Research Article Open Access
Mass Spectrometry in Clinical Diagnosis: A Preliminary Application in Tumor Cellular Proteomics for Biomarker Discovery
Ming-Hui Yang1,2,3†, Shyng-Shiou Yuan2,3,4,5, Yi-Ling Chen6†, Pei-Yu Chu7, Shiang-Bin Jong6,8, Ying-Fong Huang6,8, Chi-Yu Lu9,10, Yi-Shan Lu8, Po-Chiao Lin10,11 and Yu-Chang Tyan3,8,10,12*1Instrument Technology Research Center, National Applied Research Laboratories, Hsinchu, Taiwan
2Department of Medical Research, Kaohsiung Medical University Chung-Ho Memorial Hospital, Kaohsiung, Taiwan
3Translational Research Center, Kaohsiung Medical University Chung-Ho Memorial Hospital, Kaohsiung, Taiwan
4Department of Obstetrics and Gynecology, Kaohsiung Medical University Chung-Ho Memorial Hospital, Kaohsiung, Taiwan
5School of Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan
6Department of Nuclear Medicine, Kaohsiung Medical University Chung-Ho Memorial Hospital, Kaohsiung, Taiwan
7Department of Medical Laboratory Science and Biotechnology, Kaohsiung Medical University, Kaohsiung, Taiwan
8Department of Medical Imaging and Radiological Sciences, Kaohsiung Medical University, Kaohsiung, Taiwan
9Department of Biochemistry, College of Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan
10National Sun Yat-sen University-Kaohsiung Medical University Joint Research Center, Kaohsiung, Taiwan
11Department of Chemistry, National Sun Yat-sen University, Kaohsiung, Taiwan
12Center of Biomedical Engineering and System Biology, Kaohsiung Medical University, Kaohsiung, Taiwan
†These authors contributed equally to this work as first authors
- *Corresponding Author:
- Prof. Yu-Chang Tyan
Department of Medical Imaging and Radiological Sciences
Kaohsiung Medical University, 100
Shi-Chuan 1st Road, Kaohsiung 807, Taiwan
Tel: 886-7-3121101 ext 2357
E-mail: yctyan@kmu.edu.tw
Received date: January 27, 2014; Accepted date: February 19, 2014; Published date: February 21, 2014
Citation: Yang MH, Yuan SS, Chen YL, Chu PY, Jong SB, et al. (2014) Mass Spectrometry in Clinical Diagnosis: A Preliminary Application in Tumor Cellular Proteomics for Biomarker Discovery. J Anal Bioanal Tech S2:009. doi: 10.4172/2155-9872.S2-009
Copyright: © 2014 Yang MH, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Cancer is a group of diseases characterized by uncontrolled growth and spread of abnormal cells, which cause high mortality rates. The purpose of this study is to characterize the proteins secreted from the HepG2, MCF-7 and HT-29 cells, which may relate to cell differentiation and tumor metastasis. In the proteomic analysis, the secretome proteins were identified by reverse phase nano-high performance liquid chromatography/electrospray ionization tandem mass spectrometry (RP-nano-UPLC-ESI-MS/MS) followed by peptide fragmentation pattern analysis. Moreover, identifications of tumor cell protein expressions by proteomic approaches were indicated that several proteins may activate or enhance the PI3K/AKT/mTOR pathway. The PI3K/AKT/mTOR pathway has been shown to be related to the cancer cell metastasis and proliferation, and the ubiquitin C (UBC) may serve as potential protein diagnostic biomarker to be examined in further investigations.
Keywords
Proteomic; Biomarker; PI3K/AKT/mTOR pathway; Ubiquitin C; Mass spectrometry
Introduction
Cancer is a class of diseases characterized by abnormal cells with uncontrolled growth, which are able to invade other cells. The main categories of cancer include: carcinoma, sarcoma, leukemia, lymphoma, myeloma, and central nervous system cancers. Untreated cancer can cause serious illness and death [1,2].
Early detection of cancer greatly improves the chances for effective treatment and the survival rate. There are two major components of early detection of cancer: education to promote early diagnosis and cancer screening. More recently, screening for cancer with radiological imaging techniques such as X-rays, Computed Tomography (CT), Magnetic Resonance Imaging (MRI), Positron Emission Tomography (PET), and ultrasound, have been suggested and used regularly in order to detect where a tumor is located and what organs may be affected. However, in some studies over 50% of trial participants had an indeterminate location [3,4]. An endoscopy procedure may also be employed to check for abnormalities inside the body.
Unfortunately, early detection strategies thus far have not shown a significant reduction in cancer specific mortality, and the overall fiveyear average survival remains less than 15%. Although many insights into the molecular pathology of malignant tumors have been achieved, additional information is critical to both our understanding of the development, and progression of these malignant tumors as well as to aid in early diagnosis [5].
“Proteome” and “proteomics” are relatively new words, coined by Wilkins et al. [6]. The proteome is the entire set of proteins expressed by the genome. Proteomic analysis means a comprehensive analysis of proteins, and proteomics is the science by which proteins are comprehensively investigated with regard to their roles as functional elements. Recently, characterization of these cellular proteins by proteomic approaches has revealed a definition of protein reactivity and the protein-protein interaction for malignant tumor related proteins.
As an alternative strategy, many investigations have focused on the development of biomarkers as a noninvasive diagnostic tool. In clinical and diagnostic proteomics, it is essential to develop a comprehensive and robust system for proteome analysis [7]. Recent advances in liquid chromatography, mass spectrometry, and data analysis software enable the direct analysis of extremely complex peptide mixtures, often referred to as shotgun proteomics or multidimensional protein identification technology (MudPIT) [8]. Although multidimensional liquid chromatography/tandem mass spectrometry (MDLC-MS/MS) systems have been recently developed as powerful tools, especially for identification of protein complexes, these systems still have some drawbacks in their application to clinical research that requires an analysis of a large number of clinical samples [9].
A driving force in proteomics is the discovery of biomarkers, proteins that change in concentration or state in association with a specific biological process or disease. Determination of concentration changes, relative or absolute, is fundamental to the discovery of valid biomarkers. The objective of this study is to present proteomics profiling data of the cell secretome obtained by RP-nano-UPLCESI- MS/MS technology. A database is formed for the diversity and relative abundance of various proteins found in the cell secretome. The database provides not only information on the nature of protein present in the cell secretome but also potential protein diagnostic markers to be examined in further investigations.
Materials and Methods
Cell culture
HepG2 (liver tumor cell), MCF-7 (breast tumor cell) and HT-29 (colorectal tumor cell) cells were maintained at 37°C and 5% CO2 in RPMI 1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Hyclone Laboratories, Logan, UT), 1% penicillin/ streptomycin (Gibco, Grand Island, NY, USA) and 44 mM NaHCO3. After three days, the cells were washed with phosphate buffered saline (PBS) and the medium was replaced by serum-free RPMI 1640 medium for 12 h.
Sample preparation
After incubation with serum-free RPMI 1640 medium, the secreted proteins in the medium were centrifuged at 1500 g for 10 min at 4°C. The supernatants were filtered through 0.8 μm filters and the protein concentrations were adjusted to 1 mg/mL with 25 mM ammonium bicarbonate.
Cell secretome samples (100 μL) were transferred into 1.5 mL Eppendorf tubes and incubated at 37°C for 3 h after mixing with 25 μL of 1M dithiothreitol (DTT, USB Corporation, 15397). Then the cell secretome samples were reduced and alkylated in the dark, at room temperature, for 30 min after the addition of 25 μL of 1 M iodoacetamide (IAA, Amersham Biosciences, RPN6302V) in 25 mM ammonium bicarbonate. Approximately 10 μL of 0.1 μg/μL modified trypsin digestion buffer (Trypsin Gold, Mass Spectrometry Grade, V5280, Promega, WI, USA) in 25 mM ammonium bicarbonate were added to the cell secretome samples, which were then incubated at 37°C for at least 12 h in a water bath. Two micro liters of formic acid were added to each sample before mass spectrometric analysis for protein identification.
