Short Communication Open Access
The Role of Upregulated APOE in Alzheimer's Disease Etiology
William K Gottschalk1, Mirta Mihovilovic1, Allen D Roses1,3 and Ornit Chiba-Falek1,2*
1Department of Neurology, University Medical Center, USA
2Center for Genomic and Computational Biology, Duke University Medical Center, Durham, NC 27710, USA
3Zinfandel Pharmaceuticals, Chapel Hill, NC, USA
- Corresponding Author:
- Ornit Chiba-Falek
Department of Neurology
DUMC Box 2900
Duke University, Durham
North Carolina 27710, USA
Tel: 919 681-8001
E-mail: o.chibafalek@duke.edu
Received date: January 19, 2016; Accepted date: February 02, 2016; Published date: February 09, 2016
Citation: Gottschalk WK, Mihovilovic M, Roses AD, Chiba-Falek O (2016) The Role of Upregulated APOE in Alzheimer’s Disease Etiology. J Alzheimers Dis Parkinsonism 6:209. doi: 10.4172/2161-0460.1000209
Copyright: © 2016 Gottschalk WK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Alzheimers Disease & Parkinsonism
The first and most firmly established genetic risk factor for sporadic late onset Alzheimer’s disease (LOAD) is the e4 allele of the apolipoprotein E (APOE) gene [1]. Carrying the APOEe4 variant significantly increases the lifetime risk for LOAD, with the number of copies present indicative of level of risk [1,2] and is associated with lower age of clinical disease onset [1,3-6]. Furthermore, genomewide association studies (GWAS) for sporadic LOAD confirmed that APOE is the major susceptibility genomic region for the disease and reported significant associations with markers within the APOE linkage disequilibrium (LD) locus (contains APOE, TOMM40 and APOC1 genes). The strongest association signal (by wide margin) in these studies was found at the APOE LD region and no other LOADassociation in the human genome remotely approached the same level of significance [7-10]. However, the molecular mechanism underlying the reported genetic LOAD-associations with APOE LD region in general and APOEe4 haplotype in particular has yet to be discovered.
It has been suggested that alteration of the expression levels of specific genes may be an important mechanism in the etiology of neurodegenerative disorders including LOAD [11]. Previously, using temporal and occipital tissues obtained from APOEe3/3 donors we showed that APOE-mRNA levels are significantly increased in LOADaffected brains compared to controls [12]. In preliminary studies, we performed expression analysis in cortical neurons from the temporal cortex of 3 LOAD patients and 3 normal controls isolated by laser capture microdissection (LCM) technique. We analyzed the APOEmRNA counts relative to geometric mean of two housekeeping genes using the nCounter single cell gene expression technology and the nSolver program (NanoString). The results showed increased APOEmRNA in LOAD compared to normal (our unpublished data) and validated our published findings obtained using homogenates of brain tissue for the expression analysis [12]. Our observation was consistent with other reports of elevated levels of APOE-mRNA in LOAD brains. For example, Zarow et al. report increased APOE-mRNA levels in the hippocampus of AD cases compared to controls [13] and Matsui et al. report increased APOE-mRNA levels in temporal cortex of AD donors compared to controls [14]. Furthermore, Akram et al. have demonstrated that APOE-mRNA and protein levels in the inferior temporal gyrus and the hippocampus are strongly, positively correlated with the progression of cognitive dysfunction [15].
A recent study showed that endoplasmic reticulum (ER)- mitochondrial communication and mitochondria associated ER membranes (MAM) function-as measured by the synthesis of phospholipids and of cholesteryl esters, respectively-are increased significantly in cells treated with APOEe4-containing astrocyteconditioned media (ACM) as compared to those treated with APOEe3- containing ACM [16]. Upregulated MAM function was implicated in the pathogenesis of AD [17,18]. The new findings that APOEe4 protein upregulates the activity of MAM may explain, in part, the contribution of APOEe4 as a risk factor in the disease. Enhanced activity of APOEe4 protein in correlation to AD-related cellular phenotypes has also been described previously. In human AD brain samples, amyloid deposits correlate with gene dosage of APOEe4 [19], and APOEe4 protein more actively forms fibrils with Aβ protein than APOEe3 in vitro [20]; moreover, APOEe4 aggregates are themselves neurotoxic [21]. APOEe4 is susceptible to cleavage of the C-terminus by cellular proteases, and the C-terminal fragments are cytotoxic, in part by eliciting intracellular neurofibrillary tangle formation and in part via disruption of mitochondrial and cytoskeletal functions [22-24]. APOEe4 and APOEe3 have different lipid-binding characteristics [25], contributing to greater Aβ-elicited lysosomal leakage and apoptosis in APOEe4- producing cells [26], and affecting the respective abilities of APOEe3 and APOEe4 to support neuronal maintenance and repair.
