Research Article Open Access
Method for Discovery of Peptide Reagents Using a Commercial Magnetic Separation Platform and Bacterial Cell Surface Display Technology
Deborah A. Sarkes1, Brandi L. Dorsey2, Amethist S. Finch1 and Dimitra N. Stratis-Cullum1*
1U.S. Army Research Laboratory, Sensors and Electron Devices Directorate, Adelphi MD, USA
2Federal Staffing Resources, Annapolis MD, USA
- *Corresponding Author:
- Dimitra N. Stratis-Cullum
U.S. Army Research Laboratory
Sensors and Electron Devices Directorate
2800 Powder Mill Road Adelphi, MD 20783, USA
Tel: 301-394-0794
Fax: 301-394-0310
E-mail: dimitra.n.stratis-cullum.civ@mail.mil
Received date: June 23, 2015; Accepted date: July 04, 2015; Published date: July 11, 2015
Citation: Sarkes DA, Dorsey BL, Finch AS, Stratis-Cullum DN (2015) Method for Discovery of Peptide Reagents Using a Commercial Magnetic Separation Platform and Bacterial Cell Surface Display Technology. J Anal Bioanal Tech 6:255 doi: 10.4172/2155-9872.1000255
Copyright: © 2015 Sarkes DA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Biopanning by bacterial display has many advantages over yeast and phage display, including the speed to discovery of affinity reagents and direct amplification of bound cells without the need to elute and reinfect. However, widespread use is limited, in part due to poor performance achieved using manual Magnetic-Activated Cell Sorting (MACS) methods, and an absence of widely-available, low cost, high-performance sorting alternatives. Here, we have developed a methodology for bacterial cell sorting using the semi-automated autoMACS® Pro Separator for the first time, and have produced a complete method for sorting of bacteria displaying 15-mer peptides on their cell surface using this device, including downstream bioinformatic analysis of candidates for binding to a target of interest. Two autoMACS® programs designed for isolation of target cells with low frequency were evaluated and adapted to bacterial biopanning, using protective antigen (PA) of Bacillus anthracis as the model system. In contrast to manual MACS, the bacterial display library was preferentially enriched by autoMACS® sorting, yielding several promising candidates after only three rounds of biopanning and bioinformatic analysis. Individual candidates were evaluated for relative binding to fluorescently-labeled PA target or streptavidin negative control using Fluorescence-Activated Cell Sorting (FACS). The top thirteen peptide candidates from the autoMACS® sort demonstrate binding to PA with low cross-reactivity to streptavidin, while only two of eighteen candidates from the manual sort showed binding to PA, and both demonstrated greater cross-reactivity to streptavidin. Overall, the autoMACS® platform quickly harvested higher affinity peptide candidates with demonstrated specificity to the PA target. Peptide candidates produced with this method contained the previously reported PA consensus WXCFTC, further validating this method and the commercially available autoMACS® platform as the first low cost, semi-automated biopanning approach for bacterial display that is widely accessible and more reliable than the MACS/FACS standard protocol.
Keywords
PA; Peptide; Bacterial display; autoMACS®; Bio-threat; Affinity reagent; Biocombinatorial; eCPX
Introduction
Bacterial display sorting is a powerful emerging technology that offers a rapid, high-throughput approach to discovery of robust affinity reagents [1-3]. With increased use of peptides as therapeutics, the application space for this discovery platform has expanded in recent years [4-6]. As compared with yeast and phage display, bacterial display is ideal because of the fast doubling time of bacteria, about 20 minutes for Escherichia coli(E. coli) [7] versus about 2 hours for Saccharomyces cerevisiae (S. cerevisiae) [8,9], and direct amplification of the bound bacterial cells containing plasmid DNA encoding the displayed peptide responsible for binding, without elution and reinfection. With phage display, abrasive chemicals are often necessary to elute the tightest binders for reinfection, and low pH conditions are required at a minimum when other known ligands for the target are unavailable. Therefore, the resulting pool of candidates may be biased to lower affinity binders, or stronger binders that are able to survive the harsh conditions or that can be isolated by physical methods [10-14]. The mode of binding can also be a factor in successful elution with low pH; binders interacting by non-electrostatic interactions may not be easily eluted at low pH, and a bias for positively charged sequences can result [15]. In addition to bacterial display’s advantages of speed and direct replication, E. coli is relatively easy to manipulate with high efficiency, allowing for generation of customized libraries for development of biocombinatorial discovery methods for both biological and inorganicbinding peptide reagents [16-23].
Ideally, a semi-automated bacterial display sorting method should be used for both speed and reproducibility. It is absolutely imperative that the biopanning method allow for selective enrichment of a rare target population amongst a large population of very similar bacteria, differing only in variations across short peptide sequences displayed on the surface scaffold. Microfluidic screening of bacterial display libraries was previously demonstrated for epitope mapping [24], and we have shown that the engineered bacterial display library eCPX, displaying random 15-amino acid peptides on the bacterial cell surface [25], can be combined with Micro-Magnetic Separation (MMS) for use as a simple, semi-automated discovery platform [18,19,26]. In addition to the rapid discovery of peptide affinity reagents using the MMS method (less than one-week), the method produced a family of peptide reagents to protective antigen (PA) of Bacillus anthracis, an emerging biological threat [19]. Most recently, the eCPX E. coli display library and MMS platform were utilized in the discovery of peptide affinity reagents against staphylococcal enterotoxin B (SEB) as well, further demonstrating that this semi-automated methodology is a valuable tool for the detection of biological threats [18]. The peptides isolated by the MMS using PA as a target showed sequence consensus (WXCFTC) and exhibited similar or better peptide interaction with the PA protein target than with a streptavidin negative control, measured through Fluorescence-Activated Cell Sorting (FACS) assays and compared to peptides isolated by conventional Magnetic-Activated Cell Sorting (MACS)/FACS sorting, where a library is pre-enriched by several rounds of MACS to reduce the number of cells to be sorted using FACS [19]. FACS alone is not sufficient for earlier rounds of sorting when the diversity is above 108 members because these devices are limited to sorting 107 to 108 cells [27]. Although a single candidate with the WXCFTC consensus was isolated by MACS/FACS sorting in Kogot et al., the enrichment of peptides with this binding consensus was poor using this method as compared to the MMS. Additionally, the FACS portion of the MACS/FACS method may not be an option for some due to the higher cost of obtaining and maintaining FACS devices capable of sorting cells for downstream use, typically $350,000-$500,000 to purchase [3], not to mention that they require highly skilled personnel to operate.
