Editorial Open Access
NMR Spectroscopy Provides a Novel Bioanalytical and Biophysical Approach towards the Characterization of Protein Interactions Involved in the Integration of RAS Signaling
Reena Chandrashekar and Paul D Adams*
Department of Chemistry and Biochemistry, The University of Arkansas, Fayetteville, AR, USA
- *Corresponding Author:
- Paul D. Adams
Department of Chemistry and Biochemistry
The University of Arkansas
Fayetteville, AR, USA
Tel: 479-575-5621
E-mail: pxa001@uark.edu
Received date: August 19, 2015; Accepted date: August 23, 2015; Published date: August 31, 2015
Citation: Chandrashekar R, Adams PD (2015) NMR Spectroscopy Provides a Novel Bioanalytical and Biophysical Approach towards the Characterization of Protein Interactions Involved in the Integration of RAS Signaling. J Anal Bioanal Tech 6:e122 doi:10.4172/2155-9872.1000e122
Copyright: © 2015 Chandrashekar R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
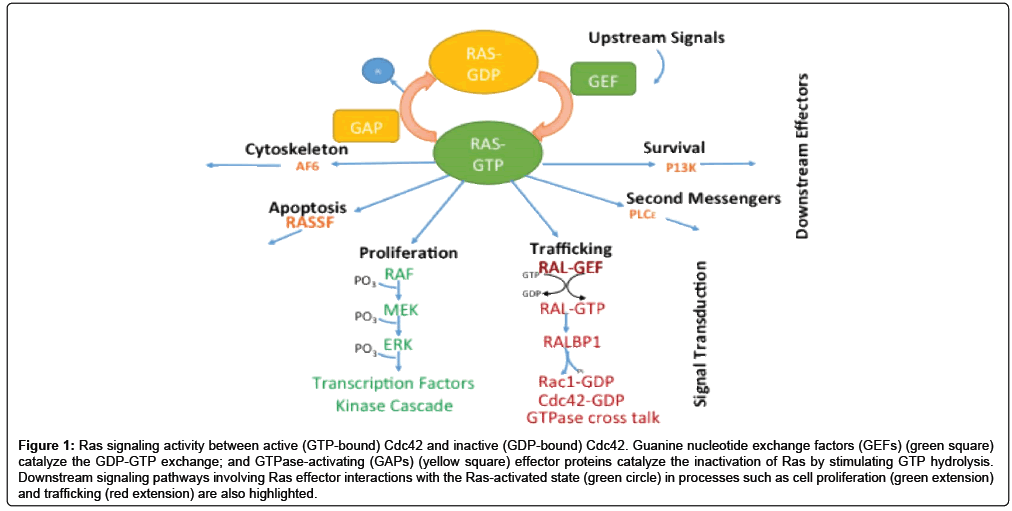
Structure-function relationships among proteins are at the center of the chemical and molecular basis of many biological processes. There remains, nevertheless, a gap in our understanding of Ras-related protein interactions which are pivotal in regulating many cell-signaling processes. The classical model that depicts how Ras-related proteins are involved in cell-signaling is dictated by the nature of the nucleotidebound state (GDP-inactive or GTP-active), which in turn is vital to processes such as cell proliferation, second messenger signaling, programmed cell death, cytoskeletal development, trafficking and differentiation (Figure 1) [1-6].
Figure 1: Ras signaling activity between active (GTP-bound) Cdc42 and inactive (GDP-bound) Cdc42. Guanine nucleotide exchange factors (GEFs) (green square) catalyze the GDP-GTP exchange; and GTPase-activating (GAPs) (yellow square) effector proteins catalyze the inactivation of Ras by stimulating GTP hydrolysis. Downstream signaling pathways involving Ras effector interactions with the Ras-activated state (green circle) in processes such as cell proliferation (green extension) and trafficking (red extension) are also highlighted.
If not properly regulated, these signaling processes involving Rasrelated proteins can result in diseases such as cancer as well as other developmental pathologies [6]. Therefore, these proteins are important models to probe structure-function relationships that mediate cell signaling and cell transformation. These cell-signaling processes are defined by how a message (or messages) may move in response to effector/regulatory protein interactions with the Ras protein. The relayed signals follow a well-regulated network of pathways. Although there has been a burgeoning rise in the analyses of the existing web of signal transduction mechanisms, a defined experimental approach to measure these signals quantitatively, still lags. Hence, strategies that can probe the competition between effectors and regulatory proteins are required to explain and quantify signals from both wild-type and oncogenic Ras proteins. Distinguishing these interactions could provide a unique approach to targeting abnormal Ras-stimulated activity with greater specificity.
Recently, Smith and Ikura used parallel Nuclear Magnetic Resonance (NMR) approaches to characterize the competitive nature of Ras binding between effector and regulatory proteins [7]. The premise for these studies lies in the fact that there has been valuable information gained from an examination of individual Ras-effector/ regulator complexes, but, in the presence of multiple effectors, details of Ras selection, as well as the order or “hierarchy” of selection is less well-known. The approach consisted of an analysis of the sequence alignment and secondary structure predictions of over fifty Ras Binding Domains (RBDs) that was used to generate a base set of ten RBDs from various Ras protein families. Two combinations of effectors, in competition with each other for Ras, either BRAF-ARAF or ARAF-RGL1, were incubated with Ras bound to a non-hydrolyzable nucleotide analog GMPPNP, to maintain active Ras status. The NMR peak intensities at characteristic chemical shifts for each effector bound to wild-type Ras individually were compared. For the BRAF-ARAF competition assay, only the BRAF-Ras complex was detected from chemical shift characteristics, suggesting that BRAF was dominant over ARAF in binding to wild-type Ras [7].
