Review Article Open Access
Glycosylphosphatidylinositol-Anchored Protein Chips for Patient-Tailored Multi-Parameter Proteomics
| Günter Müller* | |
| Department I, Genetics, Biocenter, Ludwig-Maximilians-University Munich, 82152 Martinsried near Munich, Germany | |
| Corresponding Author : | Günter Müller Sanofi Germany GmbH R & D Diabetes Industrial Park Höchst Bldg. H821.65926 Frankfurt am Main, Germany Tel: +49 69 305 4271 Fax: +49 69 305 81901 E-mail: guenter.mueller@sanofi.com |
| Received November 11, 2010; Accepted November 20, 2011; Published December 15, 2011 | |
| Citation: Müller G (2011) Glycosylphosphatidylinositol-Anchored Protein Chips for Patient-Tailored Multi-Parameter Proteomics. J Biochip Tissue chip S3:001. doi:10.4172/2153-0777.S3-001 | |
| Copyright: © 2011 Müller G. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Journal of Bioengineering and Bioelectronics
Abstract
For the past two decades, highly multiplexed microarray-based assays have had a major impact on genomic studies, including DNA microarrays for the characterization of cellular states, phenotypic differences and disease markers. However, only recently similar technologies have become widely available for the analysis of the proteome. Traditional single protein assays can yield important information, but they lack a more systematic and comprehensive view that is ultimately required for understanding of the complexity of biological regulation at the cellular and whole-body levels. Unfortunately, the majority of well-established proteomic methods, such as mass spectrometric, 2D-electrophoretic, pull-down or yeast two-hybrid technologies, address only one or two parameters simultaneously. Current efforts are focusing on miniaturizing, multiplexing and generating protein microarrays in a convenient, reproducible and cost-effective fashion. Protein microarrays can be applied for abundance-based arrays, that aim at describing and identifying relative protein amounts, and function-based assays, that manage to identify interactions between distinct proteins or proteins and small molecules, such as enzyme and substrate or inhibitor, as well as to test for post-translational modifications and binding or signalling activities. The recent development of the next-generation chips which are based on glycosylphosphatidyl-inositol-anchored (GPI-) proteins and nanoparticles may facilitate the development of abundance and function-based protein chips. Current GPI-protein chips can be designed as technology platforms designed for quantitative, functional and multiplexed determination of (protein) analytes from cells, tissues, serum and body fluids from very limited amounts of sample. In the near future the application of this class of microarray will be further enlarged ranging from basic research to clinical trials with considerably increased sensitivity, precision and reliability. Ultimately, (GPI-) protein chips may replace conventional non-array-based technologies in personalized and systems biology-based diagnosis.
| Keywords |
| Protein analytics; protein production; bioanalytical assays; personalized medicine; glycosylphosphatidylinositol; functional microarrays; multiplex analysis |
| Introduction |
| Proteins are classically analyzed by a huge variety of electrophoretic, ELISA and liquid chromatography procedures that are typically time-consuming and labor-intensive and, in general, are not compatible with the high-throughput scale. Furthermore, multiplex measurements for several protein analytes by the currently available methods necessitate multiple divisions of the original sample with multiple separate tests for each analyte which is accompanied by considerable costs. In contrast, protein chips enable the simultaneous determination of many different proteins in a single test without the need for splitting of the original sample and are therefore much faster (minutes vs. hours), more convenient (one test for the determination of multiple analytes) and less expensive (40 to 20%) than the classical, in particular ELISA-based, technologies. The basic principle of microarray technology was first introduced more than 20 years ago [1]. The underlying theory stated that a tiny spot of solid-phase purified antibody provides substantially better sensitivity than when used in conventional fluidphase immunoassay formats. Driven by large-scale genome sequencing projects, DNA microarray technology became the first application of this theory and has been widely used for gene expression profiling [2-6]. However, biological functions are preformed by proteins rather than nucleic acids. Moreover, RNA expression levels are not always correlated well to protein expression levels and it turned out to be almost impossible to predict the functional characteristics of a polypeptide encoded by a given gene simply based on its expression profiles [7]. Therefore, the focus on studies on protein structures, functions and protein-protein interactions should facilitate a more thorough characterization of the physiological function of a given gene [8-10]. |
| During the last decade a large body of evidence has accumulated that protein chips may revolutionize this area by their intrinsic capability of very rapid and simultaneous handling of many samples, and after special adaption also of multi-parameter analysis, in combination with validity, sensitivity, robustness, miniaturization and relatively low costs. Moreover, protein chips can not only be used for the evaluation of polypeptides of any size but also for the determination of small nonproteinaceous analytes, such as lipids, carbohydrates and intermediary metabolites [11-15]. This considerably expands the application profile of protein chips from mere biotechnological process analytics to various research areas in diagnosis, drug discovery and therapeutic monitoring [16-18]: (i) Personalized medicine with its scope of individualized diagnosis as well as therapy, (ii) systems biology with its aim of understanding the pathophysiology of common multifactorial diseases at the level of cells, tissues and organisms and of interacting signalling molecules and metabolic enzymes, that form complex networks rather than linear pathways and (iii) tissue engineering with its potential to provide functional organs which develop in vitro from (e.g. adipose tissue- derived) mesenchymal stem cells isolated from the corresponding patient and have to be analyzed for maintenance of the differentiated and functional state by implantable protein chips and biosensors. |
| Conventional protein chips |
| A protein microarray also termed a protein chip, is a solid surface, typically made of glass, on which thousands of different proteins, such as antigens, antibodies, enzymes, substrates, are immobilised in discrete spatial locations, forming a high-density protein dot matrix. Abundance-analyzing protein chips are typically composed of well-characterized biomolecules exerting specific binding activities, such as antibodies, for the qualitative elucidation of patterns of many to all (protein) analytes from a complex sample, such as serum and cell lysates, or for the quantitative determination of the presence of a single to a few proteins of interest. They have been applied for protein expression analysis, biomarker identification, cell surface marker profiling and clinical diagnosis [17]. Function-analyzing protein chips have been constructed by printing a large number of separately purified proteins and used primarily to comprehensively investigate the biochemical characteristics and activities of those immobilised proteins [15,19-21]. |
| On basis of the considerable differences in size, structure, charge and hydrophobicity of individual proteins, the generation and handling of protein chips is much more complicated and less straightforward and standardizable than those of DNA chips. Unlike DNA molecules, full-length polypeptides cannot be directly synthesized in vitro at high efficacy per se and since most proteins have to fold and to be post-translationally modified in correct fashion. These processes rely on complex molecular machineries consisting of a multitude of translation factors, chaperones and accessory components, which are not easily amenable for the use in microarrays. In consequence, the proteins used so far for the construction of high-content protein chips have to be individually expressed and purified. Since proteins have to fold correctly in order to remain in the active state, they are susceptible to inactivation due to loss of their native conformation if immobilised directly on a solid surface. Polypeptides vary considerably in their accessibility for direct chemical crosslinking or non-covalent adsorption or indirect binding via antibodies to a matrix or carrier surface. To complicate the matter chip surfaces can be modified by only one or two types of chemical or biological groups so far. Together these limitations pose high challenges for the optimisation of the immobilisation of (protein) analytes at the slide surface of the different protein chip configurations. |
| Fabrication – chip printing |
| For the fabrication of two-dimensional microarrays the principal and most critical step relies on the efficient dispensing process of biological fluids leading to the accessible arrangement of dense, yet spatially discrete, uniform and homogeneous nanoliter-spots on a substrate surface which guarantees the structural and functional integrity of the probe [22-27]. In addition, the dispensing process should be compatible with low costs and minimal sample volumes as well as with low risk for mutual sample contaminations and sample damage. Two basic categories of printing techniques, contact and non-contact ones, can be distinguished (Figures 1 and 2). During contact printing, the biological sample becomes deposited in course of physical contact of the printing device with the substrate. At variance, during non-contact printing there is no physical contact between the device and the substrate (e.g. laser writing, ink-jet printing, photolithography). Both of these array fabrication categories can be further classified into serial or parallel. During serial deposition, consecutively repeated operations of the printing device considerably slow down the fabrication efficacy. In this regard, parallel deposition with simultaneous printing of all spots constituting the array in a single operation is preferred for large-scale fabrication. Nevertheless, for current application the techniques for serial deposition are far more advanced than the newer and more complex parallel ones and therefore are considerably more common in practise [27]. |
| The direct contact between the printing device and the substrate is provoked by either solid pins, spit pins, nano-tips or microstamps (Figure 1). Historically, contact printing was initially performed using a single pin. Subsequently, methods were developed involving a multitude of pins that, however, do not encompass the complete array. Printed microarrays guarantee accurate quantitative analysis only, if the spots were of uniform morphology, i.e. identical spot-to-spot size, shape and surface characteristics, and precision in the positioning [27-30]. The morphology of the spots and its uniformity is predominantly affected by the sample viscosity, substrate planarity, substrate surface properties, pin surface properties and pin contact area. Moreover, the control of the robotic forward and backward movement of the pin and the regulation of environmental factors for constant humidity, air pressure and temperature as well as the minimization of contamination with dust represent the most critical factors for spot uniformity. A pin velocity of forward movement exceeding a critical threshold may cause inacceptable high inertial forces leading to the movement of considerable sample volumes out of the pin and thereby to enlargement of the spots and mutual spot-to-spot contamination due to large size [26-30]. In addition, pin printing is determined by the surface tension of the sample solution as well as its wettability on the substrate. The inherent danger of sample evaporation and drying-out from the wells and pin channels is efficiently prevented by maintenance of a high constant humidity. Sample viscosity and as a consequence also the dispensed volume is critically influenced by the temperature, which therefore has to be controlled very precisely. Finally, dust and contamination will seriously interfere with the fabrication of high-quality microarrays and have to be reduced with maximal care in order to minimize the risk of pin clogging. |
