Structure Based Discovery of Pan Active Botulinum Neurotoxin Inhibitors
Received: 05-Jan-2018 / Accepted Date: 10-Feb-2018 / Published Date: 14-Feb-2018 DOI: 10.4172/2332-0877.1000351
Abstract
Clostridium botulinum neurotoxins (BoNTs) released by the bacterium Clostridium botulinum are the most potent toxins causing the fatal disease called botulism. There are seven distinct serotypes of BoNTs (A to G) released by various strains of botulinum. They all have high sequence homology and similar three-dimensional structure. The toxicity of BoNT follows a four-step process–binding, internalization, translocation, and cleavage of its target protein, one of the three components of the SNARE complex (Soluble N-ethylmaleimde-sensitive factor attachment protein receptor) required for membrane docking and neurotransmitter release. Cleavage of one of the three proteins causes blockage of neurotransmitter release leading to flaccid paralysis. Though anyone of the above four steps could be a target for developing antidotes for botulism, the catalytic domain is the most suitable target for post exposure treatment. Of the seven serotypes BoNT/A, B, E and probably F affect humans, with BoNT/A considered to be the most potent. Development of drugs for botulism is focused on serotype specific inhibitors, but pan-active inhibitor acting on several serotypes is preferable since it is difficult to identify the serotype before the treatment, especially since there is at least a 36 h window before botulism can be diagnosed. Using structure-based drug discovery, we have developed three heptapeptides based on the SNARE proteins which inhibit BoNT/A, B and E equally well. Probable reasons for pan-activity of these peptides are discussed.
Keywords: Clostridium botulinum neurotoxins; Structure-based drug discovery; Pan-active; Peptide inhibitors; three-dimensional structure; Foot-print signature
Introduction
Clostridium botulinum toxin is the most potent neurotoxin known [1]. It predominantly affects the neuromuscular junction and autonomic synapses, and clinically it causes profound descending weakness leading to hypotonia, and ultimately to respiratory failure, if unattended. It is a Category A Biothreat agent and a large-scale terrorist attack would be associated with a sizeable immediate mortality, given that the appropriate acute care of these patients usually requires the use of relatively limited resources, e.g. respirators. Even a small-scale attack would quickly overwhelm a local medical system’s available respirators thereby causing an acute inability to care for botulinum victims. Additionally, these toxins are now being used for therapeutic and cosmetic purposes and there is a potential for accidental overdose which might lead to undesirable effects [2-9]. Botulinum toxins produced with poor quality control poses another problem [10]. In view of these it is an urgent priority to develop drugs, especially for post intoxication treatment. Although effective experimental vaccines are available against botulinum neurotoxins, pharmacological treatment in the event of exposure is still a work in progress. Therapies based on antibodies are emerging but more than one antibody may be needed to neutralize even a single serotype which is not feasible in the context of seven serotypes and new ones being discovered [11]. Furthermore, antibodies are effective only on toxins which are still in the blood stream and will not work on toxins that have entered the neuronal cell already. Pharmacological treatments that could mitigate the toxicity of the toxin once it has been internalized would be important adjuncts to existing therapeutic procedures and will be invaluable in the event of an attack by serving as antidotes to those already exposed to the toxin to prevent progression and hasten recovery. Small molecule non-peptidic inhibitors (SMNPI) are being developed [12-18]. Synthetic molecule based high-throughput drug discovery is also in progress [19]. Here, the focus is on peptide inhibitors based on substrate peptides which will have advantage over other inhibitors for reasons discussed below. In addition we also show that the peptide inhibitors could be pan active since the architecture and electrostatic properties of the active site of all serotypes of botulinum neurotoxin are nearly identical.
There are seven serotypes of botulinum neurotoxins (BoNT) produced by different strains of Clostridium botulinum [20]. Botulinum neurotoxin produced as a single chain (MW 150 kDa) undergoes proteolysis before being released as a dichain, a heavy chain (HC) of 100 kDa and a light chain (LC) of 50 kDa, held together by an interchain disulfide bond. BoNT consists of three domains. Each domain has a function associated with it. The HCC (the C-terminal half of HC) domain is involved in binding to receptor molecules at the presynaptic cell membrane and is called the receptor binding domain. The HCN (the N terminal half of HC) domain is responsible for translocation of the catalytic domain by a pH dependent process and is called the translocation domain. LC, the catalytic domain, acts as a zinc endopeptidase and cleaves its substrate, one of the SNARE proteins, once it enters the cytosol. BoNTs are unique since their substrates are large polypeptides (10–25 kDa). Moreover, each serotype cleaves a specific SNARE protein at a unique peptide bond. BoNT/A, C and E cleave synaptosomal associated protein 25kDa (SNAP-25) at specific peptide bond, while BoNT/B, D, F and G cleave vesicle associated membrane protein (VAMP) also at specific peptide bonds. BoNT/C is unique in that it cleaves syntaxin also [20].
