The Genetic Diversity of Liriodendron tulipifera Germplasm Revealed by SSR Markers
Received: 08-Feb-2018 / Accepted Date: 12-Feb-2018 / Published Date: 20-Feb-2018
Abstract
Liriodendron tulipifera is widely distributed in the world and should be studied in vary of properties. Genetic diversity is the basis and premise of biodiversity, hence, one specie with low genetic diversity would not evolve or adapt to the changing environments. Genetic diversity is a decisive factor of successful response of one specie to human disturbance, and the degree of intra specific genetic variation also has key effect on its potentiality of evolution. This study analyzes the level of genetic diversity of L. tulipifera germplasm by using SSR markers.
Keywords: SSR; Liriodendron tulipifera; Genetic diversity
Introduction
Liriodendron tulipifera is widely distributed in Eastern America, from southern Canada to central Florida peninsula, and from Atlanta coast to the Mississippi River basin. Its sister-species, L. chinense , is regarded as an endangered species. In contrast, L. tulipifera is a pioneer and dominant tree species with strong adaption in forest community in Southeast US. Genetic diversity is the basis and premise of biodiversity, hence, one specie with low genetic diversity would not evolve or adapt to the changing environments. Furthermore, genetic diversity is a decisive factor of successful response of one specie to human disturbance, and the degree of intra specific genetic variation also has key effect on its potentiality of evolution. Since genetic diversity can be inspected with various DNA markers, this study analyzes the level of genetic diversity of L. tulipifera germplasm by using SSR markers.
Materials And Methods
Plant material
All germplasm plants (Table 1) were conserved at Xiashu forest farm affiliated to Nanjing Forestry University. In the summer of 2009~2010, 2~3 fresh leaves of each plant were collected randomly and put into the ice box rapidly after they were stored in a valve bag. The fresh leaves were brought back and kept in a refrigerator under the condition of -70°C prior to DNA extraction.
| Provenance | Population code | Sample size |
|---|---|---|
| North Carolina | BK | 8 |
| Louisiana | LYS | 8 |
| Georgia | ZZY | 8 |
| Missouri | MSL | 8 |
| South Carolina | NK | 8 |
Table 1: Forty genotypes (germplasm) from five provenances of L. tulipifera were taken as plant materials
Experimental Methods
Extraction and purification of total DNA
In this experiment, the improved CTAB lysis-silica beads assay was used, in order to extract the genomic DNA of L. tulipifera [1].
SSR-PCR amplification reaction
10 μl system of SSR-PCR amplification reaction (Table 2) and touch-down PCR amplification reaction process (Table 3) were used in this experiment.
| Ingredients | Final Addition |
|---|---|
| Dd H2O | 6 μl |
| 10 × Buffer | 1 μl |
| 25 mu-MgCl2 | 1.25 μl |
| 10 mM-dNTP | 0.2 μl |
| F-Primer | 0.25 μl |
| R-Primer | 0.25 μl |
| r-Taq | 0.05 μl |
| Template DNA | 1 μl |
Table 2: System of SSR-PCR amplification. Note: dNTP and magnesium 2+ to oscillation blender before use.
| Cycle | Temperature | Time | ||
|---|---|---|---|---|
| Pre-denaturation | 94°C | 4 min | ||
| Touch-down PCR | 15 cycles | 94°C | 15 s | |
| 60°C | 15 s | (△℃=-0.7) | ||
| 72°C | 30 s | |||
| Ordinary PCR | 15 cycles | 94°C | 15 s | |
| 49.5°C | 15 s | |||
| 72°C | 30 s | |||
| Extension | 72°C | 20 min | ||
| Conservation | 4°C | ∞ |
Table 3: Procedure of PCR amplification.
Polyacrylamide gel electrophoresis of PCR products
All polyacrylamide gel electrophoresis have been carried out according to Almasizadehyaghuti et al.
Primer selection
Screening criteria of polymorphism primers followed in this experiment are as follows: amplification products should have distinct banding pattern on electropherogram of 8% polyacrylamide denaturing gel electrophoresis, various allelic variations should exist among samples, and the eliminated loci of primer should only have 1~2 bp differences. After preliminary screening, the primers which have no amplification products or undesirable polymorphisms of amplification products should be eliminated, moreover, the primers which have clear amplification bands and obvious polymorphism were selected. Finally, 22 SSR primers with obvious banding pattern, good repeatability and high stability were screened and used for this experiment.