Proteomic analysis
The complex peptide mixtures were separated by RP-nano-UPLCESI- MS/MS. The protein tryptic digests were fractionated using a flow rate of 400 nL/min with a nano-UPLC system (nanoACQUITY UPLC, Waters, Milford, MA) coupled to an ion trap mass spectrometer (LTQ Orbitrap Discovery Hybrid FTMS, Thermo, San Jose, CA) equipped with an electrospray ionization source. For RP-nano-UPLC-ESI-MS/ MS analyses, a sample (2 μL) of the desired peptide digest was loaded into the trapping column (Symmetry C18, 5 μm, 180 μm×20 mm) by an autosampler. The RP separation was performed using a linear acetonitrile gradient from 99% buffer A (100% D.I. water/0.1% formic acid) to 85% buffer B (100% acetonitrile/0.1% formic acid) in 100 min using the micropump at a flow rate of approximately 400 nL/min. The separation was performed on a C18 microcapillary column (BEH C18, 1.7 μm, 75 μm×100 mm) using the nano separation system. As peptides were eluted from the micro-capillary column, they were electrosprayed into the ESI-MS/MS with the application of a distal 2.1 kV spraying voltage with heated capillary temperature of 200°C. Each scan cycle was contained one full-scan mass spectrum (m/z range:400-2000) and followed by three data dependent tandem mass spectra. The collision energy of MS/MS analysis was set at 35%.
Database search
For protein identification, Mascot software (Version 2.2.1, Matrix Science, London, UK) was used to search the Swiss-Prot human protein sequence database. For proteolytic cleavages, only tryptic cleavage was allowed, and the number of maximal internal (missed) cleavage sites was set to 2. Variable modifications of cysteine with carboxyamidomethylation, methionine with oxidation, and asparagine/ glutamine with deamidation were allowed. Mass tolerances of the precursor peptide ion and fragment ion were set to 10 ppm and 0.5 Da, respectively. When the MOWSE score was greater than 50, the protein identification was defined as positive and considered significant (p<0.05). All Mascot results were visually confirmed. In addition, the criterion requires a readily observable series of at least four y-ions for an identified peptide [10]. When a protein was identified by three or more unique peptides, no visual assessment of spectra was conducted and the protein was considered to be present in the sample. Proteins were initially annotated by similar search conditions using UniProtKB/ Swiss-Prot databases. The analyses of protein-protein interaction pathways were performed by String 9.1 Web software.
Results and Discussions
In decades, the major leading causes of death were malignant tumors, and the three most popular cancers were liver cancers, colorectal cancers, and breast cancers. There are many kinds of cancer types, and the severity of the cancer depends on the location, the extent of cancer cell malignant growth, and whether the occurrence of metastases. The reason for out-of-control tumor cell growth may be the proto-oncogenes which activate the cells into cancerous state. It may be mainly due to the expression of abnormal cell division proteins, such as tumor suppressor gene dysfunction. In this situation, the DNA codes of the proteins were damaged, and the proteins were therefore contained the translation errors. For normal cells to translate into cancer cells, the mutations typically are required for multiple times, or translated to process protein and gene disruption.
Hepatocellular carcinoma (HCC) is the most common malignant liver tumor. It is the most or second most common cause of cancerrelated mortality, especially in Asia and Africa, with a five-year survival rate of less than 5% without treatment [1]. HCC targets hepatocytes, the major cell type of liver, leading to the deregulation of homeostasis [11,12].
Colorectal cancer, also known as colon cancer, rectal cancer, or bowel cancer, is a cancer in the colon, rectum, or appendix. It is the second leading cancer killer in the United States. Colorectal cancer symptoms include a change in bowel habits or bleeding, but often there are no symptoms. Most colorectal cancers originate from small, noncancerous (benign) tumors called adenomatous polyps that form on the inner walls of the large intestine [13-16].
The breast cancer is the most common cancer diagnosed in women. The most common types of breast cancers are lobular carcinoma and ductal carcinoma, which begin in the lobules and the lining of the milk ducts of the breast. Breast cancer can occur in both men and women, but it’s far more common in women [17-21].
In this study, we selected the three most common kinds of tumor cells as a cancer model and used an extremely sensitive technique for identification of proteins, namely, RP-nano-UPLC-ESI-MS/MS, to characterize the secretome of tumor cells. Each RP-nano-UPLC-ESIMS/ MS analysis was typically generated about 4,000 MS/MS spectra. A total around 36,000 (4,000 scans×3 repeats×3 samples) MS/MS spectra were analyzed by the database search software SEQUEST. Only a small fraction (9.65%, 3,474) of searches produced significant matches according to the inclusion criteria that we used for this study.
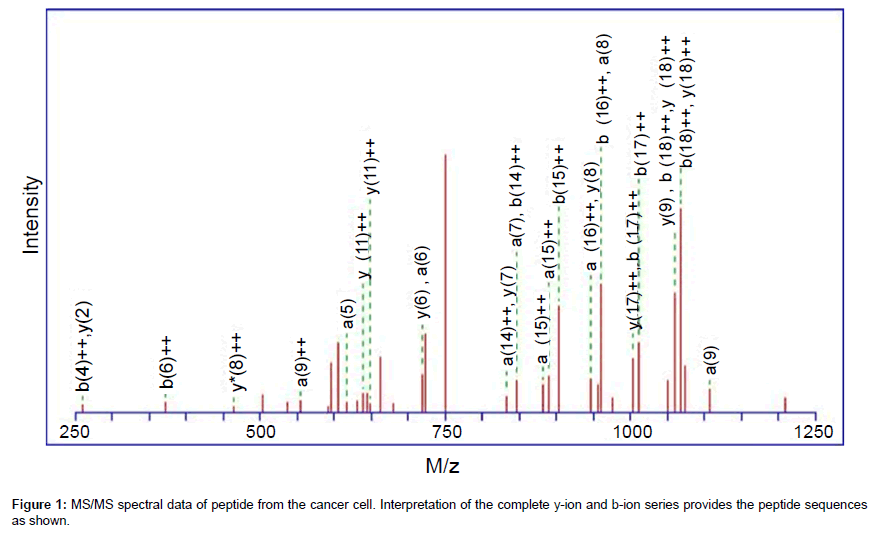
These 3,474 significant matches were assigned to 2,931 peptide fragments, which gave about 81.9% duplicate in the three replicate runs. The total number of unique peptides identified in the cell secretome samples was 619 by Mascot search software. From these short sequences, the protein composition of the composite cell secretome samples can be incurred. The tryptic peptides produced were often mapped to protein sequence entries in the UniProtKB/Swiss-Prot database. The 619 unique matched peptide sequences belonged to 415 protein sequence entries. Most of them were identified at a minimal confidence level in which only one unique peptide sequence matched, whereas 52 protein identifications showed higher confidence levels with at least three unique peptide sequences matched. Figure 1 shows the typical MS/MS spectrum of the identified peptide. The MS/MS spectrum represents the amino acid sequence of tryptic peptide, which is triply charged peptide with m/z of 761.73. The amino acid sequence of the tryptic peptide is FKEIQTQNFSLINENQSLK. The peptide originated from CASP8-associated protein 2, and the interpretation of the complete y-ion and b-ion series provides the peptide sequence as shown. In the 52 identified proteins, there were 16 proteins of special interest since they may play roles in the regulation of cell proliferation and differentiation. The information of 16 protein identifications was listed in the Table 1.