| In contrast to the serial deposition by pin printing that independently of the final practise always leads to direct contact between the substrate surface and a stamp or pin, non-contact printing technologies are of considerable heterogeneity ranging from photochemistry-based methods (that rely on the chemical treatment of the substrate and subsequent exposure with UV light using photomasks and can be categorized into photolithography and direct photochemical patterning) to laser writing to fluid droplet dispensing to microstamps [31-36]. Each of these technologies manages to deposit a large number of sample biomolecules at varying degree in parallel fashion (Figure 2). The most recent technology of nano-tip printing relies on scanning probe microscopy that enables nanoarrays of high density with spots of submicron size. Non-contact printing is characterized by two main advantages, considerably lowered risk for contamination and drastically increased throughput. The separation of the printing device and the substrate at each time during the analysis dramatically decreases the probability of cross-contamination and transfer of sample fluid from the primary spot to neighbouring ones. This makes extensive washing steps and repeated cleaning of the printing device between the individual printing operations unnecessary. Moreover, non-contact printing technologies probably have the greatest potential for further up-scaling of throughput in microarray fabrication. The majority of non-contact printers manage to deposit the sample fluids in parallel with production of the complete microarray in course of a single operation (Figure 2). |
| Taken together, the ideal profile for an array printing system, the reliable and durable generation of uniform and small-sized spots in a reproducibly dense and precise arrangement accompanied by the need for a minimal volume of solution and the avoidance of contamination and biomolecular damage as well as high costs is hardly fulfilled by the currently existing technologies. In particular, contact printing with solid and split pins suffers from tedious time and pre-printing requirements, problems of contamination, pin clogging, tip deformation, droplet uniformity and high costs, but nevertheless represents the most broadly applied technology in research laboratories in academia and industry at present on basis of its ability to provide reproducible results. At variance, non-contact printing and, in particular, the variant of inkjet technology may contribute to significant cost reduction, but with the current document printers is hampered by printing inaccuracies and the resulting formation of smeared and cross-contaminated satellite droplets of irregular shape and size. These disadvantages are also shared by the alternative technology of electrospray deposition with the additional problem of putative damage of certain biomolecules upon exposure to electric fields. This possibility of biomolecular denaturation holds also true for photochemical non-contact printing which has the potential for microarray fabrication at very high throughput. |
| Fabrication – chip surface |
| Selecting a proper surface for protein immobilization is crucial to the success of protein chips. An ideal surface should be able to retain polypeptide functionality with relatively high signal-to-noise ratios, and guarantee both high protein-binding capacity and long half-life. Different slide surfaces are in use, including aldehyde- and epoxy-derivatized glass surfaces, so-called “Fullmoon” slides, or “Schott” NHS-derivatized slides for random linkage of the proteins through amines [34,35], nitrocellulose [36,37] or gel-coated slides for coupling through diffusion and absorption [38,39] and nickel-coated slides for non-covalent binding of His6-tagged proteins [40]. In any case the immobilization step has to operate in efficient, reliable and quantitative fashion. This means that each polypeptide contained in the sample has to be retained at the chip surface, irrespective of its nature and abundance. This is complicated by the nature of the interaction of the individual sample polypeptide with the chip surface, i.e. the type and number of the amino acids involved in cross-linking or secondary bond formation, which typically is heterogeneous and can not be predicted. This may further contribute to variable and non-quantitative immobilization. |
| The typical substrate plate used are glass slides covered with polyvinylidene fluoride, nitrocellulose membrane or polystyrene. These materials are relatively soft, not excluding lateral spread of printed proteins, and hence enable only a limited density of polypeptides to be printed. Moreover, nitrocellulose membranes tend to generate high background and low signal-to-noise ratios for most purposes [41-43]. To circumvent these limitations, three-dimensional matrix arrays have been developed, in which the glass slides are coated with polyacrylamide or agarose to build a porous hydrophilic matrix, in which the proteins or antibodies are captured within the pores. Thereby, lateral diffusion is restricted and the size of the printed protein spots diminished, thus leading to elevated maximal complexity of the chip [44,45]. Protein activity is typically well preserved in those matrix arrays, and their protein-binding capacity is relatively high. In these regards as a further improvement, soft lithography has been introduced to fabricate nanowells on polydimethylsiloxane sheets fixed on top of microscope slides. These nanowell-based chips have been used for the immobilization of substrate proteins for profiling of the phosphorylation specificity of more than 100 protein kinases from budding yeast [46] and mammalian tyrosine protein kinases [47]. The open structure of the nanowells provides physical barriers and enables the consecutive addition of distinct buffers, that is critical for multi-step protein chips. The main disadvantage of this technique is the need for a specialized machinery for loading the nanowells at high density. |
| Alternatively, proteins, antigens or antibodies may be printed directly onto the plain glass slides, which are usually coated with a bifunctional cross-linker harbouring two distinct functional groups, one reacting with the glass surface, the other one with the desired protein (Figure 3). Those microarrays have been shown to have the advantages of high sensitivity, extended dynamic range and considerable spot-to-spot reproducibility. Moreover, upon immobilization of about 1000 of protein spots to aldehyde-activated plain glass surfaces, high-density protein chips have been created which enabled the detection of polypeptides belonging to completely distinct protein classes and assessed by different types of assays [41-45]. |
| After the immobilisation step, the protein chip becomes incubated with an appropriate well-characterized molecular probe which typically is a highly specific antibody (including single-chain or other variants), but may also consist of peptide-major histocompatibility complexes, carbohydrate-binding lectins, protein-interacting anti-lipocalins, protein-nucleic acids or RNA aptamers [48]. For demonstration of binding of the molecular probe to the chip surface, the probes have to be coupled to a dye, a fluorophore or an enzyme which catalyzes a luminescent reaction. The light, fluorescence and luminescence signals are detected by a multi-channel laser scanner or CCD camera with high resolution power resulting in typical patterns of (directly or indirectly) colored spots in regular arrangement. The higher the spot intensity, the larger the amount of the probe bound to the chip, the larger the amount of analyte immobilized onto the chip surface and thus contained in the sample. Data generation encompassing incubation of the chip with the probe, binding of the probe to the analyte, washing of the chip for removal of unbound probe, reading-out of the chip for the spot intensity and computer-based data transformation and calculation of the actual analyte content requires short periods of time only (typically 2 to 5 min for a single cycle). The number of samples that can be processed in parallel depends on the power of the chip printer used (see above). With the currently available microarrays up to 104 spots can be applied onto a typical light microscopic glass slide. Future nanoarrays will enable the spotting of 106 samples per slide with diameter as small as 250 nm and at distances as low as 100 nm, limited only by the resolution of the currently available scanners. |
| Categories and configurations |
| After successful printing and immobilization onto the slide surfaces, the protein analytes are evaluated for diverse parameters, which classify the protein chips into two categories, analytical configurations for the elucidation of molecular identity, structure or amount and functional configurations [49-54]. In the case of functional protein chips, a large number of proteins contained in complex biological samples, such as body fluids, or the total proteome of a cell or tissue for a systems biology approach are typically spotted. There is no need for extensive biochemical characterization of the proteins prior to immobilisation and chip analysis. The systematic screening for specific and divergent activities and functions encompasses protein-protein, protein-DNA, protein-carbohydrate, protein-lipid, protein-metabolite and protein-drug interactions as well as the identification of enzyme substrates or the detection of (undesired) immune and toxicological responses. |
| During the past decade different configurations of protein chips have been introduced into academic and industrial research applications which are basically discriminated by the mode of immobilisation and detection of the sample analyte. Each of these configurations is characterized by specific patterns of advantages and issues (Figure 3). The so-called “forward” protein chip critically depends on the quantitative immobilisation of the protein analytes by secondary bonds or covalent crosslinks at the chip surface as well as their quantitative detection by the labeled molecular probes, such as antibodies. However, even in case of quantitative recovery of the analytes, it can not be excluded that unspecific interactions and modifications involved herein will lead to masking of certain protein epitopes which are recognized by the detecting probe, e.g. the antibody. By nature, the amino acids involved in the interaction of the protein analyte with the chip surface can not be predicted and may vary with each procedure/cycle of the currently used immobilisation techniques. As a consequence, the detection of the analyte will not be quantitative leading to underestimation of the analyte content with considerable variation between distinct measurements. |
| To circumvent these problems the so-called “reverse” protein chip has been introduced which relies on the direct immobilisation of the protein analytes by binding to well-characterized immobilising probes (Figure 3). This type of analytical or functional protein chip represents the most convenient and powerful multiplexed detection platform and is commonly used for determining protein expression, cell surface markers and biomarkers as well as for clinical diagnosis. To correct for putative differences in the efficacy of the immobilisation of the analytes and of the read-out of the signal (e.g. “edge-effects” within the spots) in comparative studies (e.g. diseased vs. normal tissues), and in analogy to the typical two-colour mRNA/gene expression profiling approaches, sample and control analytes are labeled with the two distinct dyes, e.g. Cy3 and Cy5, by chemical means, then combined at equivalent ratio and finally transferred to the array. This procedure provides ratiometric quantitative data for the relative changes in protein abundance [55-57]. The immobilising probes, most often antibodies, are themselves coupled onto the chip surface by secondary interactions with coat materials (e.g. nitrocellulose) or covalent cross-linking to functional (e.g. amino) groups of the glass slide [58-60]. In contrast to “forward” chips, the “reverse” chips can be controlled and normalized for the coupling of the immobilising antibodies under standard conditions in order to compensate for eventual non-quantitative recovery of the protein analytes. This solely depends on the number of functional immobilising antibodies coupled to the chip surface. In any case, the immobilising antibodies have to be characterised for their ability to capture proteins out of the biological sample, since the arrayed antibodies may be prevented from interacting with their cognate protein analytes under the prevalent experimental conditions. |
| Within the “reverse” chip configuration the analyte can be detected by its direct labeling which, however, enables only the determination of the relative abundance between distinct samples rather than their absolute quantitative measurement. For the evaluation of the absolute amount of the protein analyte, the so-called “sandwich” configuration has to be used that relies on the specific binding of a detecting 2nd antibody that recognizes an epitope distinct from the immobilising 1st antibody and is labeled with a dye, fluorophore or luminescent enzyme. This chip configuration, which in principle represents a multiplexed version of standard ELISA immunoassays, combines the high sensitivity, accuracy and specificity of the “sandwich” approach with the throughput capability of microarray procedures [16-18](Figure 3). Determination of hundreds of proteins in complex biological samples, such as body fluids, can be performed by a single experiment using sample amounts which often enable only a single assay in the well of a microtiter plate. The specific and quantitative determination of the amount of protein analytes by abundance-based microarrays relies on two distinct analyte-specific probes, most often antibodies. The use of two probes/antibodies recognizing different epitopes of the same analyte, such as in typical ELISA sandwich immunoassays, circumvents the requirement for reliable labeling of the analyte and results in a highly specific and sensitive signal. However, as a consequence, the “reverse” configuration is less convenient for simultaneous measurement of many analytes and critically depends on the availability of two analyte-specific antibodies with non-overlapping epitopes for each analyte. Importantly, “sandwich” protein chips have the advantage of exquisite selectivity for the protein analyte due to the simultaneous operation of two different antibodies. |