Cleavage of one of the three proteins prevents formation of the complex and inhibits neurotransmitter release leading to flaccid paralysis called botulism. While all seven serotypes (A-G) of botulinum neurotoxins are poisonous, BoNT/A, B, E and probably F affect humans, with BoNT/A considered to be the most potent, persisting longest within the cell. Development of drug for botulism is focused on serotype specific inhibitors since each serotype has a specific substrate which is cleaved at a unique peptide bond. In view of this it has been difficult to design a common inhibitor for all or at least several serotypes though it will be the most desirable counter measure since often times it is difficult to identify the serotype causing botulism and it is essential the treatment commences even before the serotype is identified.
As stated earlier, our focus here is on using catalytic domain as a target for drug discovery using substrate peptides. Available substrate peptide–enzyme complex structures allow us to use this information for Structure Based Drug Discovery (SBDD). The most effective inhibitors can be designed by understanding the interaction between the substrate and the enzyme via experimental structures. This forms the basis for structure based drug discovery. The substrate-based peptides bind tightly at the active site mimicking the interactions of real substrates at the active site. The binding mode can best be understood from the crystal structure to define the pharmacophore which can be used to design molecules that will bind similarly. Though peptide-based drugs are thought to have some disadvantages compared to small molecule drugs, recent developments in pharmaceutical fields have enabled large numbers of peptide-based drugs to enter into the drug market. They are highly potent and selective, and avoid off-target side effects. Unlike small molecules, they have a large surface area of contact. They do not accumulate in organs like the kidney producing toxic effect. Peptide-based inhibitors can be improved to overcome some of their disadvantages by cyclization, burying polar groups, blocking the amide groups, reducing the number of hydrogen bonds and increasing the rigidity of the molecule by intrachain hydrogen bonds. Additionally, substitution of natural peptides with non-standard peptides will help in resistance to proteolysis. We have earlier shown that peptide inhibitors survived for more than 40 h in rat and mouse cerebellar neurons and are resistant to intracellular protease action and are good drug candidates [21].
Materials and Methods
Peptide inhibitor design: Based on earlier peptide inhibitor studies several hexa and hepta peptides were designed. The amides of the inhibitor peptides were custom synthesized from Peptide Chemical Co. (USA). One peptide GRKKRRC with the second residue corresponding to the P1´ of the scissile peptide of SNAP-25 for BoNT/A was designed. Two more peptides GFKKRRC, and GIKKRRC with P1´corresponding to scissile peptide of VAMP for BoNT/B and SNAP25 peptide for BoNT/E, respectively were also constructed.
Protein expression and purification: The botulinum neurotoxin serotype A light chain (residues 1-424aa) was expressed in E. coli and purified to homogeneity using size exclusion chromatography, as described previously [22]. The purified enzyme in 20 mM HEPES, 2 mM DTT, 200 mM NaCl, pH 7.4 was stored at -20°C until used. BoNT/E light chain (1-421aa) was expressed and purified as described before [23]. BoNT/B light chain (1-430aa) was expressed using auto induction method and purified with Ni affinity column and size exclusion columns similar to BoNT/A-LC.
Crystallization and data collection: Four peptide inhibitors were chosen for crystallographic studies. BoNT/A light chain and peptide inhibitors were co-crystallized as described earlier [22,24]. Briefly, the protein–inhibitor complex was prepared with 1:25 (protein:peptide) molar ratio and crystals were grown by sitting drop vapor diffusion at room temperature. 3 µl of the protein solution (~15 mg/ml) was mixed with an equal volume of a reservoir solution containing 20% PEG 8000, 100 mM sodium cacodylate, pH 6.5, 5% ethylene glycol and 200 mM ammonium sulfate. Thick plate-like crystals were flash frozen with liquid nitrogen using 20% ethylene glycol as cryoprotectant. The X-ray intensity data for complex crystals were collected at X29 beamline of National Synchrotron Light Source (NSLS) using ADSC QUANTUM 315 detector or at 22ID, APS SER-CAT using MAR300HS detector. All data were processed using the HKL2000 suite [25]. Crystallographic data and refinement statistics are given in Table 1.