| Protein no. * | Swiss-Prot/ TrEMBL Accession no. |
Protein name | MW (Da) |
Mascot score | Match queries | pI | Sequence coverage (%) | Match peptide |
| HepG2 | ||||||||
| LC01* | Q13148 | TAR DNA-binding protein 43 | 44711 | 91 | 10% | 5.85 | 4 | K.TSDLIVLGLPWK.T R.FGGNPGGFGNQGGFGNSR.G R.FTEYETQVK.V R.FTEYETQVK.V + Deamidated (NQ) |
| LC02* | P13693 | Translationally-controlled tumor protein | 19583 | 163 | 23% | 4.84 | 5 | R.VKPFMTGAAEQIK.H R.EIADGLCLEVEGK.M + Carbamidomethyl (C) R.VKPFMTGAAEQIK.H + Oxidation (M) R.EDGVTPYMIFFK.D K.IREIADGLCLEVEGK.M + Carbamidomethyl (C) |
| LC03* | P81605 | Dermcidin | 11277 | 139 | 16% | 6.08 | 3 | K.DAVEDLESVGK.G K.LGKDAVEDLESVGK.G R.SSLLEK.G |
| LC04 | Q9C0H9 | p130Cas-associated protein | 112397 | 63 | 2% | 9.32 | 3 | R.EMVYASRESSPTRR.L + Glu->pyro-Glu (N-term E); Oxidation (M) R.KLQLQNQESVRALLK.R + 2 Deamidated (NQ) R.KLQLQNQESVRALLK.R + Deamidated (NQ) |
| LC05 | P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein | 468501 | 598 | 3% | 6.06 | 12 | R.GSIQVDGEELVSGR.S R.GSIQVDGEELVSGR.S + Deamidated (NQ) R.RGSIQVDGEELVSGR.S R.RGSIQVDGEELVSGR.S + Deamidated (NQ) R.SLPEVPETIELEVR.T R.LVSEDPINDGEWHR.V R.LVSEDPINDGEWHR.V + Deamidated (NQ) K.GSVYIGGAPDVATLTGGR.F K.DFISLGLQDGHLVFR.Y R.TSTASGLLLWQGVEVGEAGQGK.D K.NLVLHSARPGAPPPQPLDLQHR.A + Deamidated (NQ) R.QPCQHGATCMPAGEYEFQCLCR.D + 2 Carbamidomethyl (C); Gln->pyro-Glu (N-term Q); Oxidation (M) |
| MCF-7 | ||||||||
| BC01* | P25054 | Adenomatous polyposis coli protein | 310342 | 136 | 14% | 7.05 | 27 | R.SGECSPVPMGSFPRRGFVNGSR.E + Carboxymethyl (C); Oxidation (M); 2 Phospho (ST) K.DDMSRTLLAMSSSQDSCISMR.Q + Oxidation (M); 3 Phospho (ST) R.SQTNTLAIIESGGGILRNVSSLIATNEDHR.Q + 3 Deamidated (NQ); 6 Phospho (ST) K.HKMIAMGSAAALRNLMANRPAK.Y + Phospho (ST) K.MIAMGSAAALRNLMANRPAK.Y + Deamidated (NQ); 2 Oxidation (M) K.RSSNDSLNSVSSSDGYGK.R + Phospho (ST) K.TEHNSPSSEAASAPSSNAK.R + Deamidated (NQ); 5 Phospho (ST) K.TEHMSSSSENTSTPSSNAKRQNQLHPSSAQS.N+ Deamidated (NQ); 4 Phospho (ST) R.QNQLHPSSAQS.R + 3 Deamidated (NQ); 2 Phospho (ST) R.SGQPQKAATCK.V + Deamidated (NQ) K.VSSINQETIQTYCVEDTPICFSR.C + Carboxymethyl (C); 2 Deamidated (NQ); 2 Phospho (ST) R.HKAVEFSSGAKSPSK.S + Phospho (ST) K.KPTSPVKPIPQNTEYRTR.V + Deamidated (NQ); Phospho (ST) K.ENKESEAKVTSHTELTSNQQSAN.K + 3 Phospho (ST) K.VTCHTEPSSSQQSAR.K + Carboxymethyl (C); 2 Deamidated (NQ); Phospho (ST) K.SELSPVARQTSQIGGSSKAPSR.S + Deamidated (NQ) R.NGISPPNKLSQLPRTSSPSTASTK.S + Deamidated (NQ) R.TSSPSTASTKSSGSGK.M + 4 Phospho (ST) K.SSGSGKMSYTSPGRQMSQQNLTK.Q + 4 Deamidated (NQ); Oxidation (M); 5 Phospho (ST) K.MSYTSPGRQLSQQNMSKQTGLSK.N + 3 Deamidated (NQ); Oxidation (M); 4 Phospho (ST) R.QMSQQNLTK.Q + 3 Deamidated (NQ); Phospho (ST) K.QTGLSKNASSIPRSESASK.G + 2 Phospho(ST) R.SESASKGLNQMNNGNGANKK.V+ Deamidated (NQ); Oxidation (M); 3 Phospho (ST) K.ENEFSPTNSTSQTVSSGATNGAESK.T+ 4 Phospho (ST) R.SGRSPTGNTPPVIDSVSEK.A + Deamidated (NQ); 4 Phospho (ST) R.LNSFIQVDAPDQK.G + Deamidated (NQ); Phospho (ST) R.LNSFIQVDAPDQKGTEIK.P + Deamidated (NQ); Phospho (ST) |
| BC02* | Q16644 | MAP kinase-activated protein kinase 3 | 43195 | 92 | 12% | 7.60 | 6 | M.DGETAEEQGGPVPPPVAPGGPGLGGAPGGRREP.K+ Phospho (ST) K.KQAGSSSASQGCNNQ.- + Carboxymethyl (C); 2 Phospho (ST) K.KQAGSSSASQGCNNQ.- 2 Deamidated (NQ); 3 Phospho (ST) C. I LDVYENMHHGK.R + Deamidated (NQ); Oxidation (M) R.KKQAGSSSASQGCNNQ.- + Carboxymethyl (C); 3 Deamidated (NQ); 2 Phospho (ST) R.KKQAGSSSASQGCNNQ.- + Carboxymethyl (C); Deamidated (NQ); 4 Phospho (ST) |
| BC03* | Q04721 | Neurogenic locus notch homolog protein 2 | 265191 | 127 | 9% | 5.02 | 21 | R.DGYEPCVNEGMCVTYHNGTGYCK.C + 2 Deamidated (NQ); 2 Phospho (ST); Phospho (Y) K.ECQWTDACLSHPCANGSTCTTVANQFSCK.C + Carboxymethyl (C); 2 Deamidated (NQ); 3 Phospho (ST) K.ECQWTDACLSHPCANGSTCTTVANQFSCK.C + Carboxymethyl (C); 2 Deamidated (NQ); 3 Phospho (ST) H.CQHGGTCLNLPGSYQ.C + Deamidated (NQ); Phospho (ST); Phospho (Y) G.FTGQYCDSLYVPCAPSPCVNGGTCR.Q + 3 Carboxymethyl (C); Deamidated (NQ); 3 Phospho (ST); Phospho (Y) K.GADCTEDVDECAMANSNPCEHAGK.C + Carboxymethyl (C); Deamidated (NQ); 2 Phospho (ST); Oxidation (M) K.NECLSNPCQNGGTCDNLVNGYR.C + Carboxymethyl (C); 2 Deamidated (NQ); Phospho (ST); Phospho (Y) R.CSCPLGYTGK.N + Carboxymethyl (C); 2 Phospho (ST) R.DCESGCASSPCQHGGSCHPQR.Q + Carboxymethyl (C); Phospho (ST) R.DCESGCASSPCQHGGSCHPQR.Q + Carboxymethyl (C); 2 Deamidated (NQ); 2 Phospho (ST) R.DCESGCASSPCQHGGSCHPQR.Q + Carboxymethyl (C); Deamidated (NQ); Phospho (ST) R.DCESGCASSPCQHGGSCHPQR.Q + Carboxymethyl (C); 2 Deamidated (NQ); Phospho (ST) R.DCESGCASSPCQHGGSCHPQR.Q + Carboxymethyl (C); 2 Deamidated (NQ); Phospho (ST) R.DCESGCASSPCQHGGSCHPQR.Q + Carboxymethyl (C); Deamidated (NQ); Phospho (ST) R.DCESGCASSPCQHGGSCHPQR.Q + 4 Carboxymethyl (C); Deamidated (NQ); 2 Phospho (ST) R.DCESGCASSPCQHGGSCHPQRQPPYYSCQ.C + 2 Carboxymethyl (C); 2 Phospho (ST); 2 Phospho (Y) R.SFLRALGTLLHTNLRIK.R + 2 Phospho (ST) R.GPDGCTPLMLASLR.G + Phospho (ST); Oxidation (M) R.RPSAKSTMPTSLPNLAK.E + Deamidated (NQ); 3 Phospho (ST); Oxidation (M) K.STMPTSLPNLAK.E+ Deamidated (NQ); Oxidation (M) R.GPGTHMSEPPHNNMQVYA.- + Deamidated (NQ); 2 Phospho (ST); Phospho (Y); Oxidation (M) |
| BC04* | P42345 | Serine/threonine-protein kinase mTOR | 288610 | 134 | 10% | 6.73 | 14 | R.VLDIIRAALPPKDFAHK.R K.AMQVDATVFTCISMLARAMGPGIQQDIK.E + 2 Deamidated (NQ); 2 Oxidation (M) K.ELLEPMLAVGLSPALTAVLYDLSRQIPQLKK.D + Deamidated (NQ); 3 Phospho (ST) K.DIQDGLLKMLSLVLMHKPLR.H + 2 Oxidation (M); Phospho (ST) K.MLIQILTELEHSGIGRIK.E + Deamidated (NQ); Oxidation (M); 2 Phospho (ST) R.SGQGDALASGPVETGPMKKLHVSTINLQK.A+ 2 Deamidated (NQ); Oxidation (M); 2 Phospho (ST) R.SCWALAQAYNPMAR.D + Oxidation (M); Phospho (Y) R.LALAHKTLVLLLGVDPSR.Q R.IQSIAPSLQVITSK.Q + Deamidated (NQ); 2 Phospho (ST) K.QRPRKLTLMGSNGHEFVFLLK.G + Deamidated (NQ); Phospho (ST) K.QRPRKLTLMGSNGHEFVFLLK.G + 2 Deamidated (NQ); Phospho (ST) R.TNYTRSLAVMSMVGYILGLGDR.H + Deamidated (NQ); 2 Phospho (ST); Phospho (Y) R.LTRMLTNAMEVTGLDGNYR.T + 2 Deamidated (NQ); Oxidation (M); 2 Phospho (ST); Phospho (Y) R.TRTDSYSAGQSVEILDGVELGEPAHK.K + Deamidated (NQ); 2 Phospho (ST) |