| At the end of the “sandwich” protein chip procedure, the signal is generated by chemically modified, e.g. fluorescently labeled, secondary antibodies, resulting in a convenient two-step procedure without the need for a separate staining step [16]. Alternative signal generation strategies are based on commercially available biotinylated antibodies. The detection requires the incubation of the sandwich complex with Cy3/Cy5-labeled streptavidin or other streptavidin variants, such as Texas Red conjugates [28] or streptavidin-R-phycoerythrin (SAPE)[61,62]. The reported limits of detection are in the 10-pg/ml range. A further increase in sensitivity was achieved by amplification of the fluorescent signal with the help of a second layer of SAPE linked to the first layer through an anti-SAPE antibody resulting in a four-fold elevation of the signal [63]. Moreover, signal amplification in course of horseradish peroxidase-triggered thyramide radical formation has been demonstrated to considerably increase the number of biotin labels at the antibody spot [64]. The thyramide radicals provoke the crosslinking of biotin (or a fluorophore) to all exposed tyrosine residues of any protein analyte. In contrast, classical ELISA-based technologies rely on streptavidin- horseradish peroxidase (HRP) or species-specific antibodies linked to HRP or alkaline phosphatase in combination with chemiluminescent substrates. Their enzymic cleavage results in the generation of light around the antibody spot that is recorded by a CCD camera. Both the sensitivity and the accuracy of “sandwich” configurations with chemiluminescent signal generation are typically comparable to those of plate ELISA and higher than those of standard fluorescence read-outs [65,66]. Dynamic ranges for concentration measurements of typically two to three orders of magnitude (with any of these signal generation strategies) have been reported with intra- and inter-assay coefficients of variation usually below 3 and 10%, respectively. Their multiplexing capacity is limited by the maximal spot density, only, which by no doubt will be increased in course of future instrumental progress. |
| Nevertheless so far, “reverse” protein chips of either the direct labeling or “sandwich” configuration have not reached the robustness and accuracy of classical ELISA-based assays. These problems are caused in part by technical hurdles in the generation of antibody spots during microarray production which are uniform in amount, morphology and surface characteristics [16]. In this regard, gel-type antibody spots have considerable advantages compared to flat ones since they allow larger spaces between the immobilised capturing antibodies as well as exposure of their epitope-binding domains in the gel interior within a well-hydrated environment. This seems to ensure a higher portion of antibodies remaining in the correct three-dimensional functional conformation compared to antibody spots immobilised and dried up at the spot surface. In addition to isolated gel pads, glass slides uniformly coated with “HydroGel” have also been used for the printing of “reverse” antibody microarrays [67]. This “HydroGel” is characterized by low intrinsic fluorescence background which leads to a further increase in their sensitivity. However, longer washing times are required (> 30 min) for the gel-based arrays which for sake of elimination of piercing are typically printed using a non-contact piezoelectric microarrayer. In addition, the immobilisation capability of the capturing antibody may rely on its orientation toward the chip surface with the epitope-binding domains facing the bulk solution [68]. A major portion of the Fab’ fragments which have been coupled via biotinylation of the reduced thiols at their hinge regions to a streptavidin-coated chip surface, was demonstrated to remain in the fully active state, in contrast to the drastically reduced binding activity of randomly immobilised Fab’ fragments. However, the analogous optimization of the orientation of full-length antibodies turned out to be accompanied by less pronounced advantages compared to Fab’ fragments. |
| In any case, the overall performance of antibody microarrays critically depends on the quality of the capturing antibodies used for the “reverse” configuration. Even the highest sensitivity of the signal detection system will not compensate for low antibody performance. In principal, polyclonal, monoclonal and recombinant antibodies are appropriate for “reverse” sandwich chips with high binding affinity being most important for efficient capturing antibodies. For multiplex detection polyclonal antibodies can be used which, however, have to be purified by affinity chromatography due to their potential cross-reactivity with the capturing antibodies of the different analytes. |
| The identification of a pair of antibodies directed against the protein analyte, which are specific for distinct non-overlapping epitopes, i.e. with no or very little cross-reactivity of the detection or capturing antibody of one analyte with the corresponding antibodies for the other analytes in the multiplexed assay, and do not mutually impair binding to the analyte due to steric hindrance, is often tedious, long-lasting and expensive and sometimes may even fail. Putative (partial) incompatibility of 1st and 2nd antibodies will be recognized in time during the chip development in course of assaying at varying titers in a single multiplexed experiment with minimal sample consumption. Putative cross-reactivity between the antibodies could result in false-positive signals or in reductions of the dynamic range of the array and thereby is critical for the development of the “sandwich” configuration (Figure 3). As a consequence, every “sandwich” assay has to be optimized, e.g. by decreasing the concentration of the detection antibody as much as possible to reduce cross-reactivity. The accompanying loss of signal may be compensated for by elevating the concentration of the capturing antibody. Thus, the proper adjustment of the concentrations of both detection and capturing antibodies is required for optimal performance of microarrays in the “sandwich” configuration. |
| For many secretory proteins, such as cytokines and hormones, appropriate reagents are commercially available, mostly from ELISA kits comprised of pairs of monoclonal or affinity-purified polyclonal antibodies, with demonstrated adequate performance in the microarray format. Those cytokine/hormone-specific antibody pairs developed for ELISA applications during the last two decades can be easily transferred to microarray assays. At variance, very few ELISA antibody pairs have been developed so far for intracellular proteins. The majority of the commercially available antibodies against cellular proteins were introduced for different applications, such as immunoblotting, do not operate as pairs and fail to capture the proteins out of total cell or tissue lysates, often due to the presence of detergent at high concentration. However, it is believed that antibody reagent companies will increase their efforts in the identification of antibody pairs for ELISA and bead assays, which should improve the availability of validated antibody pairs for the use in arrays. Nevertheless, in some cases compensation for partial interference and cross-reactivity by normalization may be feasible and useful for a given antibody pair in case of lack of alternatives ones. Thereby, underestimation and high variance in the determination of the analyte content would be minimized. Faced with the problems of antibody cross-reactivity, insufficient sensitivity and inadequate assay linearity during the development of multiplexed microarray assays, the generation of recombinant antibodies or antibody-like fragments will facilitate the availability of antibody reagents with adjusted affinity and specificity in course of optimization of the epitope-binding domains [69,70]. In addition, alternative capturing reagents, such as DNA or RNA aptamers [71], or alternative protein-binding scaffolds, such as anticalins and lipocalins [72-75], have been used in pilot experimental set-ups. However, their utility for assaying low-abundance proteins in complex biological samples remains to be demonstrated (Figure 3). |
| In summary, protein chips in “sandwich” configuration resemble standard ELISA immunoassays, also with regard to the lower limits of detection and accuracy, but enable measurement at high throughput and parallel scale, which is based on the considerable differences in the signal generation process. In theory, the sensitivity of miniaturized assays should be higher compared to macroscopic ones since the concentration of the signal-generating molecules on the surface of a microspot should result in a higher signal-to-background ratio at the identical analyte concentration [76]. In practice, however, the “sandwich” configuration does not dramatically improve the sensitivity compared to standard ELISA plate measurements, even in case of operation of very sophisticated signal generation technologies. Nevertheless, “sandwich” microarrays have the potential to become a valuable tool for the measurement of clinical and diagnostic markers as well as infectious agents, which are often detected with immunoassays in ELISA-plate format, on the basis of very low costs per data point, requirement of low sample volume (μl-range) and handling of complex samples (body fluids and tissues). Thus, antibody microarrays in “sandwich” configuration have huge advantages for diagnostic purposes, when the amount of sample is limited (e.g. single drop of blood) and multiple analytes (e.g. 50-100 disease markers) have to be evaluated. In addition, the random screening of general patient populations without detailed indication for these biomarkers is facilitated by “sandwich” microarrays, also on the basis of low reagent consumption. |
| At variance to the “sandwich” configuration, the “direct labeling” configuration of antibody microarrays circumvents the need for a 2nd detection antibody (and, in consequence, may be of lower selectivity than the “sandwich” chip). For this the dye, fluorophore or (luminescent) enzyme is coupled directly to the protein analyte through secondary interactions or covalent cross-linking prior to incubation of the samples with the protein chip (Figure 3). |
| A principal problem with antibody-based microarrays of each category, i.e. forward or reverse, and configuration, i.e. “direct labeling” or “sandwich”, represents the possibility of masking of the relevant epitopes that are exposed by the protein analytes and recognized by the capturing and/or detection antibodies by polypeptides also contained in the biological sample. The interaction of the protein analyte with a masking protein via secondary bonds may occur by chance or fulfil a physiological role. One example is represented by the matrix metalloproteinase- 9 (MMP-9), that is involved in the disassembly of the basement membrane and promotion of angiogenesis. MMP-9 levels are elevated in tissues, blood and urine of tumor patients, as revealed by quantitative zymography and immunoassays. This has been associated with the malignancy of various tumor types, e.g. gastric cancer, and with worse survival of the patients [77-79]. Interestingly, extra protease activity bands were detected in the zymograms of urine samples from cancer patients and most often result from complex formation of MMP-9 with lipocalin-2 [80,81]. In vitro and in vivo studies demonstrated a physiological function of lipocalin-2 in the protection of MMP-9 toward autodegradation. Most importantly, the enzymic activity of the MMP-9/lipocalin-2 complex, but not of the levels of single MMP-9 and lipocalin-2, has recently been found to be significantly correlated with the depth of tumor invasion in oesophageal squamous cell carcinomas, the survival of gastric cancer patients and the prognosis for breast cancer patients [82-84]. Together these findings suggest that urinary MMP-9/lipocalin-2 complex together with the separate constituent components may be used as novel biomarkers for various types of cancer and for the non-invasive cancer diagnosis and prognosis. |