Data management and statistical analysis
Band interpretation: SSR is co-dominant marker; the band with consistent electrophoretic mobility in the amplification product of the same primer has homology. The band data should be numbered as increasing sequences and the length of band with A, B, C, D, E…. for instance, the lowest is A, and the top most is D or E or others, the result of band reading is AB, BC, etc., besides, if one sample only has one band, the result of band reading is AA, BB, and so on.
Data analysis
Analysis of genetic diversity: The genetic parameter of populations can be obtained by using POPGENE 1.32 [2], including average number of alleles (A), effective number of allele (Ne), Kimura and Crow observed heterozygosity (Ho), expected heterozygosity (He), [3,4], Shannon diversity index (I), [5], Nei diversity index [6,7] and genetic distance (D).
Analysis of genetic relationship: Clustering analysis is conducted by utilizing software NTSYS-pc 2.10e and the clustering figure should be established with the method of UMPGA [8].
Results
Analysis of genetic diversity on different SSR loci
40 plants of L. tulipifera , 112 alleles are tested totally through 26 pairs SSR primers. The number of alleles (A) on each locus changes from two to six, the average value is 4.3077 (Table 2).
The effective number of allele (Ne) changes from 1.4369 to 4.2781, the average value is 2.9714. The maximum of Shannon diversity index (I) is 1.5162, the minimum is 0.5850 and the average value is 1.1842. The variation range of observed heterozygosity (Ho) is 0.0750~0.8718, the variation range of expected heterozygosity (He) is 0.3079~0.7759, with an average expected heterozygosity of 0.6541. Nei expected heterozygosity (Nei) ranges between 0.3041~0.7662, indicating that L. tulipifera maintains high genetic variation (Figure 1 and Table 1).
Fixation index (F) measures the degree of deviation between actual proportion of genotype and expected proportion of Hardy-Weinberg theory (Table 4). F<0 or F>0 means that heterozygosis is excessive or homozygosis is excessive respectively. When random mating of individuals in a population cannot be achieved, its actual proportion of genotype will deviate to some extent from Hardy-Weinberg equilibrium inevitably. As for LT102, LT297, LT488, and LT921 on 26 loci, their F<0, indicating that the heterozygosis are excessive on these four loci, and as for the rest loci, their F>0, showing insufficient heterozygote on the rest loci.
| Locus | A | Ne | I | Ho | He | F | Nei |
|---|---|---|---|---|---|---|---|
| LT002 | 5 | 3.3161 | 1.3085 | 0.425 | 0.7073 | 0.3991 | 0.6984 |
| LT015 | 5 | 3.4298 | 1.385 | 0.275 | 0.7174 | 0.6167 | 0.7084 |
| LT022 | 4 | 3.3578 | 1.283 | 0.225 | 0.7111 | 0.6836 | 0.7022 |
| LT026 | 4 | 3.4845 | 1.3066 | 0.2308 | 0.7223 | 0.6805 | 0.713 |
| LT049 | 3 | 2.9331 | 1.0867 | 0.175 | 0.6674 | 0.7378 | 0.6591 |
| LT056 | 4 | 2.6959 | 1.1438 | 0.375 | 0.637 | 0.4113 | 0.6291 |
| LT062 | 5 | 2.7064 | 1.115 | 0.4872 | 0.6387 | 0.2372 | 0.6305 |
| LT071 | 6 | 3.1715 | 1.3816 | 0.175 | 0.6934 | 0.7476 | 0.6847 |
| LT080 | 4 | 2.0874 | 0.9998 | 0.225 | 0.5275 | 0.5735 | 0.5209 |
| LT091 | 4 | 3.796 | 1.3582 | 0.425 | 0.7459 | 0.4302 | 0.7366 |
| LT102 | 5 | 3.3438 | 1.3682 | 0.75 | 0.7098 | -0.0566 | 0.7009 |
| LT121 | 6 | 3.5874 | 1.4001 | 0.65 | 0.7304 | 0.1101 | 0.7212 |
| LT124 | 5 | 3.0332 | 1.2752 | 0.075 | 0.6788 | 0.8895 | 0.6703 |