| BC05* | P08047 | Transcription factor Sp1 | 80723 | 72 | 3% | 6.94 | 5 | K.TSHLRAHLR.W+ Phospho (ST) K.EQSGSSTNGSNGSESSKNR.T + 3 Deamidated (NQ); Phospho (ST) K.EQSGSSTNGSNGSESSK.N + 2 Deamidated (NQ); 5 Phospho (ST) K.EQSGSSTNGSNGSESSKNR.T + Deamidated (NQ); 4 Phospho (ST) K.EQSGSSTNGSNGSESSKNR.T + 3 Deamidated (NQ); 4 Phospho (ST) |
| HT-29 | ||||||||
| CC01* | Q9UKL3 | CASP8-associated protein 2 | 222520 | 134 | 8% | 6.14 | 11 | K.DLKLSFMKK.L + Oxidation (M); Phospho (ST) R.DKSVNSHSFQDGR.C K.NISALIKTARVEINR.K + Phospho (ST) R.SDSETSKPQESFEK.N + Deamidated (NQ); 5 Phospho (ST) K.DSSAALATSTSLSAKNVIKK.K + Phospho (ST) R.SHYQVGEGSSNEDSRR.G + 2 Deamidated (NQ); 3 Phospho (ST); Phospho (Y) K.RNDNSDYCGISEGMEMK.V + Carboxymethyl (C); 2 Oxidation (M); Phospho (ST); Phospho (Y) K.FKEIQTQNFSLINENQSLK.K + 2 Deamidated (NQ) K.FKEIQTQNFSLINENQSLK.K + 2 Deamidated (NQ) K.SEENYQDQNNSSINTVK.H + 2 Deamidated (NQ); 3 Phospho (ST); Phospho (Y) K.SEPGSNCDNSELPGTLHNSHKK.R + Carboxymethyl (C); 5 Phospho (ST) |
| CC02* | P49792 | E3 SUMO-protein ligase RanBP2 | 357974 | 146 | 9% | 5.85 | 18 | K.DPNFKGFSGAGEK.L K.IIDDSDSNLSVVK.K + Deamidated (NQ) K.TFEECQQNLMK.L + Carboxymethyl (C); Deamidated (NQ); Oxidation (M) R.ITPDMTLQNMK.G + 2 Deamidated (NQ); Oxidation (M); 2 Phospho (ST) K.LRLLVQHEINTLR.A + Phospho (ST) K.TFEECQQNLMKLQK.G + Deamidated (NQ); Oxidation (M); Phospho (ST) K.SNNSETSSVAQSGSESK.V + 2 Deamidated (NQ); 3 Phospho (ST) R.GIGDIKILQNYDNKQVR.I + Phospho (Y) K.CELSKNSDIEQSSDSKVK.N + Carboxymethyl (C); 3 Phospho (ST) K.GQWDCSVCCVQNESSSLK.C + 3 Deamidated (NQ); 4 Phospho (ST) K.DPNFKGFSGAGEKLFSSQYGK.M + Deamidated (NQ); Phospho (ST) K.QNQTTSAVSTPASSETSKAPK.S + 7 Phospho (ST) K.LNQSGTSVGTDEESDVTQEEER.D + 4 Phospho (ST) K.LNQSGTSVGTDEESDVTQEEER.D + 5 Phospho (ST) K.CKFEEAQSILKAPGTNVAMASNQAVR.I + Phospho (ST) R.NTFNFGSKNVSGISFTENMGSSQQK.N + 2 Deamidated (NQ); 5 Phospho (ST) K.GEAGQNLLEMMACDRLSQSGHMLLNLSR.G + 3 Deamidated (NQ); 2 Oxidation (M); Phospho (ST) K.EGQWDCSACLVQNEGSSTKCAACQNPR.K + 4 Deamidated (NQ); 4 Phospho (ST) |
| CC03* | P13639 | Elongation factor 2 | 95277 | 53 | 5% | 6.41 | 4 | M.VNFTVDQIR.A K.STLTDSLVCKAGIIASAR.A + 2 Phospho (ST) K.EDLYLKPIQRTILMMGR.Y + Oxidation (M); Phospho (ST) K.GPLMMYISKMVPTSDK.G + Oxidation (M); 2 Phospho (ST); Phospho (Y) |
| CC04* | Q8WX93 | Palladin | 150443 | 135 | 7% | 6.67 | 8 | K.SRAGAMPQAQKK.T + Deamidated (NQ); Oxidation (M) K.LQNTGVADGYPVR.L K.TTSVSLTIGSSSPK.T + 3 Phospho (ST) R.SAPAMQSSGSFNYARPK.Q + Deamidated (NQ); Phospho (ST); Phospho (Y) R.KPAMSPLLTRPSYIRSLR.K + Oxidation (M); Phospho (Y) R.KPAMSPLLTRPSYIRSLRK.A + Oxidation (M); Phospho (ST) K.SISSPVSKRKPAMSPLLTRPSYIR.S + Phospho (ST) K.NQPSALLSASASQSPMEDQGEMER.E + 3 Deamidated (NQ); 5 Phospho (ST) |
| CC05 | Q8NF91 | Nesprin-1 | 1010398 | 137 | 3% | 5.38 | 22 | R.IQVTLRK.W + Deamidated (NQ); Phospho (ST) K.QLLQQQAKSIK.E + 2 Deamidated (NQ) K.QLLQQQAKSIK.E + 2 Deamidated (NQ) K.QVKTCKSAQASLK.T + Carboxymethyl (C); Phospho (ST) K.QVKTCKSAQASLK.T + Carboxymethyl (C); Deamidated (NQ); Phospho (ST) R.MKRIHAVANIGTALK.F + Oxidation (M) K.MIEHLNLDDKELVK.E + Deamidated (NQ) R.DLTRAQMVESNLQDK.Y + Deamidated (NQ); Oxidation (M) K.QLLQQQAKSIKEQVK.K + 4 Deamidated (NQ) K.TPTGPELDTSYKGYMK.L + Phospho (Y) R.IQNLLGTKQSEADALAVLK.K R.ELGQTWANLDHMVGQLK.I + 2 Deamidated (NQ); Oxidation (M); Phospho (ST) K.SQLEGALSKWTSYQDGVR.Q + Phospho (ST) K.IVALEEKASQLEKTGNDASK.A + Deamidated (NQ); Phospho (ST) K.LNMMLSKGELLSTLLTKEK.A + Deamidated (NQ); 2 Oxidation (M); Phospho (ST) K.RRHSELELNIAQNMVSQVK.D + Deamidated (NQ); Phospho (ST) K.QNTTASGCELMHTEMQALR.A + Carboxymethyl (C); 2 Phospho (ST) K.ALESAAVSLDGILSKAQYHLK.I + Deamidated (NQ); 2 Phospho (ST); Phospho (Y) K.HKEYFQGLESHMILTETLFRK.I + Oxidation (M) R.QLRQTVEATNSMNKNESDLIEK.D + 3 Deamidated (NQ); Oxidation (M); 2 Phospho (ST) K.IQEFLSESENGQHKLNMMLSKGELLSTLLTK.E + 3 Deamidated (NQ); 2 Oxidation (M); 2 Phospho (ST) R.GESVLQNTSPEGIPTIQQQLQSVKDMWASLLSAGIR.C + 2 Deamidated (NQ); 3 Phospho (ST) |
| CC06 | Q9BWX5 | Transcription factor GATA-5 | 41273 | 213 | 16% | 9.17 | 5 | M.YQSLALAASPR.Q + Deamidated (NQ) R.RNSEGEPVCNACGLYMK.L + Deamidated (NQ); Oxidation (M); Phospho (ST) R.NASASPSAVASTDSSAATSK.A + 4 Phospho (ST) K.MNGVNRPLVRPQKRLSSSR.R + Deamidated (NQ); Oxidation (M); Phospho (ST) R.NASASPSAVASTDSSAATSK.A + 7 Phospho (ST) |
*The proteins were involved in the PI3K/AKT/mTOR pathway.
Table 1: The 16 proteins identified with higher confidence level (at least three unique peptide sequences matched) in this study.
Identified proteins in this study were classified into different groups of molecular functions and biological processes, based on their functional categories in the SWISS-Port/TrEMBL and Bioinformatic Harvester EMBL protein database. We used the ExPASy Molecular Biology Server (Expert Protein Analysis System) of the Swiss Institute of Bioinformatics (SIB) to explore the known functions of the identified proteins, which had been reported in the literature (Table 2). The Swiss-Prot identifiers could be employed for linkages of proteins to a defined vocabulary of terms describing the biological processes, cellular components and molecular functions of known Gene Ontology (GO). The Gene Ontology Consortium provides annotation of each protein and structure to organize selected proteins into biologically relevant groups. These groupings can serve as the basis for identifying those areas of biology showing correlated protein changes [22-24].
| Protein no.* | Swiss-Prot/ TrEMBL Accession no. |
Protein name | Subcellular location |
Biological process | Molecular function | Function** |
|---|---|---|---|---|---|---|
| HepG2 | ||||||
| LC01* | Q13148 | TAR DNA-binding protein 43 |
Nucleus | 3'-UTR-mediated mrna stabilization RNA splicing Cell death Mrna processing Mitosis Negative regulation by host of viral transcription Transcription from RNA polymerase II promotor |
Double-stranded DNA binding Mrna 3'-UTR binding Microtubule binding Nucleotide binding Transcription factor activity |