| For the rapid and reliable analysis of the free MMP-9/lipocalin-2 constituents and/or complexes thereof in patient samples, it is of crucial importance that (i) the protein chip manages to unequivocally discriminate between the complex and its constituents and (ii) the data obtained are not affected by the presence of free constituents and the formation of the complex, respectively, in the biological samples. For MMP-9 and lipocalin-2 these prerequisites have been fulfiled by two microarrays reported so far for clinical use. A reverse phase protein lysate array for the measurement of sole MMP-9 in patient tumor tissue samples was constructed by robotic printing of the serially diluted protein lysates onto PVDF-coated glass slides, subsequent probing with validated (by immunoblotting) and commercially available anti-MMP-9 primary antibodies, biotinylated secondary antibodies, streptavidin-biotin-peroxidase complexes (for signal amplification) and biotinyl-tyramide/hydrogen peroxide and streptavidin-peroxidase (for signal amplification) and final development using hydrogen peroxide [82]. A microplate-based ELISA has been introduced for the independent determination of sole MMP-9, sole lipocalin-2 and MMP-9/lipocalin-2 complexes in tumor tissues from gastric cancer patients as well as the urine from breast cancer patients by immobilisation of the complexes via anti-MMP-9 antibodies followed by detection using anti-lipocalin-2 antibodies. Importantly, the MMP-9- and lipocalin-2-specific microarrays did not detect the complex and, vice versa, the complex-specific microarray did not recognize MMP-9 and lipocalin-2 in their free forms [84]. As a general conclusion, the possibility of interference of antibody-based microarrays in course of complex formation of the protein analytes, which may lead to negative or false positive results, has to be investigated rigorously prior to their application. |
| GPI-(anchored) protein chip |
| Unfortunately, the fabrication and operation of conventional protein chips of both “forward” and “reverse” configuration as described above may lead to non-quantitative detection and immobilisation, respectively, of the (antibody- or directly labeled) analytes in course of (partial) masking of the epitopes recognized by the immobilising antibody and detection antibody, respectively. The problems may be (partially) overcome by a novel type of protein chip that is based on glycosylphosphatidylinositol- (GPI-) anchored proteins (GPI-proteins). These GPI-protein chips are currently being evaluated and validated for numerous applications in “point-of-care-testing” for individualized diagnosis and health care as well as monitoring of the individual therapeutic outcome. |
| GPI-anchored molecular probes |
| Typical transmembrane proteins span the phospholipid bilayer of the cellular plasma membranes through a single or several stretch(es) of hydrophobic amino acids, the transmembrane domain(s), with large amino-terminal (carbohydrate- and disulfide bridge-harbouring) and carboxy-terminal polypeptide domains facing the cell surface and cytoplasm, respectively (Figure 4). In contrast, GPI-proteins lack (a) transmembrane domain(s) but are embedded exclusively in the outer extracellular leaflet of the phospholipid bilayer by a covalently linked GPI structure [85-87]. This glycolipid anchor consists of phosphatidylinositol and several distinct and specifically linked carbohydrate moieties, the glycan portion, and is coupled via its terminal ethanolamine residue through an amide linkage to the carboxy-terminus of the extracellular protein domain. Thus, none of the amino acids of the GPI-protein moiety is in intimate contact to the plasma membrane. Rather, the GPI-protein is associated with the cell surface solely through its GPI anchor, which can be specifically cleaved by a GPI-specific phospholipase C leading to release and solubilisation of the protein moiety. GPI-proteins are expressed in all eucaryotic cells studied so far, from yeast to man, and fulfil diverse functions as cell surface receptors, enzymes, antigens, transporters and signaling and cell adhesion molecules [86]. |
| For application in the protein chip technology, it is important that in principle each soluble passenger protein, such as a binding protein, receptor, enzyme or antibody, can be expressed ectopically as GPI-protein at the surface of eucaryotic host cells, such as the yeast Saccharomyces cerevisiae, chinese hamster ovary cells or human embryonic kidney cells, using recombinant DNA technology. For this, the relevant host cells have to be transfected with a plasmid harbouring the cDNA derived from the corresponding passenger protein gene and appropriate 5’- and 3’-regulatory elements for its inducible/repressible transcription and translation. In addition, for biogenesis of the passenger protein as GPI-protein two targeting signals, signal sequences I and II have to be placed at the 5’-/amino- and 3’-/carboxy-termini of the corresponding gene/protein constructs [88]. Signal sequence I directs the nascent GPI-protein through the endoplasmic reticulum, Golgi apparatus and secretory vesicles to the plasma membrane along the typical secretory pathway. Simultaneously, signal sequence II drives the covalent coupling of the GPI anchor, pre-fabricated at the endoplasmic reticulum by step-wise glycosylation of phosphatidylinositol, to the GPI-protein precursor in the course of removal of signal sequence II by a transamidase reaction occurring in the endoplasmic reticulum [89]. The molecular machinery including the genes involved as well as the structural features of the signal sequences I and II have been elucidated during the past two decades [90,91]. This knowledge will be helpful for the ectopic expression of GPI-modified/anchored versions of any soluble protein at the surface of host cells by preparing a chimeric cDNA in which its endogenous 5’- and 3’-terminal sequences are substituted for by those encoding signal sequences I and II, e.g. derived from the native GPI-protein. Moreover, the efficacy of the recombinant expression of any given (soluble) protein as GPI-protein variant will increase considerably upon the use of optimally engineered signal sequences I and II instead of its native counterparts derived from the authentic GPI-protein as well as of genetically engineered host yeast or mammalian cells which maximally express the rate-limiting molecular components (e.g. transamidase) of the GPI-protein biosynthetic pathway [91,92]. |
| In the case of GPI modification/anchorage of antibodies as capturing/immobilisation reagents for the construction of microarrays, intact mouse (monoclonal) antibodies suffer from the main disadvantage of their complex multi-chain structure and large size (~ 150 kDa). Therefore it has been tried to bypass these limitations of intact antibodies by the use of genetically engineered small (~ 30 kDa) single-chain variable fragment antibodies (scFv) as demonstrated first for anti-CD20 scFvs as a strategy to target CD20-positive tumor cells [93](Figure 5). They are constructed by coupling of the variable heavy chain (VH) and the variable light chain (Vテ青?) domains of the intact antibody by a short flexible peptidic linker [94]. Importantly, the exquisite specificity for the epitope-binding site and pronounced affinity for the authentic epitope has been demonstrated to be preserved in scFvs [95]. Unfortunately, due to the limited understanding of the impact of amino acid exchanges on protein folding, no a-priori method is currently available for the prediction of the capability of a selected antibody to operate with the desired selectivity and affinity when produced as an scFv. As a consequence, prior to use for immobilisation in protein chips, the scFvs in their GPI-anchored and non-anchored versions (since the former often will react non-specifically with all cellular membranes) have to be analysed and confirmed for the specific recognition of the relevant protein analyte epitope in comparison to the original antibody from the selected hybridoma. In conclusion, on basis of their size and single-chain structure with expression from a single transcript (Figure 5), scFvs are more accessible for the recombinant coupling to GPI anchors than intact immunoglobulin molecules. In fact, the strategy of modification of a scFv with a carboxy-terminal GPI structure and its efficacy were first been demonstrated for the expression of an immune modulatory signal (5H7) at the surface of T- and B-lymphoid cells as GPI-anchored 5H7 variant [96]. |
| In general, for the conversion of an intact antibody molecule selected for capturing/immobilisation of a given protein analyte during a microarray procedure into a GPI-tagged scFv by genetic engineering using standard recombinant technologies, DNA fragments encoding the VH and the Vテ青? domains were amplified from the total cDNA using degenerate primers which had been prepared from the corresponding hybridoma cells secreting the anti-analyte monoclonal antibody (Figure 6). For the design of the primers for the VH and the Vテ青? framework domains conserved between the immunoglobulin subfamily members were selected. The predicted VH and Vテ青? domains cloned from the hybridoma were coupled by a linker sequence resulting in the basic scFv construct (VH-Linker-Vテ青?) that was then modified to facilitate (i) its targeting to the typical secretory pathway (amino-terminal signal sequence I), (ii) its purification by immobilised metal affinity chromatography (carboxy-terminal His6-tag), (iii) its immunological detection (hemagglutinin [HA] 11-amino-acid peptide tag located carboxy-terminal to the His6-tag) and (iv) its anchorage at the cell surface by the post-translationally added GPI structure (signal sequence II derived from the GPI-protein alkaline phosphatase and located carboxy-terminal to the HA-tag) (Figures 5 and 6). |
| The GPI-modified scFv proteins can be expressed in High FiveTM insect cells with high yield and at relatively low costs and then easily affinity-purified in the native active state [93]. In contrast to bacterially expressed scFvs which often require the solubilisation and subsequent refolding from inclusion bodies formed during the fermentation, insect- expressed scFvs do not require renaturation [93,97,98]. In contrast to the expression of GPI-proteins in mammalian cells, insect-expressed scFvs are not inserted into detergent-insoluble lipid rafts of the endoplasmic reticulum, Golgi and plasma membranes along the secretory pathway, but can be solubilised from the membranes by non-ionic detergents at low concentration. |
| Alternatively, baker’s yeast (e.g. Saccharomyces cerevisiae) and cultured cell lines (e.g. HEK-293) represent appropriate host cells for the expression of GPI-modified scFv proteins. After the upstream processing, which includes fermentation of the host cells in appropriate bioreactors and subsequent removal of the culture medium, total GPI-proteins will be extracted from the cell surface, preferably in selective fashion without total solubilisation of the cells. For this, non-ionic detergent, such as Triton X-100 or Triton X-114, is added at low concentration at low temperature. This results in disintegration of the “non-lipid” raft domains of the host cell plasma membranes under concomitant aggregation of the GPI-proteins together with cholesterol and (glyco-)sphingolipids into the lipid rafts [99], which can be collected and enriched by sucrose gradient centrifugation on the basis of their low buoyant density. Finally, the GPI-proteins are solubilised by high concentration of detergent at room temperature and, if required, further purified by (several rounds of) conventional column chromatography. An alternative to this selective detergent extraction has recently been developed, the so-called magnetic extraction. It is based on antibodies which are directed against the glycan portion of the GPI anchor and covalently coupled to metal beads [100]. |
| In addition to the recombinant engineering of GPI-anchored scFv proteins acting as high-affinity capturing probes for protein analytes of any desired type, in principle, naturally occurring GPI-proteins with physiological receptor or binding function may be used for the immobilisation of the corresponding ligands at the protein chip surface. The number of GPI-anchored receptor and binding proteins is steadily increasing. In fact, some of them are possible candidates for the immobilisation at protein chips on the basis of their known ligands, which may serve as biomarkers of diagnostic or therapeutic relevance, such as the glypicans for the heparin-binding growth factors [101], the Ly-6/urokinase-type plasminogen activator receptors for the urokinase-type plasminogen activator [102], the FcγRIIIB low-affinity IgG receptor for the Fc portion of immunoglobulins of the IgG class [103] and glycosylphosphatidylinositol- anchored high density lipoprotein-binding protein-1, GPIHBP1, for lipoprotein lipase [104]. |
| Immobilization of GPI-(anchored) proteins |