Although eCPX bacterial display technology has been shown to be a powerful approach to biopanning and the study of genetically engineered peptides, a widely available, low cost, semi-automated method is lacking because the previously characterized MMS platform is not commercially available for routine discovery. The need for a fast, inexpensive, reliable, reproducible method for discovery of affinity reagents to emerging bio-threats necessitates investigating other platforms, such as the autoMACS® Pro Separator, commercially available from Miltenyi Biotec. The autoMACS® Pro Separator is under $50,000 and is very simple to use. Methods have been published on this platform, as well as with an earlier model that was lower-throughput, for cell sorting using yeast surface display [28-31], as well as for sorting tumor epithelial cells for downstream screening of interacting scFVs by phage display [32]. However, to the best of our knowledge, the work herein represents the first utilizing a bacterial system, let alone a bacterial display library for peptide discovery. E. coli bacteria is approximately 1 μm in diameter, 2 μm in length, and 1 μm3 in volume while Saccharomyces cerevesiae (S. cerevisiae) yeast is approximately 3-6 μm in diameter and 30-40 μm3 in volume [33-37]. The smaller size of bacteria creates unique challenges over separation of yeast in a machine primarily used for separation of eukaryotic cells, where bacteria is a contaminant to be avoided, as highlighted by the use of sodium azide in the autoMACS® Running Buffer - MACS® Separation Buffer.
In this paper, we discovered several peptide capture candidates using the autoMACS® Pro Separator and a bacterial display library for the first time. We investigated the applicability of the system for discovery of peptide reagents for PA as a model system, allowing for a benchmark of comparison to previously published work using the MMS platform [19], and included important considerations to the number of rounds of biopanning, sequence analysis, and cross-reactivity.
Materials and Methods
Biopanning bacterial display library
Four rounds of biopanning were performed using a Dynabeads® MPC®-S magnetic particle concentrator (Life Technologies, Grand Island, NY, USA) for manual MACS, or the autoMACS® Pro Separator (Miltenyi Biotec, San Diego, CA, USA) for autoMACS®, similarly to the previously described protocols for manual MACS and the MMS [18,19,26]. See supplementary protocol for detailed adaptation and optimization for our method (Biopanning bacterial display library for Protective Antigen binders using manual MACS or autoMACS®). The bacterial display peptide library used was eCPX 3.0 (Cytomx Therapeutics, San Francisco, CA, USA) and was pre-depleted of streptavidin binders to avoid direct binding of non-specific peptides to the beads [18,19,26]. The target was recombinant protective antigen (PA; List Biological Laboratories, Campbell, CA, USA) biotinylated using No-Weigh Sulfo-NHS-LC-Biotin (Thermo Scientific, Rockford, IL, USA; PA-Biotin) and tested for biotinylation using the Pierce Biotin Quantitation Kit (Thermo Scientific, Rockford, IL, USA), and the paramagnetic beads used were Dynabeads® MyOne Streptavidin T1 Beads (Invitrogen, Carlsbad, CA, USA).
Manual MACS: Each sample containing PA-Biotin-bound cells and Streptavidin T1 beads was placed in a magnetic particle concentrator and allowed to separate for 5 minutes. The supernatant was removed and the beads washed 3 times with 1 mL PBS with 0.5% w/v Bovine Serum Albumen (PBS-B) by inverting the tube 3-4 times in the absence of the magnet, then returning the sample to the magnet between washes to retain the beads and remove the supernatant. After three washes, the positive fraction containing cells and beads were processed for the next sorting round and for spot plating and FACS analysis, as described below and in the supplemental protocol. The positive fraction was the washed beads resuspended in 1 mL PBS-B.
autoMACS®: For each round, the sample containing PA-Biotinbound cells with Streptavidin T1 beads was moved to a 15 mL conical tube for sorting on the autoMACS® Pro Separator, and an additional 500 μL of PBS-B was added to the sample after using it to recover remaining beads from the microcentrifuge tube. The samples, and empty 15 mL conical tubes for positive and negative fraction collection, were placed in a cold Chill 15 rack (Miltenyi Biotec, San Diego, CA, USA), as described in the manufacturer’s instructions, and run through one of two pre-loaded separation programs, “Posseld” or “Posselds,” designed for positive selection of target cells with low frequency [38] and named “Program D” and “Program DS” herein for simplicity. PBS-B was used in place of the Miltenyi Biotec autoMACS® wash and running buffers for both methods to avoid exposing the bacteria to detergents and sodium azide, and because it worked well with the MMS [18,19,26]. A rinse step was added in between and at the end of all samples, then the system was returned to its recommended run and wash buffers and a sleep step was run with 70% ethanol before turning off the machine, to prevent bacterial growth in the tubing and pump. See supplemental protocol (Biopanning bacterial display library for Protective Antigen binders using manual MACS or autoMACS®) for more detail.
Spot plating
The positive fraction was collected after separation with either manual MACS or autoMACS® and a small amount serially diluted 1:10 for spot plating [39] on Luria Broth (LB) Agar plates containing 25 μg/ mL chloramphenicol (LB Cm25 Agar plates). In triplicate, 10 μl spots were plated for 10-2-10-7 dilutions of Colony Forming Units (CFU)/ mL, as compared to the undiluted positive fraction, such that a 10 μl spot of undiluted sample constitutes a 10-2 "dilution" of CFU/mL. The spots were allowed to dry before inverting the plates and incubating overnight at 37°C, and the colonies in each spot were counted the next day, and replicate spots averaged. A spot with about 10-20 colonies was ideal for estimating the total number of cells/mL in the positive fraction.
FACS analysis of sorting rounds
BD FACS Canto™ II and BD FACSDiva™ Software (BD, Franklin Lakes, New Jersey, USA) were used to assess the level of PA binding after each round of biopanning, using induced samples saved on ice after each round of sorting. For each sample, 5 μL of induced cells was added to 25 μL of cold PBS alone or containing 150 nM YPet Mona positive control (ARL, Adelphi, MD, USA) [21], 900 nM PA labeled with Dylight 488 NHS Ester (Thermo Scientific, Rockford, IL, USA; PA488), or 900 nm Streptavidin, R-Phycoerythrin conjugate (SAPE, Life Technologies, Grand Island, NY, USA), and incubated for 45 minutes on ice. The cells were centrifuged at 5,000 xg for 5 minutes and the supernatant removed. The pellet was resuspended in 500 μL ice cold, filtered BD FACSFlow™ (BD, Franklin Lakes, New Jersey, USA), mixed thoroughly, and read immediately. The samples were then run on the FACS and analyzed by gating and comparing to the PBS alone sample for measuring the percentage of bound cells falling outside of the gate [19,40], and by the Normalized Median Fluorescence Intensity (nMFI) of the total population for each sample, normalized to the MFI of a negative control sample expressing eCPX with no 15-mer peptide, incubated with the same fluorophore [41].
Sequence analysis of potential PA binders
For rounds 2-4 of autoMACS® and for round 4 of manual MACS, 144 bacterial colonies were sent to a local Genewiz facility (Frederick, MD, USA) for DNA sequencing using their pBad Forward primer. Resulting sequences were proof-read when necessary, and multiple sequence alignment performed using ClustalW2 (The EMBLEuropean Bioinformatics Institute, Hinxton, Cambridge, UK) and Jalview with Clustal_X windows interface [42,43], available online [44]. Gap penalties were set to 100 for both pairwise and multiple sequence alignment, but the default settings were otherwise used. This was done for all sequences in each sorting round tested, as well as for individual sets of sequences described in the results and discussion.
