2000) Wavelength-selective cleavage of photolabile protecting groups. Tetrahedron Letters 41: 6341-6346
- Beier M, Hoheisel JD (1999) New developments in light-controlled synthesis of DNA-arrays. Nucleosides & Nucleotides 18: 1301-1304
- Stengele KP, Bühler J, Bühler S, Kvassiouk E, Green R, et al. (2005) Recent highlights on photolithic oligonucleotide array in situ synthesis. Nucleosides Nucleotides Nucleic Acids 24: 891-896.
- Giegrich,H, Eisele-BuhlerS, Hermann C, Kvasyuk E and Charubala R et al.New photolabile protecting groups in nucleoside and nucleotide chemistry - Synthesis, cleavage mechanisms and applications. Nucleosides & Nucleotides 17:1987-1996
- Shum P, Kim JM, Thompson DH (2001) Phototriggering of liposomal drug delivery systems. Adv Drug Deliv Rev 53: 273-284.
- Wan Y, Angleson JK, Kutateladze AG (2002) Liposomes from novel photolabile phospholipids: light-induced unloading of small molecules as monitored by PFG NMR. J Am Chem Soc 124: 5610-5611.
- Dudia A, Koçer A, Subramaniam V, Kanger JS (2008) Biofunctionalized lipid-polymer hybrid nanocontainers with controlled permeability. Nano Lett 8: 1105-1110.
- Wu G, Mikhailovsky A, Khant HA, Fu C, Chiu W, et al. (2008) Remotely triggered liposome release by near-infrared light absorption via hollow gold nanoshells. J Am Chem Soc 130: 8175-8177.
- Bondurant B, Mueller A, O'Brien DF (2001) Photoinitiated destabilization of sterically stabilized liposomes. Biochim Biophys Acta 1511: 113-122.
- Morgan CG, Bisby RH, Johnson SA, Mitchell AC (1995) Fast solute release from photosensitive liposomes: an alternative to 'caged' reagents for use in biological systems. FEBS Lett 375: 113-116.
- Shivange AV, Marienhagen J, Mundhada H, Schenk A, Schwaneberg U (2009) Advances in generating functional diversity for directed protein evolution. Curr Opin Chem Biol 13: 19-25.
- Guven A, Dworeck T, Fioroni M, Schwaneberg U (2011) Residue K556 – A light triggerable gatekeeper to sterically control translocation in FhuA. Advanced Engineering Materials13: 324-329.
- Sawicki E, Johnson H, Fox FT, Stanley TW(1962) Sensitive new test for formaldehyde and pyruvaldehyde with 2-hydroxycarbazole. MikrochimicaActa 4:741-745
- Cherstvy AG (2006) Electrostatic screening and energy barriers of ions in low-dielectric membranes. J Phys Chem B 110: 14503-14506.
- Singh PR, Ceccarelli M, Lovelle M, Winterhalter M, Mahendran KR (2012) Antibiotic permeation across the OmpF channel: modulation of the affinity site in the presence of magnesium. J Phys Chem B 116: 4433-4438.
- Nestorovich EM, Danelon C, Winterhalter M, Bezrukov SM (2002) Designed to penetrate: time-resolved interaction of single antibiotic molecules with bacterial pores. Proc Natl Acad Sci U S A 99: 9789-9794.
- Benz R, Schmid A, Nakae T, Vos-Scheperkeuter GH (1986) Pore formation by LamB of Escherichia coli in lipid bilayer membranes. J Bacteriol 165: 978-986.
- Bajaj H,Tran QT, Mahendran KR, Nasrallah C and Colletier JP et al.(2012) Antibiotic Uptake through Membrane Channels: Role of Providenciastuartii OmpPst1 Porin in Carbapenem Resistance. Biochemistry including biophysical chemistry and molecular biology 51:10244-10249.
- De Silva AP, Uchiyama S, Vance TP, Wannalerse BA (2007) supramolecular chemistry basis for molecular logic and computation. Coordination Chemistry Reviews 251: 1623-1632.
- De Silva AP, Uchiyama S (2007) Molecular logic and computing. Nat Nanotechnol 2: 399-410.
- Szaciłowski K (2008) Digital information processing in molecular systems. Chem Rev 108: 3481-3548.