| The development of a stable and universal immobilisation method for the capturing probe, e.g. antibody, which does not grossly affect its structure, affinity and selectivity is one of the most critical aspects and challenges of microarray fabrication. So far, a number of different methods have been introduced for the immobilisation of GPI-proteins on solid chip surfaces, such as non-covalent adsorption, covalent binding and affinity capture. With regard to the adsorption technique, the GPI anchor biosynthetically attached to antibodies or scFvs, which have been raised against the desired protein analytes, manages the non-covalent immobilisation of the capturing probes at the chip surface (Figure 7). The underlying principle is based on the structure of biological membranes which represent natural nano-structures separating the intracellular components from the extracellular environment. They consist mainly of phospholipids, spontaneously assembling as a continuous spherical lipid bilayer structure where the hydrophilic polar heads shield the hydrophobic fatty acid tails from the surrounding polar cytosolic and extracellular environments. One of the major phospholipids of biological membranes is phosphatidylcholine (PC), which has a zwitterionic head group. The monomers comprising the PC heads have been synthesized with attention to the chemical structure of the phospholipid molecules, and polymers derived thereof have been applied in the preparation of cell membrane-like structures and biomaterials. A polymer family containing 2-methacryloyloxyethyl PC (MCP polymers) is of particular interest for the modification of both SiO2-based and polymer-based chip surfaces because of its high performance in the suppression of non-specific adsorption of proteins, subcellular structures, such as organelles, and cells, such as platelets. Consequently, MPC polymers have been used to form cell membrane-like interfaces for chip applications via adsorption [105]. |
| The relevant phospholipid monomers can be synthesized with high yield and excellent purity. One of the representative MPC monomers is a methacrylate with a PC unit as a phospholipid polar group. Many phospholipid polymers based on MPC chemistry have been developed and studied for functionalized surface modification. The MPC monomer can copolymerize with various vinyl monomers to form phospholipid polymers having a wide variety of molecular architectures. They can be transformed to cell membrane-like surfaces by coating the polymer, blending with the polymer or grafting to the polymer chains. Thereby they provide biointerfaces capable of suppressing many biological responses, such as non-specific interaction with proteins, organelles and cells [100,105]. In addition, with the incorporation of functional moieties enabling bioconjugation, these MPC polymers also form PC-covered surfaces capable of selectively interacting with specific biomolecules, such as GPI-proteins, among them antibodies and scFvs. |
| For the immobilisation of GPI-antibodies, the microscopic glass slide is coated with a monolayer of phospholipids, which is facilitated by hydrophobic interactions between the saturated long-chain fatty acids and the glass surface and mimics the structure of the extracellular leaflet of the plasma membrane phospholipid bilayer. Upon addition of the anti-analyte GPI-antibodies/scFVs embedded in detergent micelles to the phospholipid- or MPC-coated glass slide, which can meanwhile be managed with sufficient reliability by piezoelectric standard printers commercially available for conventional protein chips, the antibodies/scFvs become spontaneously inserted into the phospholipid or MPC monolayers, solely in response to adequate dilution. This will result in the defined orientation of the GPI-antibodies/scFvs with the epitope-binding domain facing the chip surface at high density, which resembles that of GPI-proteins at the outer face of biological membranes upon their incubation with intact cells, liposomes or reconstituted model membranes [106-108]. In comparison to covalent crosslinking, the GPI anchorage of the anti-analyte antibodies/scFvs to this special type of “reverse” protein chip has the distinct advantage of higher efficacy and selectivity in combination with lower background due to unspecific adsorption of the analyte. This often represents a problem with bifunctional chemical crosslinkers due to their hydrophobic nature and broad specificity. Moreover, the covalent coupling of the GPI anchor to the carboxy-terminus of the capturing probe, such as scFvs, does not interfere with epitope recognition, in general, and by the amino-terminal VH and Vテ青? domains, in particular. This feature will guarantee high efficacy, robustness, reliability and reproducibility of analyte immobilisation with low variance between different measurements using the same chip or different chips, which have been prepared by independent spotting procedures with different batches of capturing probes. |
| With regard to spotting of the GPI-proteins, the predominant procedures evaluated so far are the contact and the non-contact printing. Metal pins with solid or quill tips are applied in contact printers to transfer volumes in the sub-nanoliter range of the GPI-protein samples to the slide surface. Quill pins have a higher sample capacity and enable the printing of hundreds of spots continuously after each sample loading. The printed spots are typically circular with the size depending to a major degree on the dimension of the tip, the material and chemistry of its surface and the buffer used. A considerable advantage of this type of printer is their speed and high-throughput with loading of up to 48 pins and printing of up to 250 slides simultaneously. However, those pins are very fragile and expensive. Furthermore, the tip of the pins may damage the slide surface, in particular, in the case of using complex three-dimensional substrates, such as MPC polymer-coated slides and biointerfaces. In addition, GPI-proteins may expose hydrophobic surface domains, including the GPI anchor, causing unspecific adherence to metal. The typical routine washing steps may not be sufficient to get rid off them completely from the pins. Altogether, these features may result in considerable cross-contamination of GPI-protein samples and in carry-over problems. |
| To overcome these issues, non-contact dispensing techniques have been introduced for printing GPI-protein microarrays, by which a small droplet of GPI-protein sample becomes delivered to the slide surface without touching it. The droplets are generated by conventional ink-jet, piezoelectric pulsing or electrospray deposition [109-111]. Unlike contact printing, the amount of liquid deposited by non-contact printers does not rely on the surface characteristics of the slide and the morphology of the spots on hydrophobic surfaces, such as the phospholipid or MPC monolayers [112]. The major advantage of non-contact printers is their ability to print on artificial membranes, such as phospholipid and MPC monolayers, in addition to standard glass slides. However, those instrumentations typically suffer from longer printing times and from equipment with a lower number of pins, which represent major drawbacks in case of printing a large number of GPI-protein samples. In addition, non-contact printers are sometimes faced with the problem of misplacing the spots and/or the generation of satellite spots, that usually result in increased rate of failure [113]. Furthermore, in comparison to contact printers, their non-contact counterparts typically require larger sample volumes, which is challenging and often leads to considerable expenditure and costs for the high-throughput recombinant production of GPI-proteins. |
| Upon implementation of the required tools and equipment, the expenditure for the generic production of the anti-analyte GPI-antibodies/ scFvs and their immobilisation onto the chip using versatile cassette GPI-protein expression vectors, magnetic extraction and an automated standard printer for embedding into the phospholipid or MPC monolayer (Figure 7) is usually lower compared to covalent cross-linking with regard to both time and costs. Cross-linking requires intensive testing of a multitude of chemicals and reaction conditions and a careful quality control for successful coupling of functional capturing probes to the chip surface. |
| Operation of GPI-(anchored) protein chips |
| Upon addition of the sample to the GPI-protein chip, the immobilised (protein) analyte is routinely detected by binding of an analyte detection probe, e.g. antibody, labeled with a dye, fluorophore or luminescent enzyme, as is the case for conventional reverse protein chips of the “sandwich” configuration. This directly leads to corresponding light, fluorescence or luminescence signals at “positive” spots of the array. To circumvent the need for the anti-analyte detection probe that has to be identified, produced and labeled individually for each analyte and must not interfere with the immobilisation of the analyte by the analyte capturing probe, i.e. GPI-anchored scFvs, the immobilised analyte can be detected by an indirect “competitive” mode (Figure 7) [100,114]. For this, a GPI-modified version of the protein analyte is prepared by cell surface expression in recombinant immobilized yeast or adherent mammalian host cells using one of the versatile cassette GPI-protein expression vector systems. Thereafter, diacylglycerol is removed and a soluble version with phosphoinositolglycan (PIG) structure instead of GPI attached is generated by cleavage of the GPI structure with a GPI-specific phospholipase C (Figure 4). Importantly, the PIG moiety covalently coupled to the carboy-terminus of the protein analyte via a phosphodiester ethanolamine bridge harbours a terminal inositol cyclic phosphate moiety generated during the bacterial phospholipase reaction. The resulting PIG-analyte is recognized by anti-PIG antibodies which specifically react with the inositol cyclic phosphate moiety and are labeled with a dye, fluorophore or luminescent enzyme (Figure 7), similar to the 2nd anti-analyte antibodies used for conventional “reverse” protein chips of the “sandwich” configuration. |
| These labeled anti-PIG antibodies once raised can be generically used for the binding to any recombinant GPI-analyte upon its lipolytic conversion into the corresponding PIG-analyte. The labeled anti-PIG antibodies are incubated in excess with the PIG-analyte. The resulting complexes of anti-PIG antibody and PIG-analyte are then added in slight excess to the GPI-protein chip coated with a phospholipid or MPC monolayer and anti-analyte GPI-antibodies/scFvs. After removal of unbound complexes by washing, the immobilisation sites, i.e. the anti-analyte GPI-antibodies/scFvs, of the chip are all saturated which will result in uniform light, fluorescence or luminescence signals at each spot of the array. Subtle differences in signal strength between individual spots of the same chip or between the overall signals of different chips are caused by variations in spotting, embedding and binding efficacies of the anti-analyte GPI-antibodies/scFvs and will be compensated for by normalisation of the data provided by the laser scanner. Thereafter, the reference or sample probes containing the authentic, i.e. unmodified, protein analyte is added to the chip. This causes the displacement of the labelled anti-PIG antibody-PIG-analyte complexes from binding to the anti-analyte GPI-antibodies/scFvs. Thereby the signal which is emitted by these complexes at the corresponding array spot positions becomes reduced. The competition curve with the signal strength decreasing with increasing concentrations of reference analyte is calibrated similar to typical RIA/ELISA procedures and then used for calculation of the analyte concentration in the sample. |
| Single-parameter GPI-(anchored) protein chip |
| The data available so far for GPI-protein chips up to the microarray format (2-5x104 spots) demonstrate that the “competitive” and “sandwich” modes are comparable with regard to sensitivity and reliability. However, the GPI-protein chip of the “competitive” mode has the huge advantage of operating independent of a 2nd anti-analyte antibody for detection. It becomes substituted for by the labeled anti-PIG antibody-PIG-analyte complexes. The distance between the labeled anti-PIG antibody and the protein moiety of the PIG-analyte within the complex is quite large and certainly exceeds that between the analyte and the detecting anti-analyte antibody in a conventional “reverse” protein chip of the “sandwich” configuration. This topology will guarantee the absence of sterical hindrance between the immobilising anti-analyte GPI-antibody/scFvs and the anti-PIG antibody and, in consequence, ensure the immobilisation of the PIG-analyte and the authentic unmodified analyte with identical efficacy. |
| The defined modification of the protein analyte with the PIG remnant of the GPI anchor usually does not interfere with structure and function of the analyte. The carboxy-terminus is often located at the surface of a polypeptide or, if buried in its interior, the overall protein conformation will not be disturbed grossly by the relatively small and hydrophilic glycan moiety. In consequence, masking or inactivation of epitopes of the protein analyte in the course of the PIG modification with relevance for its subsequent immobilisation by anti-analyte GPI-antibodies/scFvs and detection by anti-PIG antibodies is rather unlikely. Nevertheless, moderate impairment in the immobilisation of the PIG-analytes would be acceptable since it can be determined and compensated for by normalization prior to their use. This advantage relies on the uniform labeling of each analyte molecule at the same site, i.e. at the carboxy-terminus, independent of the batch and production cycle. In contrast, chemical cross-linking as used for the conventional “reverse” protein chips of the “labeling” configuration will always result in heterogeneous modification of the various reactive amino acids and accompanying structural deteriorations, which can barely be predicted. A preliminary comparative evaluation of advantages and disadvantages of GPI-protein chips in the competitive configuration as so far available vs. conventional RIA and ELISA procedures on the basis of the determination of human insulin in a matrix of rat serum demonstrated clear-cut strengths of the GPI-protein chip with regard to selectivity, capacity, throughput and the options for multiplex and online measurements, but also revealed weaknesses in the sensitivity, dynamic range and precision (Figures 3 and 8). These will presumably be solved in near future in course of further development of the current pilot version of the GPI-protein chip fabrication and instrumentation technologies. |