FACS analysis of individual isolates
For each individual isolate of interest, including all repeats, a 5 ml culture of LB containing 25 μg/mL chloramphenicol (LB Cm25) and supplemented with 0.2% w/v D-glucose was inoculated and grown overnight at 37°C in an orbital shaker at 225 RPM. The next day, the cultures were diluted 1:50 in 3 ml LB Cm25 without glucose and grown to OD600 of 0.5-0.55, then bacterial display was induced by adding 0.04% w/v L-arabinose plus 2 mM ethylene diamine tetra acetic acid (EDTA, for facilitation of peptide display [45]) and incubating for 45 minutes at 37°C with shaking. Induced cultures were placed on ice, and 5 μL of induced cells was added to 25 μL of ice cold PBS alone or containing 150 nM YPet Mona, 250 nM PA488, or 250 nM SAPE, and incubated for 45 minutes on ice. Labeled cells were centrifuged at 5,000xg for 5 minutes and the supernatant removed. The cell pellet was resuspended in 500 μL of ice cold, filtered BD FACSFlow™ and mixed thoroughly. Each sample was then loaded on the FACS immediately after mixing and analyzed as described for sorting rounds. Four independent experiments were performed for manual MACS repeats and random colonies, and three independent experiments were performed for the autoMACS® best binders, as determined by an initial FACS screen including all repeats. The average and standard deviation were calculated and graphed using Prism Software (GraphPad, La Jolla, CA, USA).
Results and Discussion
Biopanning is a powerful method utilizing a biocombinatorial library of candidate binders, with several rounds of exposure of this library to a target of interest, isolating and amplifying the pool of interacting library members with each round. Enrichment in the percentage of the population that binds to the target is generally observed over several rounds. The end result is discovery of a smaller pool of candidates that contain the property of interest, in this case the relative binding performance of isolated peptide candidates to the model target of interest, protective antigen (PA) [3,46]. In order to investigate the feasibility of using the autoMACS® Pro Separator for biopanning a bacterial display library, it was necessary to develop a supporting method framework that considers the cell surface density of the displayed peptide library, reproducibility of isolation, enrichment throughout the discovery process, and the number of sorting rounds necessary to generate peptide candidates. However, equally important is the development of an analysis approach to guide the understanding of peptide consensus based on physio-chemical properties including cross-platform consensus, as well as a methodology framework for down-selection and analysis of the candidate pool.
Manual magnetic sorting versus autoMACS® sorting and FACS analysis of sorting rounds
To compare the enrichment of peptide binders for PA isolated from a bacterial display library using manual and semi-automated methods, biopanning via manual MACS was run in parallel and compared to the results obtained from adapting two programs for isolation of rare cells on the autoMACS® Pro Separator: “Posseld” and “Posselds,” named “Program D” and “Program DS” herein for simplicity. Both autoMACS® programs allow positive selection of labeled target cells and both use two magnetic columns to capture target cells with less than 5% frequency in the initial population. However, Program DS should be more sensitive for weak antigen expression, with a slower flow rate through column 1 to increase exposure time [38]. In order to prescreen running buffer and wash buffer compatibility and performance, the recommended manufacturer buffers were evaluated and compared to PBS-B, which was used in previous bacterial cell sorting studies [18,19,26]. Through a comparison of spot plates, both Program D and Program DS yielded similar numbers of viable cells in the positive fraction throughout the rounds of sorting using PBS-B or the autoMACS® buffers purchased from the manufacturer, about 106 cells for round 1 in both cases. However, the overnight growth in LB Cm25 solution and downstream analysis were problematic using the autoMACS® Wash and Running buffers, likely due to the presence of sodium azide and detergents in the recommended buffers. PBS-B was therefore chosen for further study, and allowed for a more direct comparison with manual MACS and the previous MMS study.