| Name/code | RRKCRLL | 14-mer | GRKKRRC | GFKKRRC | |
|---|---|---|---|---|---|
| Cell dimensions | |||||
| a (Å) | 49.11 | 49.71 | 49.71 | 51.16 | |
| b | 66.50 | 66.40 | 66.37 | 66.44 | |
| c | 64.76 | 64.76 | 64.76 | 65.03 | |
| Β (°) | 98.99 | 98.78 | 98.78 | 98.28 | |
| Space group | P21 | P21 | P21 | P21 | |
| Resolution range (Å) | |||||
| Overall | 50-1.5 | 50-1.65 | 50-1.74 | 50-1.60 | |
| Last shell | 1.59-1.5 | 1.65-1.6 | 1.78-1.74 | 1.64-1.60 | |
| #unique reflections | 63453 | 44766 | 40127 | 45143 | |
| Completeness (%) | |||||
| (Overall/Last shell) | 96.4/89.4 | 91.4/63 | 86.2/48 | 83.2/52 | |
| Rmerge1 overall/last shell | 4.7/15 | 3.9/12.7 | 5.6/22.4 | 7.5/51 | |
| <I/σ(I)> overall/last shell | 20.2/2.5 | 28.4/3 | 21.3/2 | 23.9/5.4 | |
| Refinement Statistics | |||||
| Resolution (Å) | 50-1.5 | 50-1.65 | 50-1.74 | 50.1.60 | |
| R factor2/Rfree (%) | 18.6/20.0 | 21.5/25.1 | 19.6/21.0 | 17.6/21.6 | |
| R.M.S deviations from ideality | |||||
| Bond lengths (Å) | 0.005 | 0.006 | 0.007 | 0.002 | |
| Bond angles (°) | 1.3 | 1.21 | 1.16 | 2.18 | |
| Overall B-factor (Å2) | 18.3 | 30.1 | 27.2 | 19.68 | |
| Number of atoms | |||||
| Proteins | 3492 | 3424 | 3418 | 3424 | |
| Waters | 411 | 0 | 127 | 285 | |
| Ions (Zn2+/SO42-) | 1/15 | 1/0 | 1/8 (2 EDO) | 1/0 | |
| Ligands | 65 | 54 | 44 | 24 | |
| Residues (%) in the core | |||||
| Region of Φ-Ψ plot | 90 | 86.8 | 88.7 | 90.7 | |
| 1Rmerge=∑j(Ih-<I>h∑)/ ∑Ih, where <Ih> is the average intensity over symmetry equivalents 2R-factor=∑|Fobs-Fcalc|/ ∑|Fobs. |
|||||
Table 1: Crystal data and refinement statistics of BoNT/A-LC with peptides.
Structure determination: Structures of the complexes were determined by Fourier Synthesis using the acetate bound BoNT/A light chain (Protein Data Bank id 3BWI) as model followed by rigid-body refinement and simulated annealing. The peptide inhibitors were modeled from the composite omit map and the difference Fourier maps, and were built with O or COOT and further refined with CNS or Refmac until convergence [26-28]. Models were validated with the Ramachandran plot using PROCHECK [29]. The final refinement statistics are shown in Table 1.
Fluorescent based continuous assay: Inhibitor peptides were tested using a SNAP-25-VAMP hybrid peptide similar to the one reported by Hines et al. [30] that contains a Green Fluorescent Protein (GFP) reporter. Briefly, the assay utilizes a SNAP-25-VAMP hybrid substrate containing residues Ala128-Gly206 of SNAP-25 fused to Ser2-Lys94 of VAMP, flanked by a C-terminal AviTag and N-terminal GFP. After biotinylation of the AviTag, the substrate is fixed to Streptavidin coated plates. The hybrid substrate contains cleavage sites for BoNT/A, C and E and BoNT/B, D, F and G. Subsequent cleavage of substrate by addition of buffer containing the appropriate light chain liberates GFP into bulk solution allowing for the visualization of a normal enzymatic progress curve with a maximum signal far in excess of the background. Reactions were performed in 96-well, black, streptavidin coated plates (PierceTM) in a buffer solution containing 20 mM Hepes pH 7.4, 2 mM DTT, 20–50 nM of light chain, and the inhibitor peptide. Potential inhibitor peptides were allowed to incubate with light chain for 15 min and the solutions were subsequently added to the plate wells to initiate the reactions. All reactions were conducted at 37°C and were monitored using a Tecan Saphire2 microplate reader at excitation and emission wavelengths of 460 nm and 506 nm, respectively. IC50 values were obtained by fitting the initial velocity data to a sigmoid dose-response equation using Sigma plot software (SigmaPlot® 10.0) [31].