However, for the ARAF-RGL1 competition assay for Ras binding, calculation of chemical shift intensities gave a ratio of 62% for ARAF as compared to 38% for RGL1 suggesting that, while both effector domains bound to wild-type Ras, ARAF had a stronger interaction with Ras than that of RGL1 to Ras [7]. By using this NMR-based approach for all possible combinations of effector domains produced, the authors generated a “hierarchy” of Ras-effector interactions, which showed the potential of effector selectivity by this protein.
Another interesting aspect to this work, highlighting the use of multi-dimensional NMR as a novel bioanalytical tool emerged from data analysis using a Ras variant with a “disease-causing” mutation. A binding competition assay revealed that the mutant Ras (G12V) showed a stark contrast in its ability to interact with the effectors ARAF and RGL1 as compared to wild-type Ras, as the relative binding affinities were reversed for Ras (G12V) [7] (and data therein). The results suggest that the hierarchy of Ras-signaling could be disturbed via mutation(s) that may cause conformational as well as dynamics changes in regions critical for Ras-effector binding, as has been described by this laboratory as well as others [8-11]. To rule out the possibility of concentration-dependent changes causing the inversion of effector affinities, the authors used Isothermal Titration Calorimetry (ITC) to characterize the binding interactions for the named effectors with mutant Ras and compared to wild-type. The results were consistent with the NMR data in that ARAF showed a lower Kd for wild-type Ras as compared to its Kd for Ras (G12), and RGL1 had a lower Kd for Ras (G12V) compared to its Kd for wild-type Ras. These data confirmed the hypotheses of the authors, as an oncogenic mutation indeed did alter the binding properties of key effectors of Ras and triggered the perturbation in the signaling network. The outcomes of the use of NMR as a bioanalytical tool in this approach also point to the importance in gaining a more complete understanding of the molecular details of the binding interface(s) presented to these effectors by mutant Ras in comparison to wild-type Ras.
To further quantify their results, the authors co-expressed wildtype or Ras (G12V) with the catalytic domain SOS1 to develop an in vivo system to characterize differences in effector productivity from activated wild-type or mutant Ras. After each was expressed in parallel, RALA-GTP and pERK levels were measured in a time-dependent assay. Results revealed that Ras G12V activates higher RALA-GTP levels than the wild-type RAS [7] (and data therein). However, wildtype Ras activated more pERK levels than Ras (G12V). These results strengthen the findings from the NMR competition binding assays. The combined results show that different levels of peak intensity for certain effectors correlate with a hierarchy of signaling networks which can be correlated to in vivo processes as well.
In the work cited above, novel information, acquired from traditional bioanalytical and biophysical approaches, was used to address the contributions of regulatory/effector proteins (GAPs, GEFs, etc.) in the integration of Ras signaling pathways. Indeed, a “hierarchy” of Ras-effector interactions was identified, as well as a characterization of how these networks can influence activity in vivo. Such understanding of the integration of the Ras signaling pathway can be expected to foster specific targeting of interactions that lead to aberrant cell-signaling activity, for example, by small molecule inhibitors.
Acknowledgements
We thank Dr. Roger Koeppe II for providing critical feedback and suggestions on the content and scope of this manuscript.
References
- Bos JL (1989) Ras oncogenes in human cancer: a review. Cancer Res 49: 4682-4689.
- Bos JL, Toksoz D, Marshall CJ, Verlaan-de Vries M, Veeneman GH, et al. (1985) Amino-acid substitutions at codon 13 of the N-ras oncogene in human acute myeloid leukaemia. Nature 315: 726-730.
- Barbacid M (1987) Ras genes. Annu Rev Biochem 56: 779-827.
- Bourne HR, Sanders DA, McCormick F (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349: 117-127.
- White MA, Nicolette C, Minden A, Polverino A, Van Aelst L, et al. (1995) Multiple Ras functions can contribute to mammalian cell transformation. Cell 80: 533-541.
- Smith MJ, Neel BG, Ikura M (2013) NMR-based functional profiling of RASopathies and oncogenic RAS mutations. Proc Natl Acad Sci USA 110: 4574-4579.
- Smith MJ, Ikura M (2014) Integrated RAS signaling defined by parallel NMR detection of effectors and regulators. Nat Chem Biol 10: 223-230.
- Phillips MJ, Calero G, Chan B, Ramachandran S, Cerione RA (2008) Effector proteins exert an important influence on the signaling-active state of the small GTPase Cdc42. J Biol Chem 283: 14153-14164.
- Spoerner M, Herrmann C, Vetter IR, Kalbitzer HR, Wittinghofer A (2001) Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc Natl Acad Sci USA 98: 4944-4949.
- Spoerner M, Hozsa C, Poetzl JA, Reiss K, Ganser P, et al. (2010) Conformational states of human rat sarcoma (Ras) protein complexed with its natural ligand GTP and their role for effector interaction and GTP hydrolysis. J Biol Chem 285: 39768-39778.
- Chandrashekar R, Salem O, Krizova H, McFeeters R, Adams PD (2011) A switch I mutant of Cdc42 exhibits less conformational freedom. Biochemistry 50: 6196-6207.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 15968
- [From(publication date):
October-2015 - Aug 19, 2025] - Breakdown by view type
- HTML page views : 11253
- PDF downloads : 4715