- Credi A (2007) Molecules that make decisions. Angew Chem Int Ed Engl 46: 5472-5475.
- Andréasson J, Pischel U (2010) Smart molecules at work--mimicking advanced logic operations. Chem Soc Rev 39: 174-188.
- Katz E, Privman V (2010) Enzyme-based logic systems for information processing. Chem Soc Rev 39: 1835-1857.
- Saghatelian A, Völcker NH, Guckian KM, Lin VS, Ghadiri MR (2003) DNA-based photonic logic gates: AND, NAND, and INHIBIT. J Am Chem Soc 125: 346-347.
- Ashkenasy G, Ghadiri MR (2004) Boolean logic functions of a synthetic peptide network. J Am Chem Soc 126: 11140-11141.
- Sivan S, Lotan N (1999) A biochemical logic gate using an enzyme and its inhibitor. 1. The inhibitor as switching element. Biotechnol Prog 15: 964-970.
- Sivan S, Tuchman S, Lotan N (2003) A biochemical logic gate using an enzyme and its inhibitor. Part II: The logic gate. Biosystems 70: 21-33.
- Ashkenazi G, Ripoll DR, Lotan N, Scheraga HA (1997) A molecular switch for biochemical logic gates: conformational studies. Biosens Bioelectron 12: 85-95.
- Unger R, Moult J (2006) Towards computing with proteins. Proteins 63: 53-64.
- Xu, J.; Tan, G.J., A review on DNA computing. Journal of Computational and Theoretical Nanoscience2007, 4, 1219-1230.
- Simpson ML, Sayler GS, Fleming JT, Applegate B (2001) Whole-cell biocomputing. Trends Biotechnol 19: 317-323.
- Baron R, Lioubashevski O, Katz E, Niazov T, Willner I (2006) Logic gates and elementary computing by enzymes. J Phys Chem A 110: 8548-8553.
- Privman M, Tam TK, Bocharova V, Halamek J and Wang J et al. (2011)Responsive interface switchable by logically processed physiological signals: toward “smart†actuators for signal amplification and drug delivery. ACS Applied Materials & Interfaces 3: 1620-1623
- Tokarev I, Gopishetty V, Zhou J, Pita M, Motornov M, et al. (2009) Stimuli-responsive hydrogel membranes coupled with biocatalytic processes. ACS Appl Mater Interfaces 1: 532-536.
- Motornov M, Zhou J, Pita M, Tokarev I, Gopishetty V, et al. (2009) An integrated multifunctional nanosystem from command nanoparticles and enzymes. Small 5: 817-820.
- Motornov M, Zhou J, Pita M, Gopishetty V, Tokarev I, et al. (2008) "Chemical transformers" from nanoparticle ensembles operated with logic. Nano Lett 8: 2993-2997.
- Zhou J, Tam TK, Pita M, Ornatska M, Minko S, et al. (2009) Bioelectrocatalytic system coupled with enzyme-based biocomputing ensembles performing boolean logic operations: approaching "smart" physiologically controlled biointerfaces. ACS Appl Mater Interfaces 1: 144-149.
- Privman M, Tam TK, Pita M, Katz E (2009) Switchable electrode controlled by enzyme logic network system: approaching physiologically regulated bioelectronics. J Am Chem Soc 131: 1314-1321.
- Halámek J, Bocharova V, Chinnapareddy S, Windmiller JR, Strack G, et al. (2010) Multi-enzyme logic network architectures for assessing injuries: digital processing of biomarkers. Mol Biosyst 6: 2554-2560.
- Halámek J, Windmiller JR, Zhou J, Chuang MC, Santhosh P, et al. (2010) Multiplexing of injury codes for the parallel operation of enzyme logic gates. Analyst 135: 2249-2259.
Citation: Güven A, Fioroni M, Güven G (2013) Functionalized Nanocontainers Operated as Controlled Release Systems and Bioactuators. Biosens J 2:106. DOI: 10.4172/2090-4967.1000106
Copyright: © 2013 Singh A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 19629
- [From(publication date): 12-2013 - Nov 12, 2025]
- Breakdown by view type
- HTML page views: 14760
- PDF downloads: 4869