| Multi-parameter GPI-(anchored) protein chip |
| The (GPI-) protein chips presented so far have been designed for the determination of a single parameter of the corresponding sample analyte, i.e. molecular identity and/or amount. This is based on the immobilisation and/or detection of the analyte by binding proteins, such as antibodies, scFvs or anticalins, which specifically recognize one or several epitopes of the analyte. Epitopes are typically constituted by continuous stretches of a few to up to several hundred amino acids. The successful immobilisation/detection of the analyte, i.e. its cross-reactivity with the binding protein with high affinity and selectivity, demonstrates the presence of the epitopes in the analyte polypeptide and, in consequence, strongly argues for its molecular identity with regard to the complete amino acid sequence. However, identity in amino acid sequence as indicated by this type of (GPI-) protein chip does not necessarily demonstrate the structural integrity of the protein analyte. The epitopes recognized by the capturing probes, e.g. GPI-anchored scFvs or other binding proteins, which are routinely used for protein chips,usually have been directed toward synthetic short peptides rather than large polypeptide domains with defined three-dimensional structure and therefore do not cover the complete amino acid sequence. |
| For the parallel or successive (within a short period) analysis of multiple parameters, such as identity, amount, structure, function, immunogenicity and toxicity, using (GPI-) protein chips, the microscopic glass slides have to be replaced for special gold-coated glass prisms that are exposed to light of defined wavelength in a special instrumental set-up, called “BiaCore” (Figure 9). The beam is reflected under a certain angle and with larger wavelength thereby generating a baseline signal which is composed of the reflection angle and the wavelength, the so-called surface plasmon resonance (SPR). SPR is probably the best known method for the characterization the kinetics of protein interactions [115]. SPR requires a gold surface which is derivatized with a protein layer. Binding of macromolecules to the gold surface can be detected as a change in incident angle or wavelength that is dependent on the refractive index of the interface. Mass transfer to the gold surface is detected in real-time, allowing on-rates and off-rates to be measured. SPR has been successfully integrated into microfluidic devices several times [116-120], for instance for the realization of 264 independent and simultaneous SPR measurements [119,120]. Importantly, any change in the mass that becomes intimately associated with the chip gold surface opposite to the prism will result in alterations of this baseline SPR signal. This is already caused in course of coating of the “BiaCore” chip with a phospholipid monolayer or MPC polymeric (or other bioconjugated) phospholipid analogs [121,122] and subsequent insertion of the anti-analyte, such as anti-insulin, GPI-antibodies/scFvs. The addition of the sample containing the protein analyte, such as human insulin, will lead to a further specific increase in mass at the chip gold surface that is reflected in a corresponding increment of the SPR signal. Thus, the immobilisation of human insulin by anti-insulin GPI-antibodies/scFvs and thereby demonstration of its molecular identity is monitored as upregulation of the SPR signal in real-time. Thereafter, a new equilibrium will be achieved for the immobilisation reaction resulting in a higher “baseline” value. This represents the “starting point” for the following specific associations of molecules with, i.e. mass increases at, the “BiaCore” chip gold surface. |
| Next, for analysis of the structure of the analyte, such as human insulin, which has already been immobilised at the “BiaCore”-based GPI-protein chip, one takes advantage of its specific high-affinity binding to a “biosensor”. For insulin the “biosensor” is the insulin receptor, which is a heterotetrameric transmembrane protein complex of high molecular mass [123]. Following recombinant expression, solubilisation, (partial) purification and reconstitution into detergent micelles, the structurally and functionally intact human insulin receptor is incubated in excess with the chip. Only in the case of human insulin exhibiting the authentic three-dimensional conformation, the insulin receptor will bind with high affinity and thereby cause a further increase in the “BiaCore” chip-associated mass. This is accompanied by a pronounced upshift of the SPR signal with time up to achievement of the next “baseline” value in course of equilibrium binding of insulin to its receptor. Finally, the function of the structurally intact protein analyte already immobilised onto the “BiaCore” chip gold surface is assessed. The non-covalent cross-linking of the insulin receptor αβ-subunit halves by insulin is known to stimulate its intrinsic protein kinase activity which ultimately leads to autophosphorylation of the receptor β-subunits at certain tyrosine residues in the “trans” configuration Thus, upon addition of ATP, phosphate residues will be incorporated into the human insulin receptor, but only in the case of complete functionality of the insulin analyte. The resulting increase in mass will be moderate, but yet sufficient to elicit a small but significant upregulation of the SPR signal (Figure 9). |
| Taken together, the sequential and rapid (10 to 15 min for three parameters) multi-parameter analysis of molecular identity (by successful immobilisation), three-dimensional structure (by successful receptor binding) and function (by successful receptor activation) of human insulin, e.g. derived from biotechnological production, becomes feasible with the “BiaCore”-based GPI-protein chip. A number of additional important advantages has meanwhile been recognized during the initial calibration and validation experiments, among them (i) low time requirement, (ii) high sensitivity, (iii) high precision and accuracy, (iv) high sample throughput, (v) extended dynamic range, (vi) low variance and (vii) sufficient robustness toward matrix components. Moreover, in contrast to alternative (GPI-) protein chips, e.g. those based on glass microscopic slides, the “BiaCore”-based (GPI-) protein gold chips do not necessitate any washing step for the removal of unbound ligands, such as the insulin receptor or ATP. Only ligand molecules which tightly interact with the gold surface of the chip rather than those freely floating in the incubation medium even in immediate neighbourhood to the gold surface will succeed in inducing a significant SPR signal. Altogether these features enable the reliable determination of multiple parameters for protein analytes in large number and in short time by “BiaCore”-based (GPI-) protein chips. No doubt, in future this technology will considerably facilitate process analytics for the biotechnological production of protein therapeuticals. |
| However, for multi-parameter analysis the (GPI-) protein chips have to be processed along a defined sequence of different steps. In the case of human insulin it encompasses the addition of insulin receptor and ATP, the change of buffers, several incubations etc., and does not only require the simple exposure of the chip to the sample and shortly thereafter the measurement of the light, fluorescence or luminescence signals (Figure 9). Of course, this multi-step handling, if performed with many “hands”, counteracts the efforts for acceleration, simplification and automation of process analytics, personalized diagnosis, systems biology and therapeutic monitoring. The integration of the “BiaCore”-based GPI-protein chips into a “chip-on-the-chip” represents an example for the possible automation and miniaturization of multi-parameter analysis based on the recently developed “lab-on-the-chip” technology. |
| Automation of GPI-(anchored) protein chips |
| For automated handling of all the processing steps required for multi-parameter analysis, the complete reactor device, encompassing the “BiaCore”-based (GPI-) protein chip, the SPR detector unit and the temperature control unit, have to be installed together with small reservoir chambers, which harbour all the reaction components and ingredients fuelling the reactor, within a compact glass/silicon monolith device in a so-called “(GPI-) protein chip-on-the-chip” [124-126]. In the case of analysis of human insulin for identity, structure and function (Figure 9), these reservoir chambers have to be filled with the recombinant human insulin receptor, ATP and two different buffers supporting the binding and kinase reactions (Figure 10, upper panel). The sample(s) are contained in one to several chambers, the number of which critically depends on the size of the monolith and the complexity of the multi-parameter analysis. Prior to transfer of the reaction components and samples to the reactor device, they are combined in a mixing device installed at the influx channel of the reactor device. |
| The transfer of the various reaction components to the reactor device is mediated by the continuous and constant flux of an inert carrier fluid from a corresponding reservoir chamber via the reactor device to a vacuum device. The latter operates as a combination of a membrane pump and a waste container and is placed at the efflux channel of the reactor device [100,124-128]. The distinct reservoir chambers and devices are interconnected by microfluidic channels forming a circuit network under field-effect flow control [129-131]. The channels are often fabricated in poly(dimethylsiloxane) [132] and coated with phospholipid polymer biointerfaces [133] for minimizing unspecific interactions between the microfluidic device and the bioanalyte. The carrier fluid-driven flux of samples, reaction components and buffers into the reactor device is controlled by electrically driven microvalves. These are installed at the efflux channel of each reservoir chamber and regulated in time-dependent fashion by a software-based process leading unit. For this, the microvalves as well as the devices are connected via electrical circuits to electronic chips integrated into the same monolith. In the (GPI-) protein “chip-on-the-chip” of the first generation, which was based on the pioneering work of the company Agilent Inc., the monolith harboured all reservoir chambers and devices, including the vacuum pump and waste container, and served as a one-way cartridge [134]. Only the data registration and analysis were performed by a separate central read-out unit which becomes connected with the (SPR) detector of the reactor device via an electrical interface between the one-way cartridge and the central unit (Figure 10, lower panel). |
| For reasons of sustainability, considerable efforts have been undertaken during the past decade to shift the interface between the one-way cartridge and the central read-out unit “to the left”, i.e. to leave only the reservoir chambers for the samples and reaction components in the cartridge and to install the reactor, detector, vacuum and waste devices in the central unit. With regard to the cost-intensive “BiaCore-based” GPI-protein chips it was of critical importance to implement them in the central unit for multiple cycles of regeneration and re-use. Regeneration is easily accomplished by removal of the immobilising anti-analyte GPI-antibodies/scFvs (with the analytes and detecting antibodies bound) from the phospholipid- or MPC-coated gold surface of the “BiaCore” chip in course of treatment with low concentrations of non-ionic detergent. After washing with aqueous buffer a fresh phospholipid or MPC monolayer with embedded anti-analyte GPI-antibodies/ scFvs is rapidly formed. Following control of the immobilisation, binding affinity and capacity by measurement of the SPR baseline (Figure 6), the GPI-protein chip is restored for the next round of multi- parameter analysis. Taken together, the configuration of reservoir chambers and devices installed within a (GPI-) protein “chip-on-the-chip” enables automated multi-parameter analysis of protein analytes with the considerable advantages of low time requirement (typically less than 15 min per sample), reduced sample volume, reduced reagent volume and low personal costs. Certainly, for validation (GPI-) protein “chip-on-the-chips” have to meet all the criteria typically requested for conventional bioanalytical methods, such as signal-to-noise ratio, sensitivity, resolution, precision, accuracy, selectivity, dynamic range and variance. This has to be demonstrated for each parameter and analyte, as well as for each batch of the “chip-on-the-chip” produced since the fabrication process may underlie considerable variability [135]. |