Molecular footprint comparison: A footprint signature technique implemented in the program DOCK was used to compare the interactions of the peptide inhibitor in the three BoNT-LCs [31,32]. This technique helps in providing an approximate interaction energy measurement allowing for a quantified interaction between the ligand and a given protein. In our case we used it to compare suitably positioned ligand in different but similar serotypes of BoNT-LCs. The crystal structure of GRKKRRC in complex with BoNT/A-LC was used to calculate the footprint signature and to compare the potential interactions with BoNT/B and BoNT/E-LCs. The individual electrostatic and Van der Waals (vdw) interaction energy of each ligand residue was calculated with every residue of the protein and summed up to arrive at the total energy contribution of each ligand residue. Distance measurements were calculated with standard Euclidean distance in the footprint scoring protocol. BoNT/B (PDB ID: 2ETF) and BoNT/E-LC (PDB ID: 1T3A) structures were least square fit in Chimera [33] with BoNT/A-LC to bring them to a common reference frame. The charges of the ligand were prepared in MOE and then the ligand was transferred and energy minimized [34]. The complex structures were used in footprint signature comparison using GRKKRRC in BoNT/A-LC as signature.
Results
Crystal structure of BoNT/A-LC with RRKCRLL
Previously we have studied crystal structures of tetra, hexa and hepta substrate peptides in complex with appropriate BoNT-LCs [22,24,35,36]. In our efforts to design substrate-based peptide inhibitors we continued to evaluate peptide inhibtors greater than four peptides in length. Since RRGC was a good inhibitor [22], we had tried changing the C terminal to other hydrophobic residues. As a further modification we designed a hepta peptide RRKCRLL. In addition to changing the third residue from G to K, we added RLL to the C terminal to somewhat mimic RATKML, the C–terminal residues of SNAP-25. However, the inhibitory effect of this peptide was worse than the tetra peptides and hexapeptides reported earlier [21]. The crystal structure of BoNT/A-LC with RRKCRLL was determined at a resolution of 1.50 Å and refined to R and free R values of 18.6 and 20.0%. The model includes 423aa, 1 zinc ion, 3 sulfate ion and 411 water molecules. Surprisingly, for the first time in our studies the C terminal Histag included in cloning was visible in the electron density. More than 90% of residues are within the allowed region of Ramachandran plot as per PROCHECK or PDBSUM [29].
Crystal structure of BoNT/A-LC with GRKKRRQRRRPPQC
A HIV-tat derived peptide, GRKKRRQRRRPPQC was reported earlier as a peptide inhibitor [21]. The Ki of this inhibitor was 97 nM and seemed to be better than RRGC (Ki=158 nM). This peptide was provided to us by Dr. Ahmed of USAMRIID. In order to understand the interaction of this peptide with the enzyme and to analyze the structure-activity relationship we undertook to determine the co-crystal structure with BoNT/A-LC. The X-ray diffraction data were collected to 1.65 Å resolution and the crystal structure was determind. The model refined with R and R free of 21.5 and 25.1%, respectively. The final refined model contains 423 protein residues, the six N-terminal inhibitor residues. However, the eight C-terminal residues of the inhibitor were not modeled since the electron density was very weak or absent. More than 87% of residues are within the most allowed region of the Ramachandran plot. The electron density in the residual map (Fo-Fc) was well defined for the first six N-terminal residues (P1–P5’) and could be modeled unambiguously. Most notably, the amino nitrogen and carbonyl oxygen of P1 residue (Gly196) chelate the zinc ion as in the previous BoNT/A-LC:peptide complexes [22]. The weak or absence of electron density for the C-terminal peptides was interpreted as due to the C-terminal being flappy with no real contacts with the enzyme. This prompted us to try smaller peptides like hexa and hepta peptides since we have shown earlier that hexa and hepta substrate petides bind but do not get cleaved [37]. However, they were very weak inhibitors. We tried several modifications of the truncated version of the 14-mer and finally GRKKRRC was selected for BoNT/A light chain and found to be a good inhibitor.