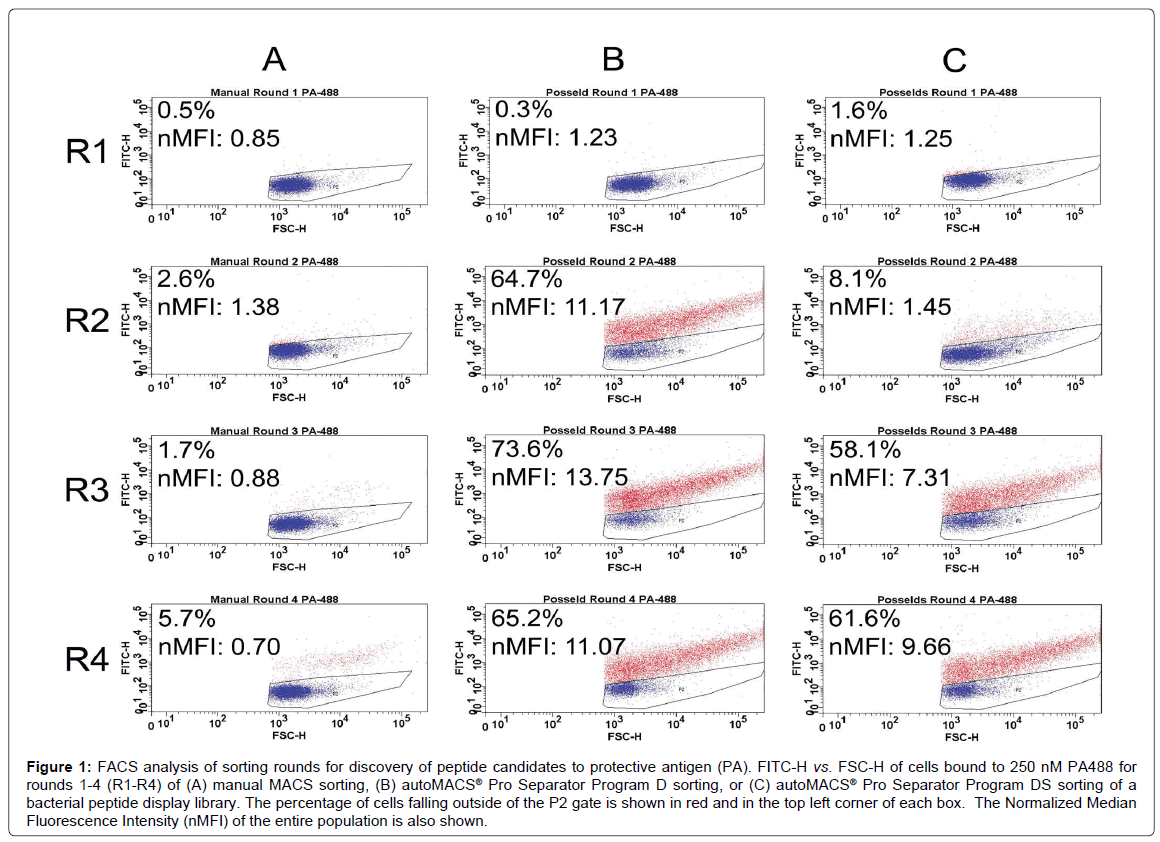
Figure 1 shows FACS analysis of each sorting round for the discovery of peptide capture candidates for protective antigen (PA) using manual MACS (Figure 1A) and autoMACS® separation programs (Figure 1B and 1C). For each round of sorting, the nMFI for binding to 900 nM PA488 using autoMACS® was found to be substantially higher than for manual MACS, with very little evidence of enrichment found using the manual methodology. Specifically, the nMFI for manual MACS reached only 0.70 by round 4, and was therefore similar to the nMFI of 0.85 reached for manual MACS in round 1. Similarly, when looking at the FACS data by percent bound, where the gated population represents unbound cells in buffer only [19,40], there was a slight increase in binding from 0.5% of the population in round 1 to 5.7% in round 4. This was substantially lower than the level of binding observed using the autoMACS®, determined by both nMFI and percent binding, but the percent binding using manual MACS is similar to the enrichment obtained using a MiniMACS™ separation column and magnet for cell sorting with bacterial display [47], although both methods may be user dependent. PA488 binding for Program D increased from nMFI of 1.23 (0.3% bound) in round 1 to nMFI of 11.07 (65.2% bound) in round 4, and for Program DS increased from nMFI of 1.25 (1.6% bound) in round 1 to nMFI of 9.66 (61.6% bound) in round 4. Differences in the rate of increase between the earlier rounds were clearly evident upon comparison of the two automated methods (Figure 1B and 1C). Program D had a substantial increase in binding between rounds 1 and 2, and quickly leveled off (nMFI of 11.17 at round 2 and 13.75 at round 3), while Program DS exhibited a slower rate of enrichment across the four rounds of biopanning (nMFI of 1.45 at round 2 and 7.31 at round 3). Program D therefore had the highest nMFI at round 3, while Program DS increased with every subsequent round and never reached the higher nMFI of 13.75 seen for Program D. It is possible that the population of cells sorted by Program DS could have reached a higher nMFI with a fifth round of sorting, further excluding low- and non-binders. Downstream analysis shows that this is not necessary to obtain candidates with affinity and specificity for PA (Figure 2). At rounds 2 and 3, the enrichment in the percentage of cells capable of binding PA using Program D was more comparable to the MMS and MACS/FACS sorting methods previously described (Figure 1B) [19].
Figure 1: FACS analysis of sorting rounds for discovery of peptide candidates to protective antigen (PA). FITC-H vs. FSC-H of cells bound to 250 nM PA488 for rounds 1-4 (R1-R4) of (A) manual MACS sorting, (B) autoMACS® Pro Separator Program D sorting, or (C) autoMACS® Pro Separator Program DS sorting of a bacterial peptide display library. The percentage of cells falling outside of the P2 gate is shown in red and in the top left corner of each box. The Normalized Median Fluorescence Intensity (nMFI) of the entire population is also shown.
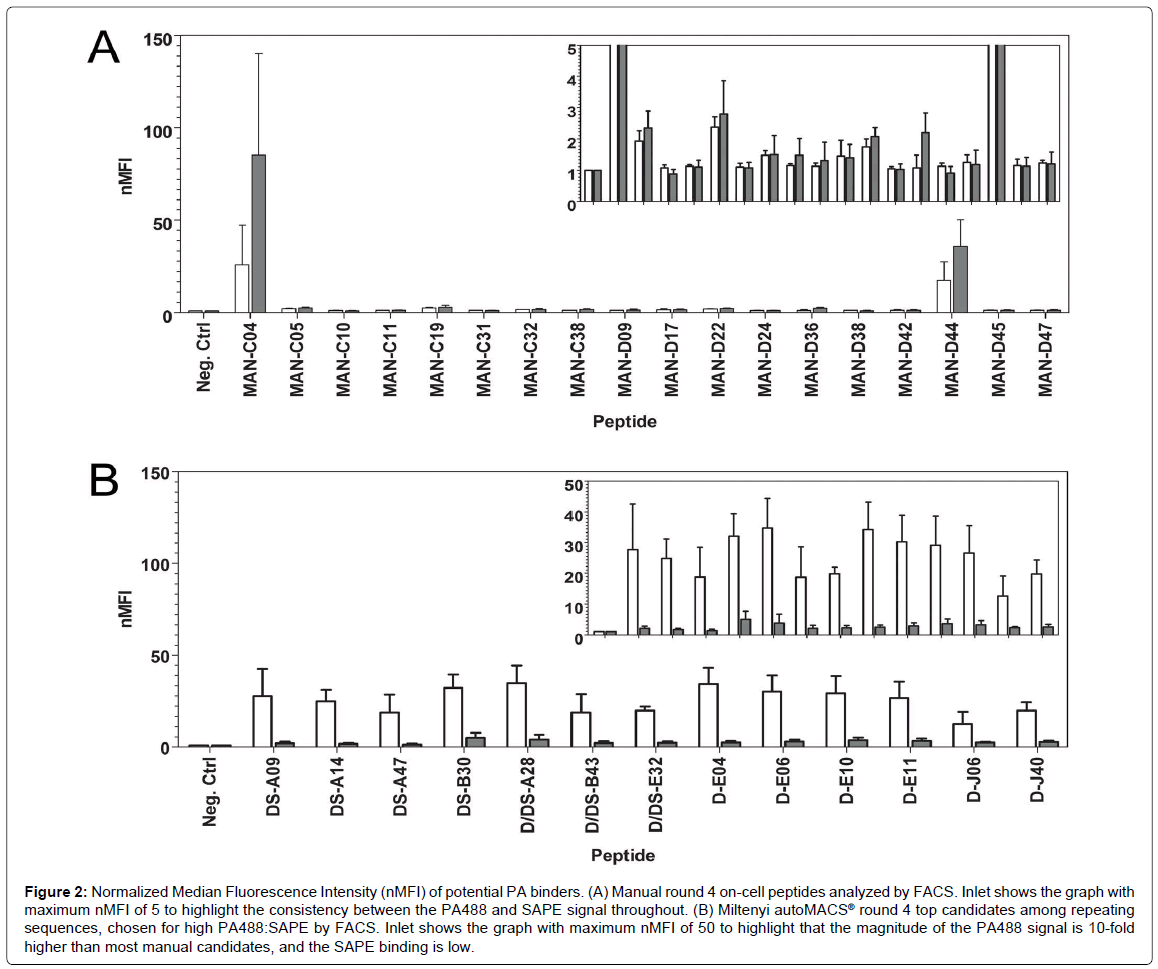
Figure 2: Normalized Median Fluorescence Intensity (nMFI) of potential PA binders. (A) Manual round 4 on-cell peptides analyzed by FACS. Inlet shows the graph with maximum nMFI of 5 to highlight the consistency between the PA488 and SAPE signal throughout. (B) Miltenyi autoMACS® round 4 top candidates among repeating sequences, chosen for high PA488:SAPE by FACS. Inlet shows the graph with maximum nMFI of 50 to highlight that the magnitude of the PA488 signal is 10-fold higher than most manual candidates, and the SAPE binding is low.