DNA and RNA-binding protein which regulates transcription and splicing. Involved in the regulation of CFTR splicing. It promotes CFTR exon 9 skipping by binding to the UG repeated motifs in the polymorphic region near the 3'-splice site of this exon. The resulting aberrant splicing is associated with pathological features typical of cystic fibrosis. May also be involved in microRNA biogenesis, apoptosis and cell division. Can repress HIV-1 transcription by binding to the HIV-1 long terminal repeat. |
| LC02* | P13693 | Translationally-controlled tumor protein |
Cytoplasm | Anti-apoptosis Calcium ion Transport Cellular calcium ion homeostasis Response to virus |
Calcium ion binding Protein binding |
Involved in calcium binding and microtubule stabilization. |
| LC03* | P81605 | Dermcidin | Secreted | Defense response to bacterium Defense response to fungus Killing of cells of another organism |
Protein binding | DCD-1 displays antimicrobial activity thereby limiting skin infection by potential pathogens in the first few hours after bacterial colonization. Highly effective against E.coli, E.faecalis, S.aureus and C.albicans. Optimal pH and salt concentration resemble the conditions in sweat. Survival-promoting peptide promotes survival of neurons and displays phosphatase activity. It may bind IgG. |
| LC04 | Q9C0H9 | p130Cas-associated protein | Cytoplasm | Exocytosis | Protein binding | Delays the onset of cell spreading in the early stages of cell adhesion to fibronectin. Also involved in calcium-dependent exocytosis from PC12 cells |
| LC05 | P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein |
Secreted | Cell adhesion Angiogenesis |
Protein C-terminus binding | Integral component of basement membranes. Component of the glomerular basement membrane (GBM), responsible for the fixed negative electrostatic membrane charge, and which provides a barrier which is both size- and charge-selective. It serves as an attachment substrate for cells. Plays essential roles in vascularization. Critical for normal heart development and for regulating the vascular response to injury. Also required for avascular cartilage development. Endorepellin in an anti-angiogenic and anti-tumor peptide that inhibits endothelial cell migration, collagen-induced endothelial tube morphogenesis and blood vessel growth in the chorioallantoic membrane. Blocks endothelial cell adhesion to fibronectin and type I collagen. Anti-tumor agent in neovascularization. Interaction with its ligand, integrin alpha2/beta1, is required for the anti-angiogenic properties. Evokes a reduction in phosphorylation of receptor tyrosine kinases via alpha2/beta1 integrin-mediated activation of the tyrosine phosphatase, PTPN6. The LG3 peptide has anti-angiogenic properties that require binding of calcium ions for full activity. |
| Mcf-7 | ||||||
| BC01* | P25054 | Adenomatous polyposis coli protein | Cell junction Cell membrane Cell projection Cytoplasm Cytoskeleton Membrane Microtubule |
Wnt signaling pathway | Microtubule plus-end binding Protein kinase regulator activity |
Tumor suppressor. Promotes rapid degradation of CTNNB1 and participates in Wnt signaling as a negative regulator. APC activity is correlated with its phosphorylation state. Activates the GEF activity of SPATA13 and ARHGEF4. Plays a role in hepatocyte growth factor (HGF)-induced cell migration. Required for MMP9 up-regulation via the JNK signaling pathway in colorectal tumor cells. Acts as a mediator of ERBB2-dependent stabilization of microtubules at the cell cortex. It is required for the localization of MACF1 to the cell membrane and this localization of MACF1 is critical for its function in microtubule stabilization. |
| BC02* | Q16644 | MAP kinase-activated protein kinase 3 |
Nucleus Cytoplasm. |
Myd88-dependent toll-like receptor signaling pathway Ras protein signal transduction TRIF-dependent toll-like receptor signaling pathway Toll signaling pathway Innate immune response Macropinocytosis Nerve growth factor receptor signaling pathway Peptidyl-serine phosphorylation Response to lipopolysaccharide Stress-activated MAPK cascade Toll-like receptor 1 signaling pathway Toll-like receptor 2 signaling pathway Toll-like receptor 3 signaling pathway Toll-like receptor 4 signaling pathway |
Kinase Serine/threonine-protein kinase Transferase |
Stress-activated serine/threonine-protein kinase involved in cytokines production, endocytosis, cell migration, chromatin remodeling and transcriptional regulation. Following stress, it is phosphorylated and activated by MAP kinase p38-alpha/MAPK14, leading to phosphorylation of substrates. Phosphorylates serine in the peptide sequence, Hyd-X-R-X(2)-S, where Hyd is a large hydrophobic residue. MAPKAPK2 and MAPKAPK3, share the same function and substrate specificity, but MAPKAPK3 kinase activity and level in protein expression are lower compared to MAPKAPK2. Phosphorylates HSP27/HSPB1, KRT18, KRT20, RCSD1, RPS6KA3, TAB3 and TTP/ZFP36. Mediates phosphorylation of HSP27/HSPB1 in response to stress, leading to dissociate HSP27/HSPB1 from large small heat-shock protein (sHsps) oligomers and impair their chaperone activities and ability to protect against oxidative stress effectively. Involved in inflammatory response by regulating tumor necrosis factor (TNF) and IL6 production post-transcriptionally: acts by phosphorylating AU-rich elements (AREs)-binding proteins, such as TTP/ZFP36, leading to regulate the stability and translation of TNF and IL6 mRNAs. Phosphorylation of TTP/ZFP36, a major post-transcriptional regulator of TNF, promotes its binding to 14-3-3 proteins and reduces its ARE mRNA affinity leading to inhibition of dependent degradation of ARE-containing transcript. Involved in toll-like receptor signaling pathway (TLR) in dendritic cells: required for acute TLR-induced macropinocytosis by phosphorylating and activating RPS6KA3. Also acts as a modulator of Polycomb-mediated repression. |
| BC03* | Q04721 | Neurogenic locus notch homolog protein 2 |
Cell membrane Membrane Nucleus |
Differentiation Notch signaling pathway Transcription Transcription regulation |
Calcium ion binding Receptor activity |
Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs .Involved in bone remodeling and homeostasis. In collaboration with RELA/p65 enhances NFATc1 promoter activity and positively regulates RANKL-induced osteoclast differentiation. |
| BC04* | P42345 | Serine /threonine-protein kinase mTOR | Cytoplasm Endoplasmic reticulum Golgi apparatus Lysosome Membrane Mitochondrion Mitochondrion outer membrane Nucleus |
T cell costimulation TOR signaling cascade Cell growth Cellular response to hypoxia Cellular response to nutrient levels Epidermal growth factor receptor signaling pathway Fibroblast growth factor receptor signaling pathway Germ cell development Insulin receptor signaling pathway Negative regulation of NFAT protein import into nucleus Negative regulation of autophagy Negative regulation of cell size Negative regulation of macroautophagy Nerve growth factor receptor signaling pathway Peptidyl-serine phosphorylation Peptidyl-threonine phosphorylation Phosphatidylinositol-mediated signaling Positive regulation of actin filament polymerization Positive regulation of endothelial cell proliferation Positive regulation of lamellipodium assembly Positive regulation of lipid biosynthetic process Positive regulation of myotube differentiation Positive regulation of peptidyl-tyrosine phosphorylation Positive regulation of protein kinase B signaling cascade Positive regulation of protein phosphorylation Positive regulation of stress fiber assembly Positive regulation of transcription from RNA polymerase III promoter Positive regulation of translation Protein autophosphorylation Protein catabolic process Regulation of Racgtpase activity Regulation of actin cytoskeleton organization Regulation of carbohydrate utilization Regulation of fatty acid beta-oxidation Regulation of glycogen biosynthetic process Regulation of protein kinase activity Regulation of response to food Response to amino acid stimulus Response to nutrient Ruffle organization |
Kinase Serine/threonine-protein kinase Transferase |
Serine/threonine protein kinase which is a central regulator of cellular metabolism, growth and survival in response to hormones, growth factors, nutrients, energy and stress signals. Functions as part of 2 structurally and functionally distinct signaling complexes mTORC1 and mTORC2 (mTOR complex 1 and 2). Activated mTORC1 up-regulates protein synthesis by phosphorylating key regulators of mRNA translation and ribosome synthesis. This includes phosphorylation of EIF4EBP1 and release of its inhibition toward the elongation initiation factor 4E (eiF4E). Moreover, phosphorylates and activates RPS6KB1 and RPS6KB2 that promote protein synthesis by modulating the activity of their downstream targets including ribosomal protein S6, eukaryotic translation initiation factor EIF4B and the inhibitor of translation initiation PDCD4. Regulates ribosome synthesis by activating RNA polymerase III-dependent transcription through phosphorylation and inhibition of MAF1 a RNA polymerase III-repressor. In parallel to protein synthesis, also regulates lipid synthesis through SREBF1/SREBP1 and LPIN1. To maintain energy homeostasis mTORC1 may also regulate mitochondrial biogenesis through regulation of PPARGC1A. mTORC1 also negatively regulates autophagy through phosphorylation of ULK1. Under nutrient sufficiency, phosphorylates ULK1 at 'Ser-758', disrupting the interaction with AMPK and preventing activation of ULK1. Also prevents autophagy through phosphorylation of the autophagy inhibitor DAP. mTORC1 exerts a feedback control on upstream growth factor signaling that includes phosphorylation and activation of GRB10 a INSR-dependent signaling suppressor. Among other potential targets mTORC1 may phosphorylate CLIP1 and regulate microtubules. As part of the mTORC2 complex MTOR may regulate other cellular processes including survival and organization of the cytoskeleton. Plays a critical role in the phosphorylation at 'Ser-473' of AKT1, a pro-survival effector of phosphoinositide 3-kinase, facilitating its activation by PDK1. mTORC2 may regulate the actin cytoskeleton, through phosphorylation of PRKCA, PXN and activation of the Rho-type guanine nucleotide exchange factors RHOA and RAC1A or RAC1B. mTORC2 also regulates the phosphorylation of SGK1 at 'Ser-422'. |
| BC05* | P08047 | Transcription factor Sp1 | Nucleus Cytoplasm. |
Host-virus interaction Transcription Transcription regulation |
RNA polymerase II core promoter proximal region sequence-specific DNA binding RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity involved in positive regulation of transcription RNA polymerase II core promoter sequence-specific DNA binding RNA polymerase II repressing transcription factor binding Bhlh transcription factor binding Double-stranded DNA binding Enhancer binding Protein homodimerization activity Sequence-specific DNA binding transcription factor activity Zinc ion binding |
Transcription factor that can activate or repress transcription in response to physiological and pathological stimuli. Binds with high affinity to GC-rich motifs and regulates the expression of a large number of genes involved in a variety of processes such as cell growth, apoptosis, differentiation and immune responses. Highly regulated by post-translational modifications (phosphorylations, sumoylation, proteolytic cleavage, glycosylation and acetylation). Binds also the PDGFR-alpha G-box promoter. May have a role in modulating the cellular response to DNA damage. Implicated in chromatin remodeling. Plays a role in the recruitment of SMARCA4/BRG1 on the c-FOS promoter. Plays an essential role in the regulation of FE65 gene expression. In complex with ATF7IP, maintains telomerase activity in cancer cells by inducing TERT and TERC gene expression. |
| Ht-29 | ||||||
| CC01* | Q9UKL3 | CASP8-associated protein 2 | Cytoplasm Mitochondrion Nucleus |
Apoptosis Cell cycle Transcription Transcription regulation |
DNA binding SUMO polymer binding Cysteine-type endopeptidase activator activity involved in apoptotic process Death receptor binding Transcription corepressor activity |