Crystal structure of BoNT/A-LC with GRKKRRC
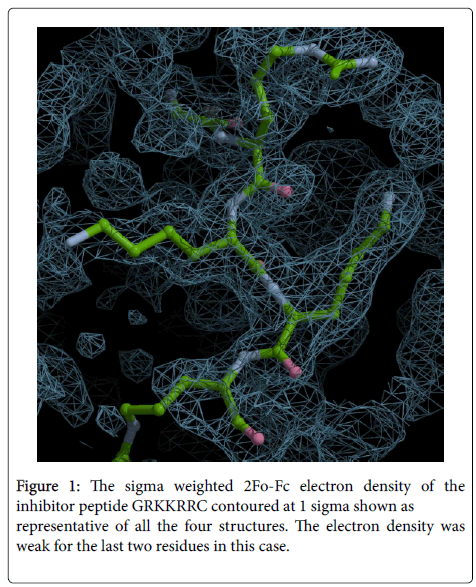
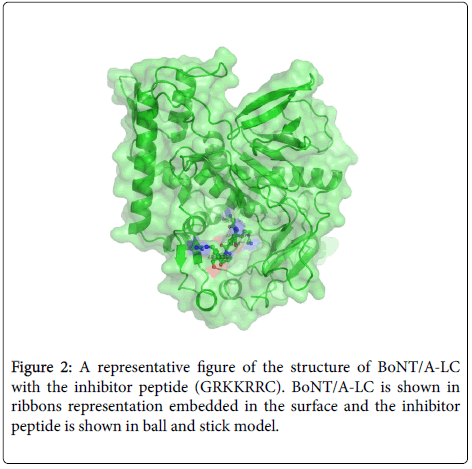
Since only the first six residues of GRKKRRQRRRPPQC were observed we designed a hepta (GRKKRRC) peptide as inhibitor to check if shortening the length affected the inhibitory effect. As shown later it had good IC50 value with BoNT/A-LC. The crystal structure of BoNT/A-LC with GRKKRRC was determined at a resolution of 1.74 Å and refined to R and free R values of 19.4 and 22.0%. The model includes 423aa, 1 zinc ion, 2 ethylene glycol (EDO), five N terminal residues of the inhibitor peptide and 127 water molecules. More than 87% of residues are within allowed region of Ramachandran plot as per PROCHECK. In this complex structure the last two C terminal residues of the inhibitor peptide had very weak electron density (Figure 1). However, other structural details were similar to the 14-mer complex. The mode of binding of GRKKRRC is presented in Figure 2 as a representation of all four structures.
Crystal structure of BoNT/A-LC with GFKKRRC
Since our inhibitor studies showed that GFKKRRC and GIKKRRC also inhibit BoNT/A-LC equally well, the crystal structure of BoNT/A-LC with GFKKRRC was also studied to analyze the mode of binding, with the focus particularly on the P1’ residue (being F instead of R). The structure was determined to a resolution of 1.60 Å and refined to R and free R of 17.6 and 21.4, respectively. The model includes 423aa, 1 zinc ion and at least three N terminal residues of the inhibitor peptide and 284 water molecules. More than 87% of residues are within allowed region of Ramachandran plot as per PROCHECK. In this complex structure the density was very weak for the residues P3’-P6’ and were not modeled.
Inhibitor assay
Before testing the inhibitor peptides with 96-well streptavidin plates, in-solution digest of SNAP-25-VAMP-GFP hybrid substrate with BoNT/A-F was performed to verify that proteolysis occurs at the expected location for each serotype. The SDS-PAGE analysis showed characteristic bands corresponding to appropriate cleavage sites for each serotype (data not shown). With the 96-well plate, each serotype was tested and the progress curve was plotted. The six serotypes produced a robust fluorescence signal as compared to a blank control with buffer only demonstrating that the progress curves are due to BoNT proteolysis. Further, two known serotype specific inhibitors of BoNT/E and F were cross checked. An inhibitor 2-(9H-fluorene-2-carbonyl) benzoic acid, designated NSC-77053 in the National Cancer Institute repository and proved to inhibit BoNT/E-LC was tested with BoNT/F-LC and showed no inhibition. Similarly a BoNT/F inhibitor, VAMP22-58(Q58/D-C) was tested with BoNT/E-LC and showed no inhibition [38,39].
The designed peptides (except RRKCRLL and the 14-mer) were tested against BoNT/A, BoNT/B and BoNT/E and BoNT/F light chains to test if the inhibitor peptides are cross reactive. RRKCRLL had been tested earlier with BoNT/A-LC and gave IC50 value of 28.65 µM. Since the IC50 was high it was not tried with other BoNTs. The fluorescent assay described earlier was used for assaying these peptides and the results are presented in Table 2. The IC50 were calculated using SigmaPlot and are presented in Figure 3.
| GRKKRRC | GFKKRRC | GIKKRRC | |
| BoNT/A-LC | 96 (30) nM | 93 (20) nM | 113 (20) nM |
| BoNT/B-LC | 418 (82) nM | 113 (80) nM | 265 (106) nM |
| BoNT/E-LC | 803 (130) nM | 964 (319) nM | 438 (80) nM |
Table 2: IC50 values from fluorescent assay. The values are average of three measurements for GIKKRRC and average of two measurements for GRKKRRC and GFKKRRC. Standard errors are given within parentheses.
Discussion
Structural studies
The mode of inhibitor binding is similar in all the four structures and the following discussion is restricted to GRKKRRC inhibitor complex. The carbonyl oxygen and the nitrogen atom of P1 residue (Gly196) coordinate with Zn atom as in earlier structures. His223, His227 and Glu262 provide other coordination bonds to Zn. The peptide is stabilized by the following interactions with the enzyme residue (the inhibitor residue is given first in italics followed by the enzyme residue): Gly196 N–Glu224 OE1, Gly196 O–Try366 OH, Lys198 O–Tyr366 OH, Lys199 NZ–Tyr366 O, Arg200 NH2– Glu257 OE1 and Lys198 NZ–Tyr366 OH. In addition, Arg197 forms a salt bridge with Asp370 (Figure 4). These strong interactions contribute to the high binding energy. These electrostatic interactions are also obvious in Footprint signature calculations. The highly basic peptide counters the electronegative residues providing a very strong binding (Figure 5). In the complex structure with GFKKRRC, Phe197 of the inhibitor peptide stacks with Phe194 of the enzyme.