Bioinformatic analysis of isolated PA binders and further characterization by FACS
In order to investigate individual candidate binders, single bacterial colonies were plated for sequencing. A bioinformatics analysis on the resulting peptide sequences was performed, and relative binding of isolated binders was accomplished via FACS analysis. Since the early rounds of manual MACS showed little promise by FACS analysis (Figure 1A), only candidates from round 4 were sequenced. For each of the autoMACS® programs, however, rounds 2 through 4 were sequenced. Any sequences that repeated at least once in round 4 were screened by FACS using 250 nM PA488 and 250 nM SAPE. Screening against SAPE was essential to assess specificity for the target since streptavidin is used in the process of isolating peptides from the bacterial display library through the use of streptavidin-conjugated paramagnetic beads. From this initial screen, the top PA binders from each autoMACS® method, thirteen in total, were selected for their high target (PA488) to background (SAPE ratio). The sequence, frequency, and PA488 nMFI to SAPE nMFI ratio of these top candidate PA binders are shown in Table 1. The prefixes “D” or “DS” in the peptide names signify that they were discovered using Program D or Program DS, respectively. Three of these sequences, D/DS-A28, D/DS-E32, and D/DS-B43, were present in the sequenced colonies from both autoMACS® methods. None of these thirteen sequences were present when sequencing the round 4 pool of candidates isolated using manual MACS.
| Peptide Sequence | Peptide Name | Program D | Program DS | PA: SAPE | ||||
|---|---|---|---|---|---|---|---|---|
| Round 2 | Round 3 | Round 4 | Round 2 | Round 3 | Round 4 | |||
| WFCFTCPSSSDVIKG | DS-A09 | 0 | 0 | 0 | 0 | 1 | 2 | 13.2 |
| YTDFVCFTCTMPQLQ | DS-A14 | 0 | 0 | 0 | 0 | 1 | 2 | 14.8 |
| WSCFTCDHGAETLVS | DS-A47 | 0 | 0 | 0 | 0 | 1 | 2 | 13.9 |
| TWFCFTCYKAPVKHD | DS-B30 | 0 | 0 | 0 | 0 | 9 | 10 | 6.5 |
| SYWSCFTCTTLSGFS | D/DS-A28 | 0 | 4 | 13 | 0 | 3 | 4 | 8.9 |
| FTNWSCFTCSSSTNA | D/DS-B43 | 0 | 3 | 1 | 0 | 1 | 3 | 8.9 |
| SNWICFTCAFPRETA | D/DS-E32 | 0 | 9 | 5 | 0 | 2 | 0 | 9.0 |
| PGISEVQWSCFTCIV | D-E04 | 0 | 12 | 12 | 0 | 0 | 0 | 13.8 |
| VVWIPWTVWTVAPET | D-E06 | 0 | 2 | 2 | 0 | 0 | 0 | 10.5 |
| STLFYCFTCLSSVGS | D-E10 | 2 | 13 | 23 | 0 | 0 | 0 | 7.9 |
| SSWLCFTCLQAPAIS | D-E11 | 3 | 6 | 5 | 0 | 0 | 0 | 8.1 |
| YWHCWTCNSVNTDSR | D-J06 | 0 | 5 | 2 | 0 | 0 | 0 | 5.4 |
| PFSYLGTLYIPWESF | D-J40 | 0 | 1 | 2 | 0 | 0 | 0 | 7.6 |
| E. coli cells displaying peptides on an eCPX scaffold were sorted using the autoMACS® separation programs shown and 144 colonies were sequenced for sorting rounds 2 through 4. The number of sequenced colonies expressing each top PA binding peptide candidate is shown for each round. Consensus sequence WXCFTC is shown in bold. PA:SAPE is the ratio of the average nMFI for each, as determined by FACS from 3 independent experiments. | ||||||||
Table 1: Frequency of top PA binding candidates sorted by autoMACS®.
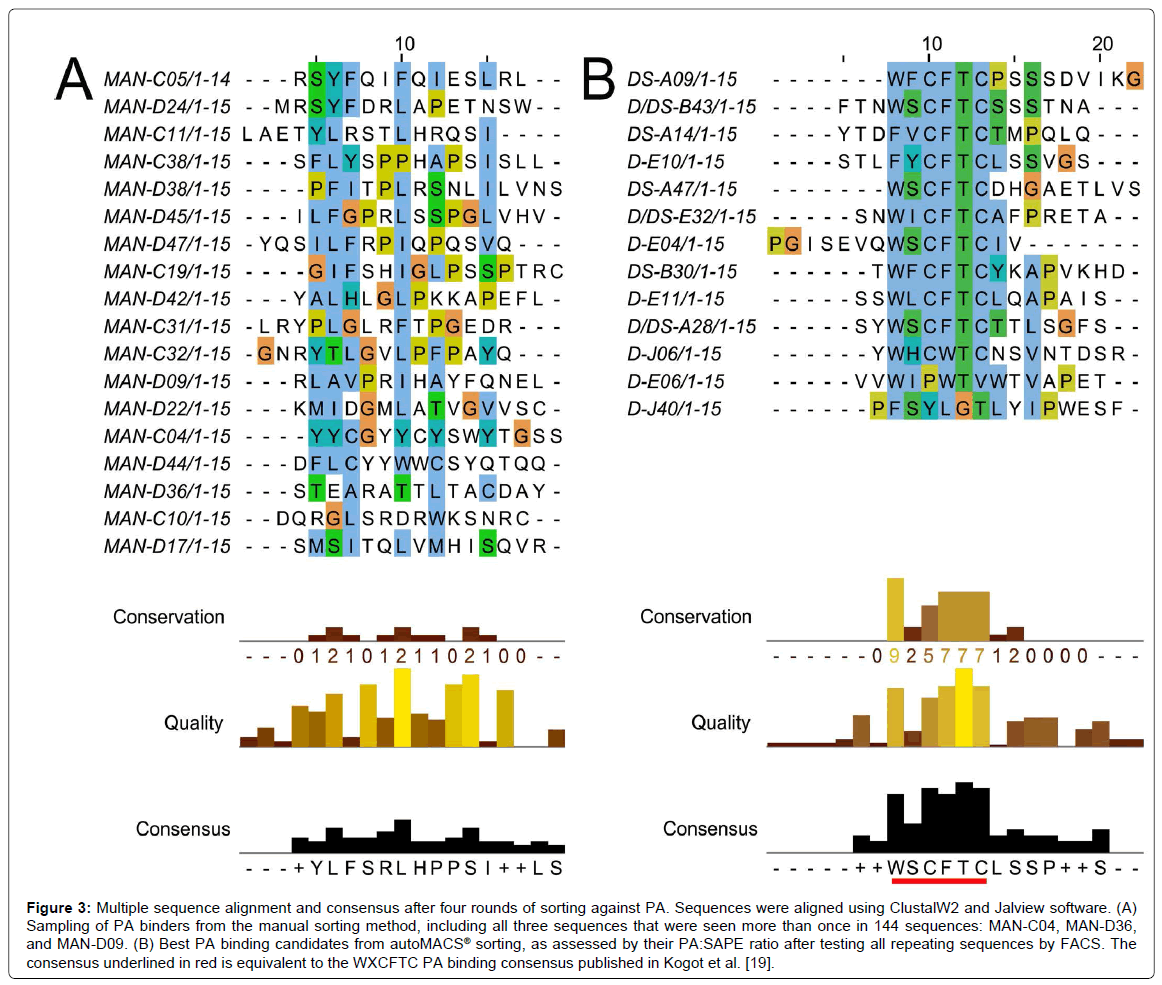
In general, all of the top candidates listed in Table 1 were represented in round 3 of autoMACS® sorting, although their presence continued to increase in round 4. Therefore, three rounds of sorting are sufficient to obtain PA binders with potential for future applications, using either Program D or Program DS. This is consistent with the result obtained using the MMS [19], and means that peptide candidates are available for study after only three days. The PA488 signal quickly saturated in Program D but not Program DS (Figure 1), so it is not surprising that some of these sequences were already evident in round 2 for Program D, but not Program DS (Table 1). The sequences of the top candidates from Table 1 were aligned using the online proteomics tools ClustalW2 and Jalview, and these results are shown in Figure 3. Note the high level of conservation leading to the consensus WXCFTC, the same consensus previously shown for binding to PA488 using the MMS platform [19], further demonstrating the utility of this approach. Phenylalanine and tryptophan are found interchangeably in both the first and fourth positions of this consensus, although tryptophan is usually in the first and phenylalanine in the fourth position. All sequences obtained from each manual or autoMACS® round tested were also aligned using ClustalW2 and Jalview, and the same WXCFTC consensus was seen in the total sequenced population (144 sequences for each) after 3 or 4 rounds of sorting using either autoMACS® program, but not for either autoMACS® program at round 2 or for the manual sort at round 4 (data not shown), further indicating that 3 rounds of sorting was sufficient with either autoMACS® program. Two of the top PA binders, D-E06 and D-J40, did not contain the entire WXCFTC consensus, but did contain tryptophan and phenylalanine residues present in the consensus. The most notably different of these was D-J40, which had a high nMFI for PA488 at 19.8 and low nMFI for SAPE at 2.6, with very little similarity to the consensus aside from the tryptophan and