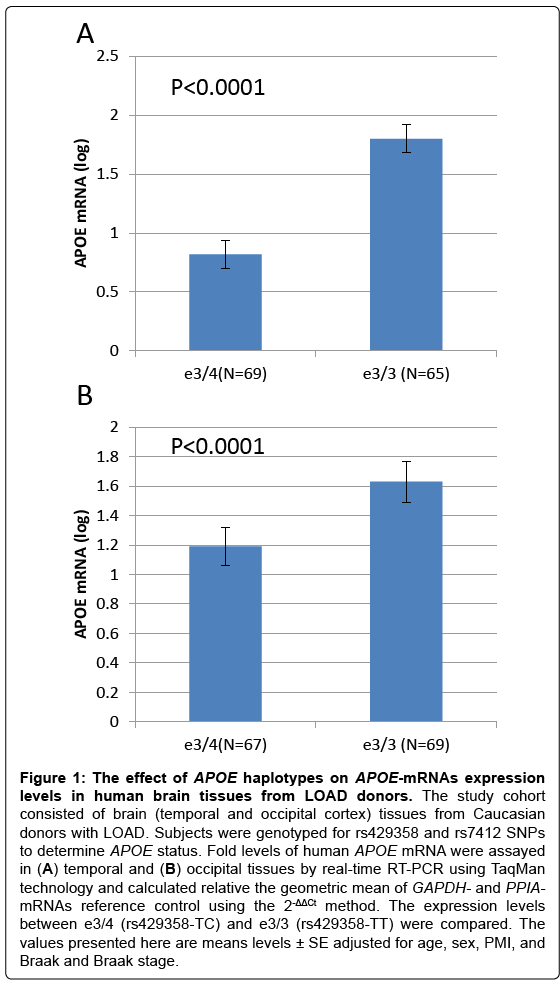
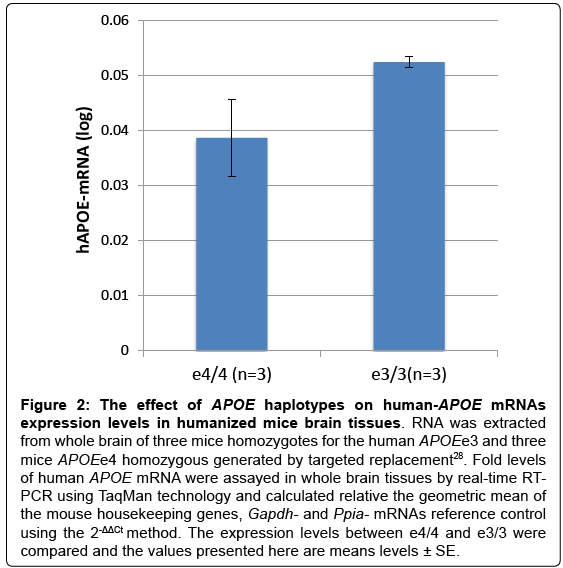
Interestingly, we showed that SNP rs429358, that defines the APOEe4 haplotype, has a significant effect on APOE-mRNAs levels in temporal cortex obtain from LOAD cases. We demonstrated that the level of APOE mRNA was significantly higher in the APOEe3/3 genotype group compared to APOEe3/4-genotype (Figure 1). In unpublished work, we measured APOE- mRNA levels in whole brains from humanized –APOEe3 and –APOEe4 homozygous mouse models generated by targeted replacement [27,28]. We found that human APOEmRNA levels are>35% higher in brains of APOEe3 homozygous mice compared to mice homozygotes to APOEe4 (Figure 2). The analysis of humanized- APOE mice support the findings in LOAD-human brains, suggesting that while the effect of e4 variant is putatively on increased activity of the APOE protein, the effect of the e3 background is possibly executed via regulation of APOE gene expression that determines the steady state amount of the protein.
Figure 1: The effect of APOE haplotypes on APOE-mRNAs expression levels in human brain tissues from LOAD donors. The study cohort consisted of brain (temporal and occipital cortex) tissues from Caucasian donors with LOAD. Subjects were genotyped for rs429358 and rs7412 SNPs to determine APOE status. Fold levels of human APOE mRNA were assayed in (A) temporal and (B) occipital tissues by real-time RT-PCR using TaqMan technology and calculated relative the geometric mean of GAPDH- and PPIA- mRNAs reference control using the 2-ΔΔCt method. The expression levels between e3/4 (rs429358-TC) and e3/3 (rs429358-TT) were compared. The values presented here are means levels ± SE adjusted for age, gender, PMI, and Braak and Braak stage.
Figure 2: The effect of APOE haplotypes on human-APOE mRNAs expression levels in humanized mice brain tissues. RNA was extracted from whole brain of three mice homozygotes for the human APOEe3 and three mice APOEe4 homozygous generated by targeted replacement28. Fold levels of human APOE mRNA were assayed in whole brain tissues by real-time RT-PCR using TaqMan technology and calculated relative the geometric mean of the mouse housekeeping genes, Gapdh- and Ppia- mRNAs reference control using the 2-ΔΔCt method. The expression levels between e4/4 and e3/3 were compared and the values presented here are means levels ± SE.