Inhibition studies
Three hepta peptides which differ only in the second position (from the N-terminal) were tested on light chains of botulinum serotypes A, B and E, the three that affect humans most (Table 2). Use of small peptides as inhibitors for BoNT/A has been extensively studied. Using CRATKML (P1 to P5´) as the basis, residues at each position from P1´ to P5´ were altered and found that except P1 and P1´, modification of other residues did not change the inhibition effect significantly [40,41]. It is generally believed that the P1´ of the scissile peptide plays a key role in catalytic activity and accordingly on inhibition also. In our studies we found that substituting basic residues at positions P2´ to P5´increases the inhibitory effect, basically creating a shortened modified form of the 14-mer peptide. In addition to this we found that changing P1´ residue to F or I did not alter significantly the inhibitory effect of these peptides on BoNT/A. This may be because the active site cavity is large in BoNTs and highly electronegative. Our studies found that reasonable changes to P1’ does not affect the inhibitory effect adversely as long as the peptide has a net positive charge to effectively counter the electronegativity of the active site region (Figure 5). Earlier we have shown that the stacking of aliphatic chain of arginine (P1´) helps in binding [36]. Such stacking interactions are maintained in every peptide with all the three BoNT-LCs. The inhibitory effect of these three peptides is similar for BoNT/A, B and E.
Cross reactivity of these heptapeptides
All three peptides were tested on serotypes A, B, E and F of BoNTs. Each one of them inhibits BoNT/A, B and E similarly except BoNT/F. The reason may be similar to what we have discussed in the previous section. It is important to note that the inhibitor peptide must be basic and the P1´ has minimal effect on inhibition. However, since these peptides were not good inhibitors for BoNT/F, they were not tested extensively.
Using Footprint signature calculations we compared the interaction energy of each ligand peptide with the entire protein of all three BoNT-LCs. Footprint calculations were done in two ways–(1) using the inhibitor peptide as ligand and (2) using the protein as ligand. In the first case the total energy contribution due to the whole inhibitor peptide to each residue of the protein is calculated and in the second the total energy contribution to each ligand amino acid from the entire protein is calculated (Figures 6 and 7). The strong total negative energy contributions to the Arg in the P1’ position in all three scenarios (Figure 7) highlights the importance of the basic residues of the inhibitor peptide. Similarly, in scenario 1 (Figure 6), the negative residues lining the binding site of each BoNT have the largest energy contribution to the footprint score, primarily through electrostatic interactions.
Although the authors recognize that the energy states reported here for BoNT/B and BoNT/E are derived from homology modeling and not values derived from crystal structures, the vdw scores for the footprint scores are within reasonable values indicating minimal vdw clashes. Each structure, including the crystallographically derived BoNT/A with GRKKRRC, contained ≈2-3 residues with significant positive vdw forces on the order of 50% the maximum electrostatic potential. It should be noted that for BoNT/E in particular, the footprint score required relieving the vdw clash manually between Tyr349, Gly341, and the P1’ site on the ligand since initial energy minimization failed to account for vdw clashes, probably because no molecular dynamics simulations were used. A different rotamer for the P1’ residue was selected which minimized the vdw clash, but maintained a similar conformation. Further minimizations or a different rotamer on the P1’ residue could reduce this clash, and specifically for the P1’ residue and Tyr349 create a much lower vdw clash. Similarly, different rotamers were tested on the P3’ site (data not shown) in the BoNT/E footprinting, with the observed vdw force improving at the expense of reduced electrostatic interaction. For both of these adjustments, rotamers were chosen that most closely mimicked the original rotamer of the BoNT/A crystallographic structure, while still minimizing the vdw clash, however a crystallography derived structure is recommended for a more complete footprint score comparison. Even with these few positive vdw values, all three BoNT-LCs reflect a similar trend with the positively charged residues of the ligand driving overall footprint score. The sequence alignment of the LCs of the three botulinum serotypes is presented in Figure 8. All residues in each serotype with favorable total energy of interaction (negative energy, Figure 6) are marked in red font. These residues with favorable energy are aligned both in sequence and structure enabling similar binding of the basic inhibitor peptide in all three serotypes. Most of these residues are common in all three, but individual BoNTs have extra residues with favorable energy adding to total binding energy. We hypothesize that the structural and sequence alignment of residues with favorable energy is important for the inhibitor to be panactive. This suggests that a suitable pharmacophore could be modeled for improving the inhibitor or for designing a non-peptide inhibitor.