phenylalanine residues (Table 1, Figures 2 and 3B). For comparison, the DS-A14 peptide showed the highest signal (24.9 nMFI) to SAPE background (1.7 nMFI) with a ratio of 14.8. The peptide with the highest PA488 overall, D/DS-A28, had nMFI for PA488 of 34.7 and nMFI for SAPE of 3.9 with a similar signal to SAPE background of 8.9. Peptide D-J40 demonstrated similar PA and streptavidin binding, even without the entire consensus. The same was true of D-E06, with PA488 nMFI of 30.2 and SAPE nMFI of 2.9. D-E06 was lacking the cysteine residues of the WXCFTC consensus but otherwise followed the same pattern, although it contained a second tryptophan instead of phenylalanine. These aromatic residues, spaced two residues apart, appear to be the most important part of the WXCFTC consensus.
Figure 3: Multiple sequence alignment and consensus after four rounds of sorting against PA. Sequences were aligned using ClustalW2 and Jalview software. (A) Sampling of PA binders from the manual sorting method, including all three sequences that were seen more than once in 144 sequences: MAN-C04, MAN-D36, and MAN-D09. (B) Best PA binding candidates from autoMACS® sorting, as assessed by their PA:SAPE ratio after testing all repeating sequences by FACS. The consensus underlined in red is equivalent to the WXCFTC PA binding consensus published in Kogot et al. [19].
To further compare manual MACS to autoMACS®, the three repeat sequences from the manual round 4 sort, MAN-C04, MAN-D36, and MAN-D09, and fifteen additional, non-repeating sequences, were tested by FACS for binding to 250 nM PA488 and 250 nM SAPE. Most of the fifteen additional sequences were chosen at random, other than avoiding stop codons, due to a lack of repeating candidates and the WXCFTC consensus. However, several of these sequences were also selected for a noted trend of arginine richness, and MAN-D44 was specifically selected for its double tryptophan residues, a trend also present in two αPA peptides previously reported [48,49]. All manual sequences tested by FACS are listed in the peptide alignment in Figure 3A. Note that there is poor consensus when using the manual method, unlike the autoMACS® sequences that exhibited the strong consensus WXCFTC. Many of the individual sequences, and the manual “consensus” (or lack thereof) determined by Jalview, have a high frequency of leucine and serine residues, most likely due to the higher number of codons available for translating these amino acids rather than interactions with PA. The most abundant sequence in the manual sort, MAN-C04, repeated 12 times and was tyrosine-rich, with 6 tyrosine residues out of 15 total amino acids. Tyrosine residues are aromatic like tryptophan and phenylalanine, so it seemed a promising candidate. This sequence was the best PA-binding candidate from the manual sort but was non-specific, with greater than 3 fold higher binding to SAPE (nMFI 85.2) than to PA488 (nMFI 25.7) (Figure 2A). MAN-D36, which repeated 3 times in the 144 sequenced colonies, was actually a previously isolated sequence from biopanning bulk aluminum: the best performing aluminum binding peptide in that study, “DBAD1” [16]. This was likely present in the manual PA sort due to the use of iron oxide-containing MyOne Streptavidin T1 beads, since DBAD1 could potentially bind to other metal oxides in addition to aluminum oxide. MAN-D36/DBAD1 was a poor PA binder, with nMFI of 1.1. The third repeating sequence from manual MACS, MAN-D09, repeated only twice and was also a poor PA binder, with nMFI of 1.1 as well. Other than MAN-C04, the only candidate examined from the manual sort with nMFI greater than 3, including the upper bound of standard deviation, was MAN-D44. This peptide, which contained two adjacent tryptophan residues, had nMFI of 17.5 for PA488, but a 2-fold higher nMFI of 35.8 for SAPE. This is not surprising since, as noted in Sarkes et al., the double tryptophan motif seems to have lower specificity and affinity than the WXCFTC consensus when the “X” is also an aromatic residue [50]. The PA488 nMFI to SAPE nMFI ratio for all 18 manual MACS isolates tested was 1.3 or lower, so specificity was problematic. Not one of the sequences obtained from the manual round 4 sort was found to contain the full WXCFTC consensus (Figure 2A and data not shown), or to demonstrate both affinity and specificity for PA (Figure 2A). One sequence, MAN-C05, did contain two phenylalanine residues with the proper separation of two residues, but the nMFI for PA488 was only 2.0. It was therefore concluded that no promising PA binding candidates were obtained from the manual MACS, even when employing knowledge from previous sorting to select potential binders. It may have been possible to find a good candidate with further sequencing of round 4 candidates, or further rounds of sorting, but the time required to fully characterize all candidates is impractical for routine isolation and study. The characterization bottleneck with manual MACS further necessitates a reliable and reproducible biopanning approach, such as the approach presented herein using the autoMACS® Pro Separator.