| LT151 | 4 | 2.7997 | 1.1734 | 0.3 | 0.6509 | 0.5391 | 0.6428 |
| LT199 | 4 | 2.917 | 1.2135 | 0.575 | 0.6655 | 0.136 | 0.6572 |
| LT233 | 5 | 2.6711 | 1.1644 | 0.45 | 0.6335 | 0.2897 | 0.6256 |
| LT274 | 4 | 3.0651 | 1.174 | 0.55 | 0.6823 | 0.1939 | 0.6738 |
| LT297 | 5 | 3.3725 | 1.3351 | 0.8718 | 0.7126 | -0.2234 | 0.7035 |
| LT298 | 2 | 1.9577 | 0.6823 | 0.2059 | 0.4965 | 0.5853 | 0.4892 |
| LT316 | 5 | 4.2781 | 1.5162 | 0.45 | 0.7759 | 0.42 | 0.7662 |
| LT371 | 5 | 2.1277 | 0.983 | 0.15 | 0.5367 | 0.7205 | 0.53 |
| LT414 | 3 | 2.558 | 0.9981 | 0.4 | 0.6168 | 0.3515 | 0.6091 |
| LT446 | 3 | 1.4369 | 0.585 | 0.1 | 0.3079 | 0.6752 | 0.3041 |
| LT488 | 4 | 3.0756 | 1.1972 | 0.7368 | 0.6839 | -0.0774 | 0.6749 |
| LT921 | 4 | 2.926 | 1.1517 | 0.7632 | 0.667 | -0.1442 | 0.6582 |
| LT978 | 4 | 3.1281 | 1.2023 | 0.35 | 0.6889 | 0.4919 | 0.6803 |
| Mean | 4.3077 | 2.9714 | 1.1842 | 0.3998 | 0.6541 | 0.3888 | 0.6458 |
Table 4: Genetic diversity of 26 SSR loci within 5 provenances of L. tulipifera. Note: An average number of alleles; Ne Effective number of alleles; I Shannon’s Information index; Ho observed heterozygosity; He average expected heterozygosity. F Fixation index; Nei Nei’s expected heterozygosity.
Genetic variation among five provenances of L. tulipifera
From Table 5, it can be found that, among five provenances, the effective number of alleles (Ne) of provenance LYS is the highest (2.4031), while provenance ZZY is the lowest (1.9382). The variation range of Nei diversity index ranges from 0.4330 (ZZY) to 0.5643 (LYS); the maximum of Shannon diversity index (I) is 0.9532 (LYS), and the minimum is 0.7009 (ZZY). Generally, LYS has the highest level of genetic diversity, while ZZY the lowest genetic diversity. As for the fixation index, the degree of deviation of NK from equilibrium is largest, although it has maximal fixation index (F=0.4187), however, the degree of deviation of ZZY from equilibrium is smallest, although it has minimal fixation index (F=0.0623).
| Population code | A | Ne | I | Ho | He | F | Nei |
|---|---|---|---|---|---|---|---|
| BK | 2.962 | 2.2073 | 0.8579 | 0.409 | 0.5425 | 0.247 | 0.509 |
| MSL | 2.577 | 2.076 | 0.7491 | 0.414 | 0.4968 | 0.168 | 0.465 |
| ZZY | 2.5 | 1.9382 | 0.7009 | 0.433 | 0.4622 | 0.062 | 0.433 |
| LYS | 3.115 | 2.4031 | 0.9532 | 0.424 | 0.6026 | 0.297 | 0.564 |
| NK | 2.962 | 2.3408 | 0.8904 | 0.325 | 0.5589 | 0.419 | 0.523 |
| Mean | 2.823 | 2.1931 | 0.8303 | 0.401 | 0.5326 | 0.238 | 0.499 |
Table 5: The genetic diversity among 5 provenances of L. tulipifera. Note: An average number of alleles; Ne Effective number of alleles; I Shannon’s Information index; Ho observed heterozygosity; He average expected heterozygosity. F Fixation index; Nei Nei’s expected heterozygosity.
Genetic structure among five provenances of L. tulipifera
Fis and Fit represent the degree of deviation of single population and total population from Hardy-Weinberg equilibrium. As for five L. tulipifera provenances, Fis=0.1965, the average Fit=0.3789, which shows that heterozygote is insufficient not only in total level but also in individuals of each provenance. Among five L. tulipifera provenances, gene flow Nm=0.8513 and genetic differentiation coefficient Fst=0.2270, which means that 22.70% of genetic variation exist among provenances, and 77.30% of genetic variation exist within provenance.