Participates in TNF-alpha-induced blockade of glucocorticoid receptor (GR) transactivation at the nuclear receptor coactivator level, upstream and independently of NF-kappa-B. Suppresses both NCOA2- and NCOA3-induced enhancement of GR transactivation. Involved in TNF-alpha-induced activation of NF-kappa-B via a TRAF2-dependent pathway. Acts as a downstream mediator for CASP8-induced activation of NF-kappa-B. Required for the activation of CASP8 in FAS-mediated apoptosis. Required for histone gene transcription and progression through S phase. |
| CC02* | P49792 | E3 SUMO-protein ligase RanBP2 | Membrane Nuclear pore complex Nucleus |
Protein transport Translocation Transport Ubl conjugation pathway Mrna transport |
Isomerase Ligase Rotamase |
E3 SUMO-protein ligase which facilitates SUMO1 and SUMO2 conjugation by UBE2I. Involved in transport factor (Ran-GTP, karyopherin)-mediated protein import via the F-G repeat-containing domain which acts as a docking site for substrates. Could also have isomerase or chaperone activity and may bind RNA or DNA. Component of the nuclear export pathway. Specific docking site for the nuclear export factor exportin-1. Sumoylates PML at 'Lys-490' which is essential for the proper assembly of PML-NB. |
| CC03* | P13639 | Elongation factor 2 |
Cytoplasm. | Protein biosynthesis | Elongation factor | Catalyzes the GTP-dependent ribosomal translocation step during translation elongation. During this step, the ribosome changes from the pre-translocational (PRE) to the post-translocational (POST) state as the newly formed A-site-bound peptidyl-tRNA and P-site-bound deacylatedtRNA move to the P and E sites, respectively. Catalyzes the coordinated movement of the two tRNA molecules, the mRNA and conformational changes in the ribosome. |
| CC04* | Q8WX93 | Palladin | Cell junction Cell projection Cytoplasm Cytoskeleton |
Cytoskeleton organization | Muscle alpha-actinin binding | Cytoskeletal protein required for organization of normal actin cytoskeleton. Roles in establishing cell morphology, motility, cell adhesion and cell-extracellular matrix interactions in a variety of cell types. May function as a scaffolding molecule with the potential to influence both actin polymerization and the assembly of existing actin filaments into higher-order arrays. Binds to proteins that bind to either monomeric or filamentous actin. Localizes at sites where active actin remodeling takes place, such as lamellipodia and membrane ruffles. Different isoforms may have functional differences. Involved in the control of morphological and cytoskeletal changes associated with dendritic cell maturation. Involved in targeting ACTN to specific subcellular foci. |
| CC05 | Q8NF91 | Nesprin-1 | Cytoplasm Cytoskeleton Membrane Nucleus |
Golgi organization Cell death Cytoskeletal anchoring at nuclear membrane Muscle cell differentiation Nuclear matrix anchoring at nuclear membrane |
Actin binding Protein homodimerization activity |
Multi-isomeric modular protein which forms a linking network between organelles and the actin cytoskeleton to maintain the subcellular spatial organization. Component of SUN-protein-containing multivariate complexes also called LINC complexes which link the nucleoskeleton and cytoskeleton by providing versatile outer nuclear membrane attachment sites for cytoskeletal filaments. May be involved in the maintenance of nuclear organization and structural integrity. Connects nuclei to the cytoskeleton by interacting with the nuclear envelope and with F-actin in the cytoplasm. May be required for centrosome migration to the apical cell surface during early ciliogenesis. |
| CC06 | Q9BWX5 | Transcription factor GATA-5 |
Nucleus | Transcription Transcription regulation |
Activator | Binds to the functionally important CEF-1 nuclear protein binding site in the cardiac-specific slow/cardiac troponin C transcriptional enhancer. May play an important role in the transcriptional program(s) that underlies smooth muscle cell diversity |
*The proteins were involved in the PI3K/AKT/mTOR pathway. **The protein functions in Table 2 were reproduced from the UniProtKB web site.
Table 2: Subcellular location and protein function of 16 proteins with higher confidence levels identified in cancer.
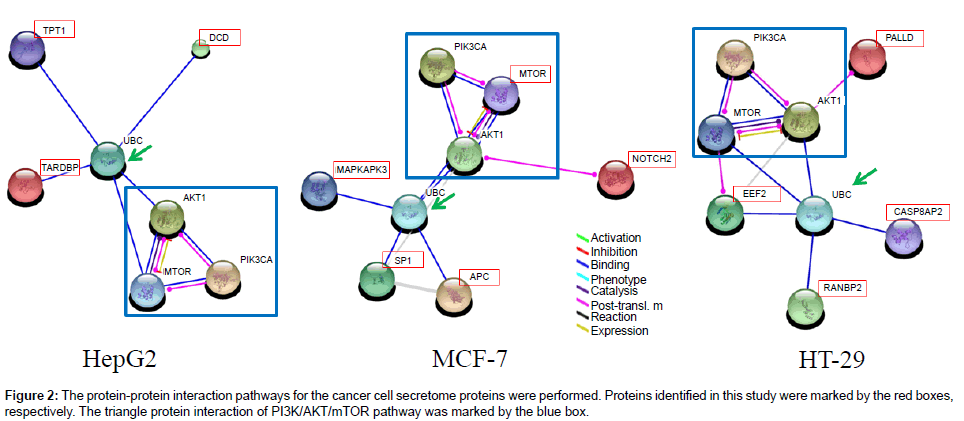
In this study, the protein-interaction networks were analyzed by String 9.1 Web software, which showed the interacted proteins in cells. Thus, the biological significance between increase in the secretion of those proteins and tumor progression could be further studied accordingly. Utilizing the String 9.1 Web software, 12 of 16 proteins were connected by protein-protein interaction and marked in the Figure 2, respectively. In Figure 2, the identified proteins in liver (3), colorectal (5), and breast (4) cancers were showed and involved in the PI3K/AKT/mTOR pathway. Those proteins were also marked astral symbols in the Tables 1 and 2.
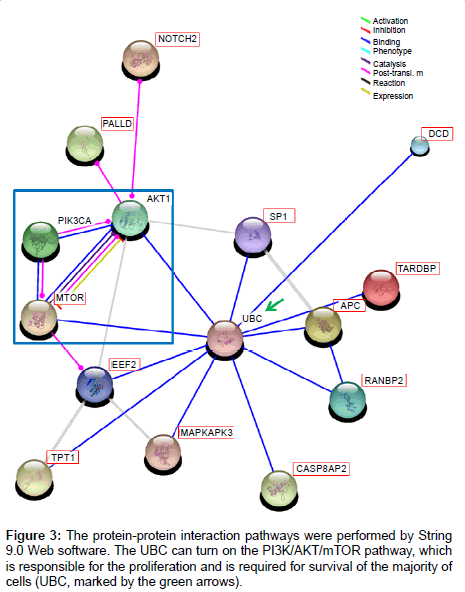
In the experimental results, the main finding was the secretome proteins of tumor cells may activate or enhance the PI3K/AKT/mTOR pathway, which may result in tumor metastasis and cell proliferation. In Figure 3, the protein-interaction network was identified and the key protein was the ubiquitin C (UBC, marked by the green arrows). The UBC can turn on the PI3K/AKT/mTOR pathway, which is responsible for the proliferation and is required for survival of the majority of cells.
The mammalian target of rapamycin pathway (mTOR pathway, also known as FRAP, RAFT1 and RAP1 pathway) has been identified as a key kinase acting downstream of the activation of phosphoinositide- 3-kinase (PI3K) [25]. The hypothesis of the mTOR pathway is that it acts as a master switch of cellular catabolism and anabolism, thereby determining the growth and proliferation of tumor cells [26,27]. Activation of PI3K/AKT/mTOR signaling through the mutation of pathway components as well as through the activation of upstream signaling molecules, occur in a majority of cancer cells contributing to deregulation of proliferation, resistance to apoptosis, and changes in the metabolism characteristic of transforming cells [28].