Comparison of autoMACS® to (non-commercial) micromagnetic separation (MMS) platform
To further validate the autoMACS® biopanning approach, the top candidates from round 4 were analyzed by flow cytometry in a manner that more closely resembles the analysis used in Kogot et al., using percentage of cells falling outside a gated population of cells incubated with PBS buffer alone, for candidates isolated using a noncommercial microfluidic system with the same target, PA [19]. This was the standard FACS analysis method used with candidates isolated by bacterial display for many years [17-23], but nMFI is preferred because the percent binding can saturate at 100%, and the nMFI can vary between populations with the exact same percent binding due to the extent of binding [41]. Using percent binding, the affinity and specificity of the autoMACS® candidates for PA binding herein are similar to those discovered by MMS sorting, but were discovered using a widely - available platform. Specifically, the autoMACS® yielded a tighter range of 65.1%-87.4% for PA binding (Supplemental Table 1 and Supplemental Figure 1) when compared to 44.1%-89.8% obtained using the MMS [19]. When comparing the average PA binding for the candidates tested, both approaches yielded similar results: 78.1% and 71.5% for autoMACS® and MMS, respectively. A tighter range of binding to the negative control is also evident upon comparison of the percent binding to streptavidin in this study versus the literature. Specifically, the streptavidin negative control binding range was 0.7%- 13.5% for autoMACS® (Supplemental Table 2 and Supplemental Figure 1), while the streptavidin binding range was 0.1%-39.5% for MMS [19]. While these studies were performed at similar (but not exactly the same) concentrations of target and negative control, it is clear that the autoMACS® biopanning method described here yielded more consistent and reliable results. Furthermore, it is important to note that the autoMACS® yields better results than the MACS/FACS standard protocol for bacterial display sorting but in a significantly lower cost and simpler platform. For example, the MACS/FACS sorting method, demonstrated for comparison to MMS in Kogot et al., had a range of 1.2%-62.0% and average of 20.4% for PA binding and range of 0.4%- 16.3% and average of 3.5% for streptavidin binding [19]. The range for both target and negative control were tighter using autoMACS®, and the average PA binding was almost 4x greater for autoMACS® than for MACS/FACS, at a fraction of the cost. Overall the commercial availability of the autoMACS® Pro Separator gives it an advantage over the MMS, and when combined with the analysis approach herein is extendable to other cell surface display applications.
Conclusion
In this work, we successfully demonstrated bacterial display sorting using the commercially-available autoMACS® Pro Separator for the first time. Several new PA peptide reagent candidates were discovered as a result, with the same consensus sequence, WXCFTC, as candidates discovered using a non-commercial platform previously tested by our group [19]. Both of these semi-automated platforms are preferred over manual MACS or MACS/FACS sorting due to cost and/or sorting capability, since a FACS capable of sorting cells for downstream use is more expensive, and both MACS/FACS and manual MACS yielded lower affinity binders. Only a single candidate discovered by MACS/FACS yielded the WXCFTC consensus for PA [19], while this consensus was completely absent in the manual MACS study herein. For autoMACS®, whether Program D or Program DS is preferable may depend on the target of interest, the round of sorting (due to sheer number of cells to sort), or how well the peptide is displayed on the cell surface. In this test case, both programs led to PA binders with the same consensus and similar specificity after only three rounds of sorting, although Program D demonstrated more promise after only two rounds of sorting and Program DS yielded more unique sequences containing the consensus. Pushing to a fourth round of sorting led to a high frequency of repeats and is therefore recommended to simplify bioinformatics analysis, and testing these repeat sequences by FACS led to inclusion of candidates that did not quite follow the consensus but that had similar affinity and specificity to candidates containing the consensus. This approach could therefore enable discovery of candidates that bind to different epitopes, and enable rapid discovery of binders in cases where there is no obvious consensus for a particular target, without extensive, brute force characterization of candidates. The specificity obtained without further maturation is notable for a small peptide. The affinity and specificity obtained with minimal effort and relatively low cost using this approach gives this method strong potential for discovery of robust peptide capture candidates and peptide therapeutic agents that can be further matured by incorporation into protein catalyzed capture agents, for instance [49]. We are confident in this instrument’s ability as a semi-automated tool for bacterial display sorting due to the strong enrichment and consensus, and future work will include continued use of this platform to screen for peptide capture candidates against existing and future bio-threat agents, and further automation of the biopanning protocol, which could become the new workhorse for bacterial display. This method should be extendable to use with other aerobic bacteria and the autoMACS® Pro Separator is small enough that it may be used inside of a large anaerobe chamber if needed for a specific application. Although the current study was for biopanning of rare cells expressing peptides which bind to a biothreat agent, the platform could be used to isolate bacteria from a variety of mixtures, such as whole blood. The key requirement for extending this protocol to other applications with bacteria is obtaining a binding partner for the target of interest that can be attached covalently or indirectly to a magnetic bead, and that specifically recognizes the cell surface of the bacteria directly, or through a signaling molecule or displayed peptide.
Acknowledgements
The authors would like to thank Dr. Bryn Adams and the Daugherty Lab (UCSB) for their help with cloning the YPet Mona reagent. This research was primarily supported by the U.S. Army Research Laboratory. The content does not necessarily reflect the position or the policy of the United States Government, and no official endorsement should be inferred.
References
- Bessette PH, Rice JJ, Daugherty PS (2004) Rapid isolation of high-affinity protein binding peptides using bacterial display. Protein Eng Des Sel 17: 731-739.
- Georgiou G, Stathopoulos C, Daugherty PS, Nayak AR, Iverson BL, et al. (1997) Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat Biotechnol 15: 29-34.
- Stratis-Cullum D, Kogot JM, Sarkes DA, Val-Addo I, Pellegrino PM (2011) Bacterial display peptides for use in biosensing applications. In: Pramatarova L (ed) On Biomimetics. InTech, Washington DC, USA.
- Uhlig T, Kyprianou T, Martinelli FG, Oppici CA, Heiligers D, et al. (2014) The emergence of peptides in the pharmaceutical business: From exploration to exploitation. EuPA Open Proteom 4: 58-69.
- Fosgerau K, Hoffmann T (2015) Peptide therapeutics: current status and future directions. Drug Discov Today 20: 122-128.
- Daugherty PS (2007) Protein engineering with bacterial display. Curr Opin Struct Biol 17: 474-480.
- Sezonov G, Joseleau-Petit D, D'Ari R (2007) Escherichia coli physiology in Luria-Bertani broth. J Bacteriol 189: 8746-8749.
- Boder ET, Wittrup KD (1997) Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol 15: 553-557.
- Jagadish MN, Carter BL (1978) Effects of temperature and nutritional conditions on the mitotic cell cycle of Saccharomyces cerevisiae. J Cell Sci 31: 71-78.
- Donatan S, Yazici H, Bermek H, Sarikaya M, Tamerler C, et al. (2009) Physical elution in phage display selection of inorganic-binding peptides. Mater Sci Eng C 29: 14-19.
- Gaskin DJ, Starck K, Turner NA, Vulfson EN (2001) Phage display combinatorial libraries of short peptides: ligand selection for protein purification. Enzyme Microb Technol 28: 766-772.
- Nixon AE (2002) Phage display as a tool for protease ligand discovery. Curr Pharm Biotechnol 3: 1-12.