Genetic relationship of five L. tulipifera provenances
From Table 6, it can be found that the variation range of genetic identity (I) of these five L. tulipifera provenances is 0.5333~0.8135, and the variation range of genetic distance (D) is 0.2064~0.6286, with an average genetic distance of 0.4574, thus it can be deduced that these five L. tulipifera provenances have great genetic variation, and genetic relationship is relative remote. Among which, genetic identity (I) of MSL and BK is highest, which is 0.8135, showing that genetic relationship of these two populations is much closer than other ones. However, genetic identity (I) of NK and ZZY is the lowest (0.5333), indicating that genetic relationship of these two populations is much remoter than other ones.
| pop ID | BK | MSL | ZZY | LYS | NK |
|---|---|---|---|---|---|
| BK | **** | 0.8135 | 0.5554 | 0.6844 | 0.678 |
| MSL | 0.2064 | **** | 0.5618 | 0.5866 | 0.577 |
| ZZY | 0.5881 | 0.5766 | **** | 0.7094 | 0.5333 |
| LYS | 0.3793 | 0.5334 | 0.3433 | **** | 0.684 |
| NK | 0.3887 | 0.5499 | 0.6286 | 0.3798 | **** |
Table 6: Nei's genetic identity and genetic distance in 5 provenances of L. tulipifera. Nei's genetic identity (above diagonal) and genetic distance (below diagonal).
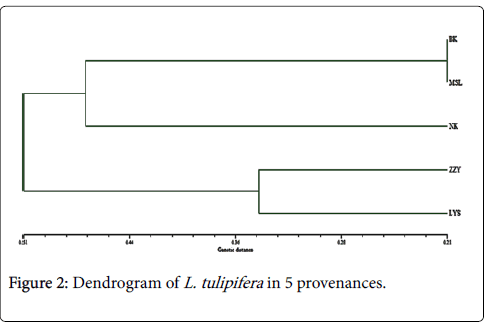
According to genetic distance and genetic similarity calculated by Nei method, the clustering figure of five L. tulipifera provenances can be established with the method of UMPGA. As shown in Figure 2, when the threshold value is 0.51, these five L. tulipifera provenances can be divided into two categories, among which, ZZY and LYS belong to one category and the rest three provenances belong to the other category. Meanwhile, when the threshold value is 0.47, these three provenances can be divided in two subgroups further: namely, BK and MSL belong to one category, and NK constitutes one category alone.
Discussion
Genetic diversity of five L. tulipifera provenances
Genetic diversity is the result of long-term evolution of the species and represents the potential of evolution of species, which is the premise for the survival and development of populations. In order to survive and develop in an unstable environment, the species are obliged to generate and accumulate genetic variation. On the basis of the results of this research, the effective number of allele of five L. tulipifera provenances is 2.9714, the variation range of Nei expected heterozygosity (Nei) is 0.3041~0.7662, which show that L. tulipifera has high level of genetic variation. It can further declare that L.tulipifera is one pioneer tree species with high adaption and wide distribution range, moreover, continues geographical distribution increases the pollen flow and seed dispersal among populations. Meanwhile, the large gene flow among populations also results in high level of allelic richness and genetic diversity of L.tulipifera.
Acknowledgements
We would like to thank all professors’ Huogen Li and Qiang Zhuge lab crews and students that helped us to carry out experiments and prepare manuscript.
References
- Almasizadehyaghuti A, Movahedi A, Mohammadi K, Zhuge Q, Li H (2017) Genetic diversity among a collection of L. chinense germplasm analyzed with SSR Markers. J Biochem Microb Toxicol 1:102.
- Yeh FC, Yang RC, Boyle T (1999) POPGENE software package version 1.31 for population genetic analysis, University of Alberta.
- Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Journal of Genetics 49: 725-738.
- Levene H (1949) On a matching problem arising in genetics. J Ann Math Statist 20: 91-94.
- Lewontin RC (1972) The apportionment of human diversity. J Evol Biol 6: 381-398.
- Nei M (1973) Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Science 70: 3321-3323.
- Rohlf FJ (2002) NTSYS-pc. Numerical Taxonomy and Multivariate Analysis System, Version 2.10. Exeter Software, New York.
- Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York.
Citation: Almasizadehyaghuti A, Movahedi A, Mohammadi K, Zhuge Q, Li H (2018) The Genetic Diversity of Liriodendron tulipifera Germplasm Revealed by SSR Markers. J Biochem Microb Toxicol 2: 105.
Copyright: © 2017 Almasizadehyaghuti A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Open Access Journals
Article Usage
- Total views: 4640
- [From(publication date): 0-2018 - Dec 08, 2025]
- Breakdown by view type
- HTML page views: 3672
- PDF downloads: 968