| Nanoparticle-based (GPI-anchored) protein chips |
| The chip technologies described so far rely on the immobilisation of the protein analyte at the chip surface as well as on the use of anti-analyte antibodies/scFvs or other specific capturing probes for their immobilisation and/or detection. Therefore, intensive efforts have been made during the past decade to establish alternative protein chips which operate with all components, including the analyte, in solution and do not depend on antibodies or specific capturing probes [136]. Instead, these innovative methods are based on nanoparticles (NPs) and soluble reporter enzymes [137-143]. In 2007, a sensor array consisting of six cationic functionalized gold NPs and an anionic PPE polymer was introduced by the Rotello group [144], which manages to identify seven distinct polypeptides with high sensitivity and reliability. The gold NPs provoke quenching of the polymer fluorescence. This NP-polymer interaction and thus the quenching are abrogated by the presence of the protein analytes which thereby results in unique fluorescence response patterns relying on the differential NP-analyte affinity. The efficacy of these “chemical noses” has been attributed to both the quenching characteristics of the gold NPs as well as the “molecular wire” effect of the PPE polymer [145]. On the basis of the dependence of the analyte-NP interactions on their respective structural properties, such as surface charge, hydrophobicity/hydrophilicity and hydrogen-bonding sites, the differential affinities will result in differential fluorescence response fingerprint patterns for distinguished proteins [146] which are quantitatively evaluated by linear discriminant analysis (LDA) of the raw data obtained [147]. The limit of detection reported for the NP-PPE system, that succeeded in the correct elucidation of 52 out of 55 unknown protein analytes, was 4-215 nM depending on their size [144]. |
| As an alternative to PPE-NPs as scaffolds for the sensor construction, a separate class of polymeric NPs has been introduced with a polymeric micellar nanosystem which manages to respond toward electronic complementarity and thereby guarantees selectivity for metalloproteins [148]. For this eight different fluorescence dyes were non-covalently coupled to the micellar interior of an homopolymer eliciting a specific fluorescence pattern that enables the discrimination of four different metalloproteins with high sensitivity. |
| Another approach is based on a micellar disassembly process for signal transduction [149]. For this five different non-covalently assembled receptors were used for analysis of the disassembly by following the liberation of the encapsulated dye in response to five different non-metalloproteins. The resulting shift in fluorescence due to the disassembly of the receptor-analyte complexes resulted in analyte-specific patterns with a limit of detection of 8μM. In extension of this principle of differential response by a single polymer-surfactant complex, two novel variants relied on the disassembly and analyte-induced release pathways and the photo-induced quenching of charge/energy transfer, i.e. on excited state quenching [150]. By variation of non-metalloprotein or metalloprotein transducers the limits of detection for the non-metalloproteins and metalloproteins were lowered to 8μM and 80nM, respectively. In addition, the application of a fluorescent anthracene-core dendrimer system with carboxylic acid residues on the particle periphery enabled a differential protein pattern in response to differential binding energy transfer steps at analyte concentrations ranging from 1 to 5μM [151]. In course of binding, the energy transfer step induced led to quenching of the fluorescent core. This interchange between quenching and binding triggered differential responses enabling the unequivocal identification of three different metalloproteins. |
| The most recent variation of the “chemical nose” approach takes advantage of the amphiphilic nature of GPI-proteins. For this a reporter enzyme has to be chosen that catalyzes an easily detectable reaction and exhibits a large surface area with exposed polar as well as hydrophobic amino acid side chains and/or post-translational modifications of heterogeneous nature (Figure 11). The bacterial hydrolase, β-galactosidase, converts the dye precursor, 4-methylumbelliferyl-β-D-galactopyranoside, into the dye, methylumbelliferone, which can be followed by simple photometric measurement. It is characterized by a negative net surface charge at physiological pH due to a multitude of negatively charged amino acids protruding into the aqueous environment. To increase the apolar surface area of β-galactosidase without drastically decreasing its (in the wildtype form high) solubility or causing its aggregation, the GPI-anchored β-galactosidase was generated by recombinant expression in mammalian host cells (e.g. HEK-293, CHO) and purification using magnetic extraction (see above). In contrast to the wildtype form, GPI-β-galactosidase displays amphiphilic character (according to its partitioning between a Triton X-114 detergent and an aqueous phase). This is caused by the carboxy-terminal GPI structure harbouring two saturated long-chain fatty acids and the glycan moiety, which protrude from the core of the polypeptide chain onto its surface, in combination with the negatively charged surface-exposed amino acids. Importantly, the GPI modification does not significantly impair the hydrolytic activity of β-galactosidase and is compatible with its solubility in the absence of detergent at the concentrations required for the assay. |
| The NPs which are used for the antibody-free protein chips and in a certain sense replace the antibodies will serve as “chemical noses”. They have to manage the smelling of the surfaces of both the reporter enzyme and the protein analyte in differential fashion [152]. For this, the NPs of 2 nm core size and about 10 nm hydrodynamic diameter consist of a gold core and an outer shell of covalently coupled organic molecules. These shell structures are built by a terminal positively charged ammonium residue with different functional groups of varying hydrophobicity as substituent and bound to the gold core via a constant hydrophobic spacer. In consequence, the NPs bind with high selectivity and avidity, rather than affinity, to protein surfaces through electrostatic and hydrophobic/apolar interactions to the major part and hydrogen and van der Waal bonds to the minor part. In fact, upon incubation those cationic/hydrophobic NPs bind to the surface of the GPI-β-galactosidase through salt bridges between the terminal ammonium groups and the negatively charged amino acids as well as hydrophobic interactions between the spacer/functional groups and the fatty acids of the GPI moiety. These multiple interactions between the NPs and the GPI-β-galactosidase lead to its complete inhibition. Importantly, binding of the NPs to GPI-β-galactosidase does not cause its denaturation and is reversible. Upon release of the NPs, e.g. in course of high dilution of the binding reaction, the hydrolase activity of GPI-β-galactosidase was shown to be completely restored. |
| Protein analytes added to an incubation mixture which consists of the reporter enzyme and the NPs under appropriate binding conditions have two options (Figure 11): (i) They do not interact with the NPs and thus the NPs remain bound to the reporter enzyme and continue in blocking its activity, i.e. the generation of the corresponding light signal. (ii) They interact with the NPs with certain selectivity and avidity and thereby cause displacement of the NPs from the reporter enzyme, thereby leading to its activation, i.e. the generation of the corresponding light signal. The signal critically depends on the relative strength of the interactions of the NPs and the reporter enzyme vs. the protein analyte, which is determined by avidity and selectivity rather than mere affinity. Thus low and high signals, i.e. β-galactosidase activities measured, indicate relatively high and low interaction, respectively, between the NPs and the GPI-β-galactosidase and vice versa low and high interaction, respectively, between the NPs and the protein analyte. In consequence, the signal strength is positively and negatively correlated to the efficacy of the “smelling” of the surface characteristics of the protein analyte and GPI-β-galactosidase, respectively, by the NPs. These “chemical noses”, here constituted by the ammonium and functional groups of the shell structures and by the spacer of the NPs, “smell” the negatively charged amino acids and the GPI moiety of the reporter enzyme, here the GPI-β-galactosidase. |
| It is easily conceivable that a single type of NPs enables only limited differentiation between the different protein analytes contained in a complex biological sample, such as human plasma. This is exemplified by the analysis of human reference insulin vs. insulin variants vs. the serum protein albumin (Figure 11). NPs equipped with the distinct functional group, NP5, allowed the discrimination of human insulin from albumin as well as unfolded and mutant insulin, but not from degraded insulin, with NP4 from degraded, unfolded and mutant insulin, but not from albumin, NP2 from albumin as well as degraded and unfolded insulin, but not from mutant insulin, etc. It was the combination of the six distinct types of NPs equipped with the functional groups, NP1-6, that enabled the unambiguous discrimination of the four distinct insulins and albumin from one another on the basis of the unique signatures provided by NP1-6. |
| The minimal number of different types of NPs required for the unambiguous discrimination of a given, but defined, number of protein analytes very critically depends on the nature of the analytes and the NPs used and can therefore hardly be predicted. Certainly, gross structural differences between the analytes on one side and between the NPs on the other side will facilitate the analysis and limit the expenditure to the use of a rather low number of NPs. In contrast, subtle deviations in the biophysical/biochemical characteristics of the analytes will necessitate the use of a higher number of NPs of more intricate structural variation. The currently available data already indicate that four to eight NPs may enable the differentiation of protein analytes at similar number [152,153]. It remains to be clarified whether the determination of a distinguished analyte out of a complex sample originating form a body fluid, such as plasma, and harbouring several hundred proteins, among them several high-abundant ones, will be feasible at all and, if so, will necessitate high expenditure, i.e. very many NP types for the unequivocal identification of a single low-abundance protein. An interesting possibility to increase the number and types of interaction between the NPs and the reporter enzyme and, in consequence, to support the competition between the protein analytes and the reporter enzyme for binding to the NPs relies on the carboxy-terminal modification of the reporter enzyme with a GPI structure (see below). Its amphiphilic nature determined by the saturated long-chain fatty acids and the core glycan component may facilitate or impair the interaction of the GPI-modified reporter enzyme with the NPs and thereby enhance its accessibility for differential competition by the different protein analytes. |
| Nevertheless, taking into consideration the current experimental experience with non-GPI coated NPs, the convenient, straightforward, rapid and inexpensive determination of protein analytes in complex biological samples on the basis of NP-based protein chips using a limited set of NP types seems to be feasible. It may be driven in the near future by considerable progress in the development of parallel synthesis of NP materials starting from a common core or scaffold structure and varying the functional building groups at the NP surface by chance. Combinatorial chemistry may turn out as efficient tool for the generation of a huge variety of NP types around a common scaffold structure as has already been demonstrated in medicinal chemistry during the last two decades. In addition, it seems feasible to identify NP types for a given protein analyte that manage to discriminate between the analyte in the free and complexed state, e.g. MMP-9 unbound and bound to lipocalin- 2 (see above), as has been reported previously as a potential drawback for typical (non-NP based) protein chips (see above). From an experimental point of view, the hurdles for the development of protein chips with the capability to discriminate between free and complexed protein analytes are presumably significantly lower for NP-based compared to conventional protein chips. |