- Goldman ER, Pazirandeh MP, Mauro JM, King KD, Frey JC, et al. (2000) Phage-displayed peptides as biosensor reagents. J Mol Recognit 13: 382-387.
- New England BioLabs Inc (2014) Ph.D. Phage Display Libraries Instruction Manual, Version 1.2.
- Puddu V, Perry CC (2012) Peptide adsorption on silica nanoparticles: evidence of hydrophobic interactions. ACS Nano 6: 6356-6363.
- Adams BL, Finch AS, Hurley MM, Sarkes DA, Stratis-Cullum DN (2013) Genetically Engineered Peptides for Inorganics: Study of an Unconstrained Bacterial Display Technology and Bulk Aluminum Alloy. Adv Mater 25: 4585-4591.
- Kenrick SA, Daugherty PS (2010) Bacterial display enables efficient and quantitative peptide affinity maturation. Protein Eng Des Sel 23: 9-17.
- Kogot JM, Pennington JM, Sarkes DA, Kingery DA, Pellegrino PM, et al. (2014) Screening and characterization of anti-SEB peptides using a bacterial display library and microfluidic magnetic sorting. J Mol Recognit 27: 739-745.
- Kogot JM, Zhang Y, Moore SJ, Pagano P, Stratis-Cullum DN, et al. (2011) Screening of peptide libraries against protective antigen of Bacillus anthracis in a disposable microfluidic cartridge. PLOS ONE 6: e26925.
- Little LE, Dane KY, Daugherty PS, Healy KE, Schaffer DV (2011) Exploiting bacterial peptide display technology to engineer biomaterials for neural stem cell culture. Biomaterials 32: 1484-1494.
- Rice JJ, Schohn A, Bessette PH, Boulware KT, Daugherty PS (2006) Bacterial display using circularly permuted outer membrane protein OmpX yields high affinity peptide ligands. Protein Sci 15: 825-836.
- Thomas JM, Daugherty PS (2009) Proligands with protease-regulated binding activity identified from cell-displayed prodomain libraries. Protein Sci 18: 2053-2059.
- Dane KY, Chan LA, Rice JJ, Daugherty PS (2006) Isolation of cell specific peptide ligands using fluorescent bacterial display libraries. J Immunol Methods 309: 120-129.
- Bessette PH, Hu X, Soh HT, Daugherty PS (2007) Microfluidic library screening for mapping antibody epitopes. Anal Chem 79: 2174-2178.
- Rice JJ, Daugherty PS (2008) Directed evolution of a biterminal bacterial display scaffold enhances the display of diverse peptides. Protein Eng Des Sel 21: 435-442.
- Kogot JM, Pennington JM, Sarkes DA, Stratis-Cullum DN, Pellegrino PM (2011) Population Enrichment and Isolation with Magnetic Sorting. US Army Research Laboratory, Adelphi, MD Sensors and Electron Devices Directorate.
- An Z (2011) Therapeutic monoclonal antibodies: from bench to clinic. John Wiley & Sons, New Jersey, USA.
- Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, et al. (2006) Isolating and engineering human antibodies using yeast surface display. Nat Protoc 1: 755-768.
- Miller KD, Pefaur NB, Baird CL (2008) Construction and screening of antigen targeted immune yeast surface display antibody libraries. Curr Protoc Cytom Chapter 4: Unit4.
- Schuijt TJ, Narasimhan S, Daffre S, DePonte K, Hovius JW, et al. (2011) Identification and characterization of Ixodes scapularis antigens that elicit tick immunity using yeast surface display. PLoS One 6: e15926.
- Puri V, Streaker E, Prabakaran P, Zhu Z, Dimitrov DS (2013) Highly efficient selection of epitope specific antibody through competitive yeast display library sorting. MAbs 5: 533-539.
- Monaci P, Luzzago A, Santini C, De Pra A, Arcuri M, et al. (2008) Differential screening of phage-ab libraries by oligonucleotide microarray technology. PLoS One 3: e1508.
- Ahmad MR, Nakajima M, Kojima S, Homma M, Fukuda T (2008) The effects of cell sizes, environmental conditions, and growth phases on the strength of individual W303 yeast cells inside ESEM. IEEE Trans Nanobioscience 7: 185-193.
- Kubitschek HE, Friske JA (1986) Determination of bacterial cell volume with the Coulter Counter. J Bacteriol 168: 1466-1467.
- Nelson DE, Young KD (2000) Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J Bacteriol 182: 1714-1721.
- Tyson CB, Lord PG, Wheals AE (1979) Dependency of size of Saccharomyces cerevisiae cells on growth rate. J Bacteriol 138: 92-98.
- Grossman N, Ron EZ, Woldringh CL (1982) Changes in cell dimensions during amino acid starvation of Escherichia coli. J Bacteriol 152: 35-41.
- Miltenyi Biotec GmbH (2007) autoMACS™ Pro Separator User Manual, Version 1.1.
- Miles AA, Misra SS, Irwin JO (1938) The estimation of the bactericidal power of the blood. J Hyg (Lond) 38: 732-749.
- Getz JA, Schoep TD, Daugherty PS (2012) Peptide discovery using bacterial display and flow cytometry. Methods Enzymol 503: 75-97.
- Chan LY, Yim EK, Choo AB (2012) Normalized Median Fluorescence: An Alternative Flow Cytometry Analysis Method for Tracking Human Embryonic Stem Cell States During Differentiation. Tissue Eng Part C Methods 19: 156-165.
- Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20: 426-427.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876-4882.
- Multiple Sequence Alignment.
- Henriques ST, Thorstholm L, Huang YH, Getz JA, Daugherty PS, et al. (2013) A novel quantitative kinase assay using bacterial surface display and flow cytometry. PLoS One 8: e80474.
- Stratis-Cullum DN, Finch AS (2013) Current Trends in Ubiquitous Biosensing. J Anal Bioanal Tech S7-009.
- Christmann A, Walter K, Wentzel A, Kratzner R, Kolmar H (1999) The cystine knot of a squash-type protease inhibitor as a structural scaffold for Escherichia coli cell surface display of conformationally constrained peptides. Protein Eng 12: 797-806.
- Kogot JM, Sarkes DA, Pennington JM, Pellegrino PM, Stratis-Cullum D (2014) Binding specificity and affinity analysis of an anti-protective antigen peptide reagent using capillary electrophoresis. Adv Biosci Biotechnol 5: 40-45.
- Farrow B, Hong SA, Romero EC, Lai B, Coppock MB, et al. (2013) A chemically synthesized capture agent enables the selective, sensitive, and robust electrochemical detection of anthrax protective antigen. ACS Nano 7: 9452-9460.
- Sarkes DA, Dorsey BL, Stratis-Cullum DN (2015) Analysis of protective antigen peptide binding motifs using bacterial display technology. Proceedings of the International Society for Optics and Photonics, SPIE Defense+ Security.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 15677
- [From(publication date):
August-2015 - Sep 04, 2025] - Breakdown by view type
- HTML page views : 10997
- PDF downloads : 4680