Figure 8: Sequence comparison. Residues contributing favorably to the ligand binding are marked in red font. Residues with favorable energies are all aligned both in sequence and structure. The energy values were taken from Footprint calculations (Figure 6) using the inhibitor peptide as ligand. In each serotype there are extra residues with favorable energy contributing to the binding energy.
Acknowledgment
Research was supported by an award from DTRA BO742081 under DOE prime contract No. DEAC02-98CH10886 (PI: SS) with Brookhaven National Laboratory. We gratefully acknowledge the data collection facilities at the NSLS beamline X29A and SER-CAT of APS. This article has been authored by Brookhaven Science Associates, LLC under contract number DE-AC02-98CH10886 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. Authors thank Prof R. Rizzo and Y. Zhou of Department of Applied Mathematics and Statistics, Stony Brook University for helping us with Footprint calculations. We are grateful to Dr. K. Ichtchenko of New York University for providing us the expression vector of SNAP-25-VAMP-GFP hybrid substrate.
References
- Gill DM (1982) Bacterial toxins: a table of lethal amounts. Microbiol Rev 46: 86-94.
- Bhidayasiri R, Truong DD (2008) Evidence for effectiveness of botulinum toxin for hyperhidrosis. J Neural Transm 115: 641-645.
- Bhidayasiri R, Truong DD (2005) Expanding use of botulinum toxin. J Neuro Sci 235: 1-9.
- Caya JG, Agni R, Miller JE (2004) Clostridium botulinum and the clinical laboratorian: a detailed review of botulism, including biological warfare ramifications of botulinum toxin. Arch Pathol Med 128: 653-662.
- Cheng CM, Chen JS, Patel RP (2006) Unlabeled uses of botulinum toxins: a review, part 2. Am J Health Syst Pharm 63: 225-232.
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, et al. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497-3500.
- Foster KA (2004) The analgesic potential of clostridial neurotoxin derivatives. Expert Opin Investig Drugs 13: 1437-1443.
- Foster KA (2009) Engineered toxins: new therapeutics. Toxicon 54: 587-592.
- Foster KA, Bigalke H, Aoki KR (2006) Botulinum neurotoxin-from laboratory to bedside. Neurotox Res 9: 133-140.
- Cote TR, Mohan AK, Polder JA, Walton MK, Braun MM (2005) Botulinum toxin type A injections: adverse events reported to the US Food and Drug Administration in therapeutic and cosmetic cases. J Am Acad Dermatol 53: 407-415.
- Marks JD (2004) Deciphering antibody properties that lead to potent botulinum neurotoxin neutralization. Mov Disord Suppl 8: S101-S108.
- Burnett JC, Henchal EA, Schmaljohn AL, Bavari S (2005) The evolving field of biodefence: therapeutic developments and diagnostics. Nat Rev Drug Discov 4: 281-297.
- Burnett JC, Opsenica D, Sriraghavan K, Panchal RG, Ruthel G, et al. (2007) A refined pharmacophore identifies potent 4-amino-7-chloroquinoline-based inhibitors of the botulinum neurotoxin serotype A metalloprotease. J Med Chem 50: 2127-2136.
- Burnett JC, Ruthel G, Stegmann CM, Panchal RG, Nguyen TL, et al. (2007) Inhibition of metalloprotease botulinum serotype A from a pseudo-peptide binding mode to a small molecule that is active in primary neurons. J Biol Chem 282: 5004-5014.
- Burnett JC, Schmidt JJ, McGrath CF, Nguyen TL, Hermone AR, et al. (2005) Conformational sampling of the botulinum neurotoxin serotype A light chain: implications for inhibitor binding. Bioorg Med Chem 13: 333-341.
- Burnett JC, Schmidt JJ, Stafford RG, Panchal RG, Nguyen TL, et al. (2003) Novel small molecule inhibitors of botulinum neurotoxin A metalloprotease activity. Biochem Biophys Res Commun 310: 84-93.
- Lai H, Feng M, Roxas-Duncan V, Dakshanamurthy S, Smith LA, et al. (2009) Quinolinol and peptide inhibitors of zinc protease in botulinum neurotoxin A: effects of zinc ion and peptides on inhibition. Arch Biochem Biophys 491: 75-84.
- Roxas-Duncan V, Enyedy I, Montgomery VA, Eccard VS, Arrington MA, et al. (2009) Identification and biochemical characterization of small- molecule inhibitors of Clostridium botulinum neurotoxin serotype A. Antimicrob Agents Chemother 53: 3478-3486.