| Taken together, protein chips based on NPs rather than on antibodies or other capturing/detection probes and operating with soluble rather than immobilised analytes have the potential for sensitive, quantitative, selective and reliable determination of protein analytes from complex sample mixtures. The feasibility, in particular their resolution, critically depends on the availability of appropriate “chemical noses”, i.e. NPs, capable of “smelling” subtle differences at the surface structure of protein analyte(s) vs. the reporter enzyme. The functional groups required can hardly be predicted. In fact, there is no need for elucidation of the specific structural determinants that may be appropriate for highly selective and avid interactions between the analyte and the NPs. In most cases, the set of appropriate NPs will be identified just by trial and error. For this, functional groups are synthesized by high-throughput combinatorial chemistry, subsequently automatically coupled to the gold core and finally assayed for interaction with the reporter enzyme by simply assessing its activity under throughput mode. In any case, the probability for finding appropriate NPs should considerably increase, i.e. the number of NPs to be synthesized and tested decrease, upon the introduction of the GPI moiety at the carboxy- terminus of the reporter enzyme and the consequent increase in its surface hydrophobicity. No doubt, so far NP-based (GPI-) protein chips are less far advanced with regard to the (automated) rapid analysis of many samples under routine conditions, e.g. for the personalized measurement of biomarkers. Certainly, for validation and calibration NP-based (GPI-) protein chips have to fulfil the same requirements as conventional protein chips. |
| In addition to assaying the surface integrity of the protein analyte, which typically reflects the combination of molecular and structural identity, NP-based (GPI-) protein chips are capable of detecting any post-translational modifications with great sensitivity, in particular those which alter the surface hydrophobicity of the protein analyte, such as myristoylation, acylation, prenylation, phosporylation, glycosylation, denaturation and aggregation. |
| Moreover, during the past decade NP-based protein chips have already been successfully used for the “smelling” of the surfaces of various bacterial and mammalian cell types for different purposes [138,153]. For instance, pathogenic bacteria were detected with extremely high sensitivity at 102 particles per ml drinking water. Human tumor and healthy cells were discriminated from one another by NPs on the basis of subtle yet adequate differences in their surface characteristics. These differential surface patterns are most likely caused by the combination of many distinct small structural features rather than the predominance of one or a few striking markers protruding from the bacterial cell wall and plasma membrane, respectively [140]. In fact, in case of tumor cells, the critical surface antigens or markers often remain ill-defined. Yet the tumor cells may be identified without knowledge about primary or secondary consequences for the plasma membrane structure on the basis of NPs which succeed in “smelling” the overall cell surface landscape. Nevertheless, in any case the cell types to be discriminated have to be clearly defined (however often a tremendous problem in tumor biology) prior to efforts for their differential recognition by (a set of) NPs. |
| At variance with the non-NP based chip-on-the-chip described above (Figure 10), the reactor for the NP-based (GPI-reporter) protein chip is just a temperature-controlled incubation chamber that becomes filled with binding buffer, reporter enzyme, NPs, sample and dye in a defined sequence. The chamber walls have to be coated with materials compatible with the sample analyte and the matrix [154,155]. The dye formed during the incubation is then measured in a distinct detection unit equipped with a photometer and installed at the efflux channel. This configuration enables the sequential filling and emptying of the reactor with the various types of NPs for each cycle in rapid (~ 2 min per cycle) and precisely controlled fashion (Figure 11). Eventually, the reactor may be washed between two successive cycles with carrier fluid to get rid of the GPI-reporter protein or the NPs adhering to the chamber walls. Again, for maximal sustainability only the reservoir chambers for the samples, NPs and consumables are installed together into the one-way cartridge, whereas the instrumentation encompassing the mixer, reactor, detector, vacuum pump and electronic chips are combined in the central unit. Again, one-way cartridge and central unit are interconnected via fluidic and electrical interfaces [156]. |
| Typically, the determination of the unambiguous signature for a protein analyte will be completed within 15 to 45 min, corresponding to 5 to 15 cycles with distinct types of NPs each. The time requirement depends on the complexity of the sample mixture and the number and quality, i.e. selectivity and avidity, of the NPs used. The full automation, including the filling of the samples into the one-way cartridge, its connection with the central unit and the reading-out of the central unit, enables (i) the parallel measurements of the same sample with distinct types of NPs or of distinct samples with the same type of NPs and then (ii) the successive measurements of the next samples or with the next types of NPs. Central units equipped with up to 96 reading channels are currently being under validation for pilot process analytics of biotechnological production and have already demonstrated the reliable and sensitive determination of human insulin in plasma with low time requirements (~200 samples per hour) and acceptable running costs. |
| Conclusions and Perspectives |
| The standard approach in biology has mainly been described as reductionist procedure, where a biological system is broken down into the components and these components in turn are analysed separately to interpret their effect on the system’s behaviour [157]. Various genetic and pharmacological tests are then carried out to confirm that this analysis is correct. While this reductionism has turned out to be powerful, it is rarely quantitative and the explanations are only descriptive or qualitative. Recent advances in network analysis enabled the investigation of biological systems from a new perspective, relying on the integration of multiple parameters or factors into larger scale models. These become very powerful when they are combined with standard mechanistic studies. While systems approaches per se are limited by measurement and computational tools, they have the potential to generate a physiological dimension that with mechanistic analyses per se can hardly be obtained. |
| The next generation of drugs for common multifactorial diseases, such as cancer, type II diabetes, obesity and neurodegenerative diseases, will approach a novel level of efficacy if the therapy is tailored to the molecular characteristics and composition of each patient’s aberrant cells and affected tissues. Over the past six years significant progress has been achieved in the application of genetic and genomic profiling to individualize and stratify drug administration, such as for chemotherapy during the treatment of cancer based on kinase inhibitors, that target multiple signaling pathways for the regulation of growth and cell division [158-160]. This should lead to the incorporation of protein microarray procedures into future clinical trials (Figure 12). Importantly, molecular therapy will require a novel class of technologies for proteomic profiling since genomics typically fails to decipher the activation state of specific signaling pathways that harbour the relevant target of the newly identified inhibitor or activator. Protein chips and, in particular, those relying on GPI-proteins represent a broadly adopted technology which meets the requirement for profiling of the functional state of cellular signaling pathways and networks. The (GPI-) protein chip has graduated from the realm of basic science to the level of clinical trial design and application. |
| The systems approaches critically depend on proteomic analysis at large scale and multi-parameter level [161,162]. Whereas the chemical tools for this analysis are still primitive, different combinations of (GPI-) protein chips and printed proteins enable the assessment of binding interactions, such as antibody-epitope, receptor-ligand and enzyme-substrate, on a genome-wide scale [163]. While major progress in the area of conventional protein chips has been achieved during the past decade, the future development of microarrays based on GPI-proteins and NPs offer hope for better quantitative and multi-parameter analyses at acceptable expenditure in time, costs, skills and equipment. In a broader sense, (GPI-) protein chip technology may fill a critical missing component of molecular profiling, i.e. the quantitative evaluation of the function of signaling proteins and their post-translationally modified versions. Measurement of this class of protein analytes, which includes important drug targets, provides information about the disease state of cells that can not be obtained by genetics or genomics. The need to measure those analytes from small biopsy samples will continue and expand in the future. New signaling molecules and drug targets are continuously identified and become validated. Consequently, the complexity of the analyte repertoire will increase as is true for the demands on sensitivity, precision, reproducibility and versatility of (GPI-) protein microarrays. They will utilize nanotechnology to an increasing degree, as already indicated here for the NP-based GPI-protein chip, the 3rd-generation amplification technologies, and the novel detection procedures. Ideally the ultimate clinical embodiment will be a one-step technology from the user’s perspective, with completely electronic readout. It will not rely on an array imaging step if the construction of a multi-parameter protein chip with high sensitivity and precision in a homogenous (solution phase) NP-and/or GPI-based format were successful. |
| Thus the utility of (GPI-) protein chip technology resides in signaling pathway analysis and profiling. The outcome of this knowledge will be a new form of diagnostic report for individualized therapy and its monitoring. So far, the signaling network map of a given cell could not be delineated to a fixed circuit board which unequivocally hints to abnormally abundant or functional network proteins like a marked route on a street map. Instead, signaling network proteins coalesce into pathways following a stimulus. Once the stimulus is removed, the interconnected proteins disperse and the network organization dissolves. As more will become known about how signaling networks are continuously remodeled under the influence of the tissue microenvironment, it will turn out that the diagnostic report of the future will be an individualized network map or the equivalent of a distinct street map for each patient. The most meaningful measurements may rely on the strength of connections and functional interactions between nodes in the network, not just the amount of each node or protein in the network. In course of the integration of genomics, transcriptomics, proteomics, metabolomics and fluxomics into a systems biology approach, dynamic individualized pathway maps will provide improved visualization of the cell signaling networks for designing a rationale personalized therapy. |
| The ambitious goal of in vivo multi-parameter analysis should enable or at least facilitate the reliable tracking of spatial and temporal determinants of cell function encompassing protein expression, structure, activity and interactions at the level of tissues, organs and whole organisms [164]. Ultimately, the medical application of protein chips may not be limited to in vitro routine diagnosis and to pharmacological and preclinical research but rather be extended to in vivo diagnosis on the basis of implantation of the microarrays into patients as physiological probes [165,166]. The data about the patients’ physiology measured continuously and collected in real-time by the implanted protein chip will considerably support the diagnosis with improved reliability and compliance compared to routine hospital visits and accompanying time-consuming invasive tests. |
| Acknowledgements |
| The author would like to thank the reviewers for very valuable comments and suggestions which were all considered for the final version of the text with considerable gain in information and quality. |
| References |
|
Figures at a glance
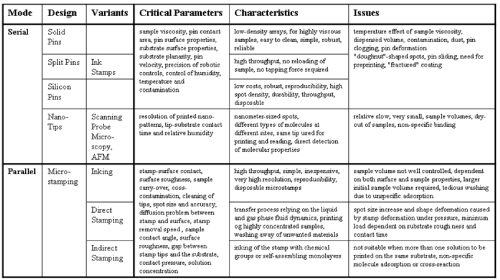 |
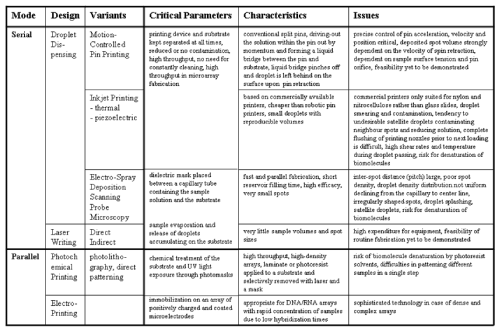 |
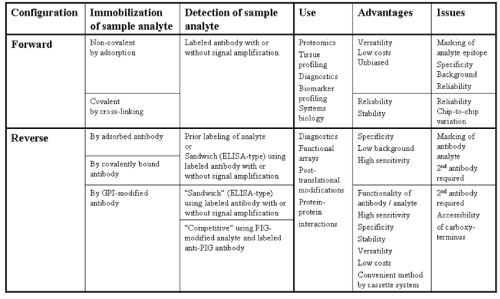 |
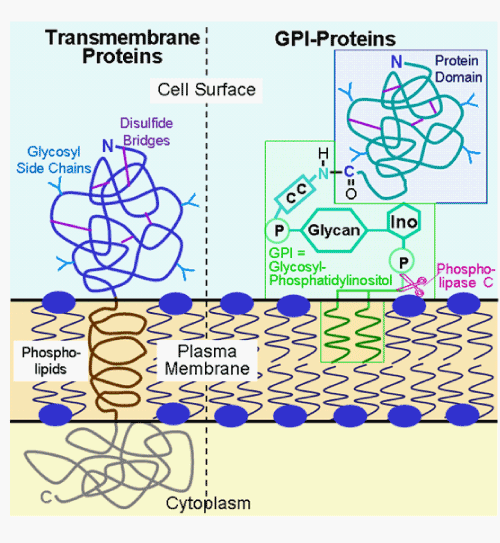 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
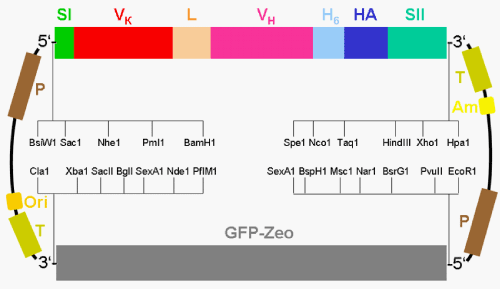 |
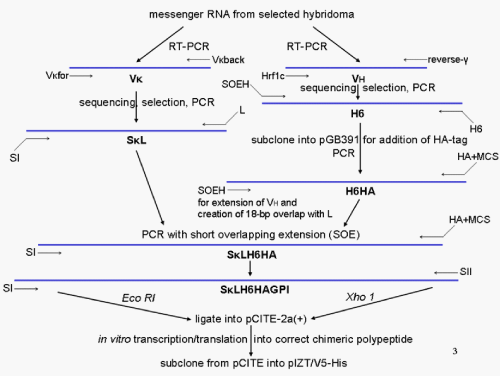 |
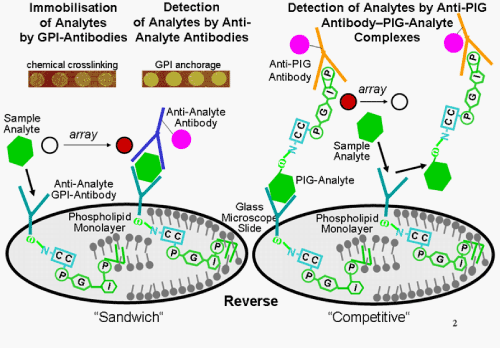 |
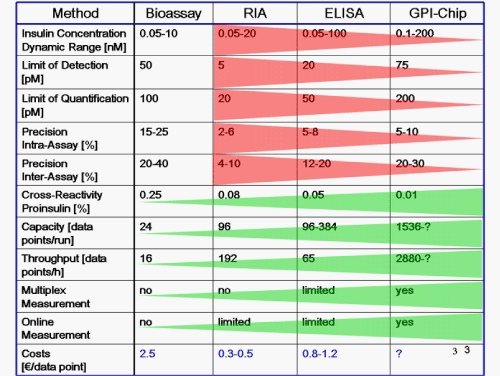 |
| Figure 5 | Figure 6 | Figure 7 | Figure 8 |
`
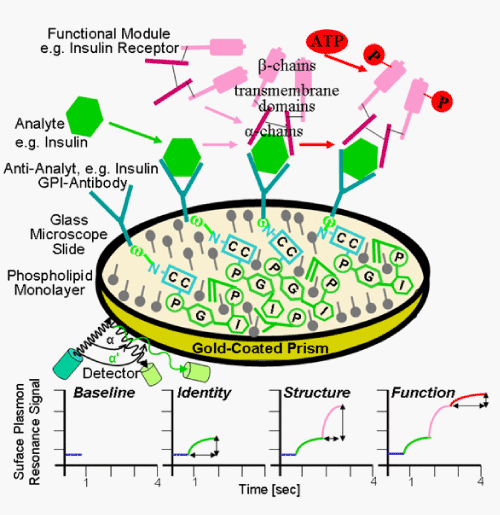 |
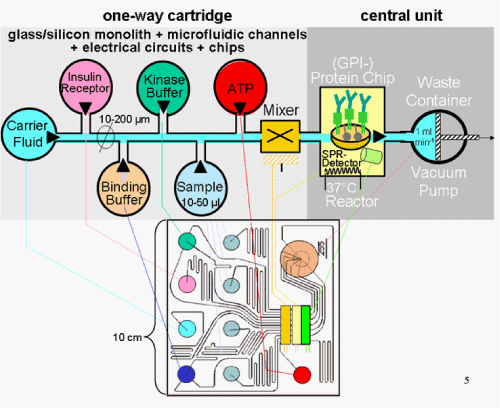 |
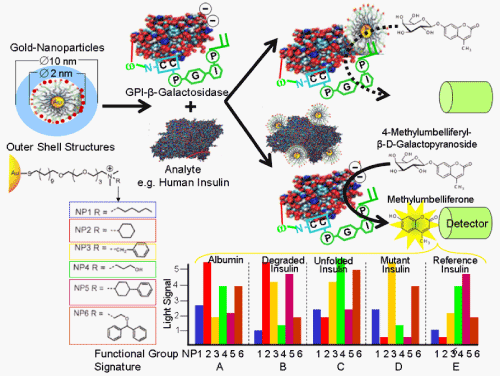 |
 |
| Figure 9 | Figure 10 | Figure 11 | Figure 12 |
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 15160
- [From(publication date):
specialissue-2011 - Aug 29, 2025] - Breakdown by view type
- HTML page views : 10603
- PDF downloads : 4557