Conclusion
The protein constitutively secreted by cells in vitro is referred to as the secretome. In this study, we used the mass spectrometry technique as an analytical method for determining cancer biomarkers on tumor cell secretome. The cell model was used to identify the tumor progression related proteins, which are secreted into the serum-free culture medium. Secreted proteins may coordinate various cellular processes in tumor cells, including growth, division, differentiation, apoptosis, migration, metastasis, angiogenesis and adhesion, and thereby contribute to the growth and spread of the tumor cells. Thus, the identification of secreted proteomics in tumor cells provides potential biomarkers closely related to carcinogenesis. Moreover, disease-specific protein biomarkers allow us to define the prognosis of the disease and gain deep insight into disease mechanisms by which proteins play a major role. Studies on secretome of different tumor cells may find novel biomarkers.
Our data demonstrate that the development of strategies to isolate proteins can expand the tumor cell proteomic map. With the bioinformatics analysis, the tumor related proteins may help discern the origin of proteins in cancer. This approach is a potentially powerful tool to discover new biomarkers and/or causative factors of disease-related proteins in cancer clinical studies; in addition, its sensitivity may also make it applicable to direct ultra-micro analysis of other bio-samples.
In this study, of these 16 proteins, 12 were presented that closely related to PI3K/AKT/mTOR pathway and may serve as cancer biomarkers. In addition, the correlation of UBC in the cancer cells was greatly indicated. The potential utility of UBC as an early cancer biomarkers merits study in the future. The database we generated provides information both on the identities of proteins present in tumor cells and a potential diagnostic biomarker for cancer.
Acknowledgements
The authors thank the Center of Excellence for Environmental Medicine, Kaohsiung Medical University for the assistance in protein identification, and S. Sheldon MT (ASCP, Retired) of Oklahoma University Medical Center Edmond for fruitful discussions and editorial assistance before submission. This work was supported by research grants 94-KMUH-ND-017 from Kaohsiung Medical University Hospital, NSC-102-3114-Y-492-076-023, NSC-100-2320-B-037-007- MY3 from the National Science Council, NSYSUKMU 102-P006 from NSYSUKMU Joint Research Project, and MOHW103-TD-B-111-05 from Ministry of Health and Welfare, Taiwan, Republic of China.
References
- Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63: 11-30.
- Ulmer SC (2000) Hepatocellular carcinoma. A concise guide to its status and management. Postgrad Med 107: 117-124.
- Howard BA, Wang MZ, Campa MJ, Corro C, Fitzgerald MC, et al. (2003) Identification and validation of a potential lung cancer serum biomarker detected by matrix-assisted laser desorption/ionization-time of flight spectra analysis. Proteomics 3: 1720-1724.
- Swensen SJ, Jett JR, Sloan JA, Midthun DE, Hartman TE, et al. (2002) Screening for lung cancer with low-dose spiral computed tomography. Am J RespirCrit Care Med 165: 508-513.
- Chen G, Gharib TG, Huang CC, Thomas DG, Shedden KA, et al. (2002) Proteomic analysis of lung adenocarcinoma: identification of a highly expressed set of proteins in tumors. Clin Cancer Res 8: 2298-2305.
- Wilkins MR, Sanchez JC, Williams KL, Hochstrasser DF (1996) Current challenges and future applications for protein maps and post-translational vector maps in proteome projects. Electrophoresis 17: 830-838.
- Hamler RL, Zhu K, Buchanan NS, Kreunin P, Kachman MT, et al. (2004) A two-dimensional liquid-phase separation method coupled with mass spectrometry for proteomic studies of breast cancer and biomarker identification. Proteomics 4: 562-577.
- Wolters DA, Washburn MP, Yates JR 3rd (2001) An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem 73: 5683-5690.
- Fujii K, Nakano T, Kawamura T, Usui F, Bando Y, et al. (2004) Multidimensional protein profiling technology and its application to human plasma proteome. J Proteome Res 3: 712-718.
- Yang MH, Jong SB, Lu CY, Lin YF, Chiang PW, et al. (2012) Assessing the responses of cellular proteins induced by hyaluronic acid-modified surfaces utilizing a mass spectrometry-based profiling system: over-expression of CD36, CD44, CDK9, and PP2A. Analyst 137: 4921-4933.
- Harris CC (1994) Solving the viral-chemical puzzle of human liver carcinogenesis. Cancer Epidemiol Biomarkers Prev 3: 1-2.
- Harris CC (1996) The 1995 Walter Hubert Lecture--molecular epidemiology of human cancer: insights from the mutational analysis of the p53 tumour-suppressor gene. Br J Cancer 73: 261-269.
- Vilar E, Tabernero J (2013) Molecular dissection of microsatellite instable colorectal cancer. Cancer Discov 3: 502-511.
- Lee SY, Lim TG, Chen H, Jung SK, Lee HJ, et al. (2013) Esculetin suppresses proliferation of human colon cancer cells by directly targeting β-catenin. Cancer Prev Res (Phila) 6: 1356-1364.
- Lynch HT, de la Chapelle A (2003) Hereditary colorectal cancer. N Engl J Med 348: 919-932.
- Pickhardt PJ, Hassan C, Halligan S, Marmo R (2011) Colorectal cancer: CT colonography and colonoscopy for detection--systematic review and meta-analysis. Radiology 259: 393-405.
- Brennan SF, Cantwell MM, Cardwell CR, Velentzis LS, Woodside JV (2010) Dietary patterns and breast cancer risk: a systematic review and meta-analysis. Am J Clin Nutr 91: 1294-1302.
- Colonna SV, Douglas Case L, Lawrence JA (2012) A retrospective review of the metabolic syndrome in women diagnosed with breast cancer and correlation with estrogen receptor. Breast Cancer Res Treat 131: 325-331.
- Autier P, Boniol M, La Vecchia C, Vatten L, Gavin A, et al. (2010) Disparities in breast cancer mortality trends between 30 European countries: retrospective trend analysis of WHO mortality database. BMJ 341: c3620.
- Amir E, Freedman OC, Seruga B, Evans DG (2010) Assessing women at high risk of breast cancer: a review of risk assessment models. J Natl Cancer Inst 102: 680-691.
- Shin HR, Boniol M, Joubert C, Hery C, Haukka J, et al. (2010) Secular trends in breast cancer mortality in five East Asian populations: Hong Kong, Japan, Korea, Singapore and Taiwan. Cancer Sci 101: 1241-1246.
- O'Donovan C, Martin MJ, Gattiker A, Gasteiger E, Bairoch A, et al. (2002) High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Brief Bioinform 3: 275-284.
- Pandey A, Podtelejnikov AV, Blagoev B, Bustelo XR, Mann M, et al. (2000) Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc Natl Acad Sci U S A 97: 179-184.
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, et al. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31: 3784-3788.
- Faivre S, Kroemer G, Raymond E (2006) Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov 5: 671-688.
- Khan N, Afaq F, Khusro FH, Mustafa Adhami V, Suh Y, et al. (2012) Dual inhibition of phosphatidylinositol 3-kinase/Akt and mammalian target of rapamycin signaling in human nonsmall cell lung cancer cells by a dietary flavonoid fisetin. Int J Cancer 130: 1695-1705.
- O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, et al. (2006) mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res 66: 1500-1508.
- Zhou HY, Huang SL (2012) Current development of the second generation of mTOR inhibitors as anticancer agents. Chin J Cancer 31: 8-18.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 16204
- [From(publication date):
specialissue-2014 - Aug 30, 2025] - Breakdown by view type
- HTML page views : 11402
- PDF downloads : 4802