- Capkova K, Salzameda NT, Janda KD (2009) Investigations into small molecule non- peptidic inhibitors of the botulinum neurotoxins. Toxicon 54: 575-582.
- Schiavo G, Matteoli M, Montecucco C (2000) Neurotoxins affecting neuroexocytosis. Physiol Rev 80: 717-766.
- Hale M, Oyler G, Swaminathan S, Ahmed SA (2010) Basic tetrapeptides as potent intracellular inhibitors of type A botulinum neurotoxin protease activity. J Biol Chem 286: 1802-1811.
- Kumaran D, Rawat R, Ludivico ML, Ahmed SA, Swaminathan S (2008) Structure and substrate based inhibitor design for clostridium botulinum neurotoxin serotype A. J Biol Chem 283: 18883-18891.
- Agarwal R, Eswaramoorthy S, Kumaran D, Dunn JJ, Swaminathan S (2004) Cloning, high level expression, purification and crystallization of the full length Clostridium botulinum neurotoxin type E light chain. Protein Expr Purif 34: 95-102.
- Kumaran D, Rawat R, Ahmed SA, Swaminathan S (2008) Substrate binding mode and its implication on drug design for botulinum neurotoxin A. PLoS Pathog 4: e1000165.
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307-326.
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M (1991) Improved methods in building protein models in electron density map and the location of errors in these models. Acta Cryst A47: 110-119.
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr D54: 905-921.
- CCP4 (1994) CCP4 Suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr D50: 760-763.
- Laskowaski RA, Jablonska J, Pravda L, Varekova RS, Thronton JM (2017) PDBsum: Structural summaries of PDB entries. Protein Scl 27: 129-134.
- Hines HB, Kim AD, Stafford RG, Badie SS, Brueggeman EE, et al. (2008) Use of a recombinant fluorescent substrate with cleavage sites for all botulinum neurotoxins in high-throughput screening of natural product extracts for inhibitors of serotypes A, B, and E. Appl Environ Microbiol 74: 653-659.
- Zhou Y, McGillick BE, Teng YG, Haranahalli K, Ojima I, et al. (2016) Identification of small molecule inhibitors of botulinum neurotoxin serotype E via footprint similarity. Bioorg Med Chem 24: 4875-4889.
- Balius TE, Mukherjee S, Rizzo RC (2011) Implementation and evaluation of a docking-rescoring method using molecular footprint comparisons. J Comput Chem 32: 2273-2289.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. (2004) UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 25: 1605-1612.
- Montreal (2010) Molecular Operating Environment [MOE 2009.10] Montreal, Quebec, Canada, H3B 3X3).
- Agarwal R, Swaminathan S (2008) SNAP-25 substrate peptide (residues 180-183) binds to but bypasses cleavage by catalytically active Clostridium botulinum neurotoxin E. J Biol Chem 283: 25944-25951.
- Kumar G, Kumaran D, Ahmed SA, Swaminathan S (2012) Peptide inhibitors of botulinum neurotoxin serotype A: design, inhibition, cocrystal structures, structure-activity relationship and pharmacophore modeling. Acta Crystallogr D Biol Crystallogr 68: 511-520.
- Kumar G, Kumaran D, Ahmed SA, Swaminathan S (2011) Discovery of peptide and peptidomimetic inhibitors of botulinum neurotoxin serotype A: Inhibition, co-crystal structures, SAR and pharmacophore modeling. PLoS Pathog Submitted.
- Â Kumar G, Agarwal A, Swaminathan S (2016) Small molecule non-peptide inhibitors of botulinum neurotoxin serotype E: Structure-activity relationship and a pharmacorphor model. Bioorg Med Chem 24: 3978-3985.
- Agarwal R, Schmidt JJ, Stafford RG, Swaminathan S (2009) Mode of VAMP substrate recognition and inhibition of Clostridium botulinum neurotoxin F. Nat Struct Mol Biol 16: 789-794.
- Schmidt JJ, Stafford RG (2002) A high-affinity competitive inhibitor of type A botulinum neurotoxin protease activity. FEBS Lett 532: 423-426.
- Schmidt JJ, Stafford RG, Bostian KA (1998) Type A botulinum neurotoxin proteolytic activity: development of competitive inhibitors and implications for substrate specificity at the S1' binding subsite. FEBS Lett 435: 61-64.
Citation: Vieni C, McGillick B, Kumaran D, Eswaramoorthy E, Kandavelu P, et al. (2018) Structure Based Discovery of Pan Active Botulinum Neurotoxin Inhibitors. J Infect Dis Ther 6: 351. DOI: 10.4172/2332-0877.1000351
Copyright: © 2018 Vieni C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 6893
- [From(publication date): 0-2018 - Dec 18, 2025]
- Breakdown by view type
- HTML page views: 5823
- PDF downloads: 1070