Different factors may regulate APOE gene expression including, but not limited to, genetic [12,29-31] and epigenetic [32] mechanisms. Cis-genetic variably on the background of the e3 haplotype contributes to differential APOE gene expression. We reported data showing that 523- polyT genotype, located upstream of APOE within the adjutant TOMM40 locus, affects expression of genes in APOE LD region [12]. We demonstrated that the LOAD risk allele, very long (‘VL’), is associated with increased levels of APOE transcripts in normal and LOADaffected human brain tissues and with higher luciferase expression in a cell-based reporter system, compared to the short (‘S’) allele [12]. These observations provide a possible explanation for the genetic association of the 523-polyT locus with age of LOAD onset [33,34] and other disease related phenotypes [35-38]. Our observations were recently reproduced by Payton, et al. They showed that the shorter length poly-T variants act as a repressor of luciferase gene expression in reporter gene constructs, whereas expression was reduced to approximately half of that observed for the ‘VL’ variant [39].
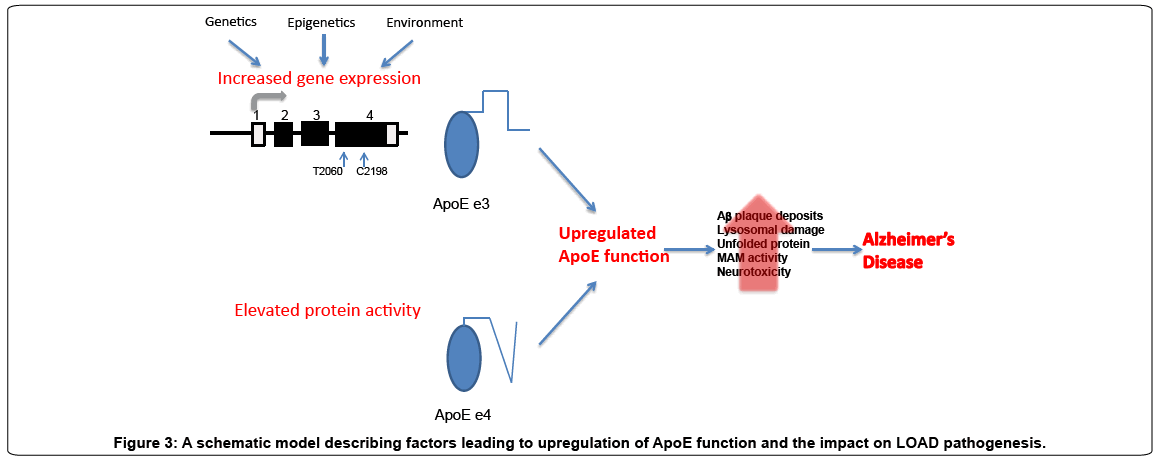
Collectively the studies reviewed here suggest that up-regulated function of APOE due to either enhanced protein activity or increased APOE expression levels may contribute, in part, to the etiology of LOAD. Figure 3 summarizes our proposed model. While this model suggests the triggering event, the biochemical and cell biological pathways that mediate the consequences of this event are still being determined. Our perception of increased APOEe3 protein levels as a LOAD-pathogenic mechanism agrees with the concept that changes in expression levels of ‘normal’ protein in the brain can lead to neurodegenerative diseases. In conclusion, genetic heterogeneity across the APOE-LD region may lead, through different molecular mechanisms, to elevated (‘pathogenic’) ApoE function and possibly explains the extremely strong genetic association of the APOE-LD region with increased LOAD-risk and related phenotypes.
Funding
This work was funded in part by the National Institute on Aging (NIA) [R01AG040370 to AR].
References
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, et al. (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261: 921-923.
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, et al. (1997) Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 278: 1349-1356.
- Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, et al. (2008) A genome-wide association study for late-onset Alzheimer's disease using DNA pooling. BMC Med Genomics 1: 44.
- Yu CE, Seltman H, Peskind ER, Galloway N, Zhou PX, et al. (2007) Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer's disease: patterns of linkage disequilibrium and disease/marker association. Genomics 89: 655-665.
- Li H, Wetten S, Li L, St Jean PL, Upmanyu R, et al. (2008) Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol 65: 45-53.
- Waring SC, Rosenberg RN (2008) Genome-wide association studies in Alzheimer disease. Arch Neurol 65: 329-334.
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, et al. (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet 41: 1088-1093.
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, et al. (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet 41: 1094-1099.
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, et al. (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 45: 1452-1458.
- Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, et al. (2007) A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J Clin Psychiatry 68: 613-618.
- Singleton A, Myers A, Hardy J (2004) The law of mass action applied to neurodegenerative disease: a hypothesis concerning the etiology and pathogenesis of complex diseases. Hum Mol Genet 13 Spec No 1: R123-126.
- Linnertz C, Anderson L, Gottschalk W, Crenshaw D, Lutz MW, et al. (2014) Thecis-regulatory effect of an Alzheimer's disease-associated poly-T locus on expression of TOMM40 and apolipoprotein E genes. Alzheimers Dement 10: 541-551.
- Zarow C, Victoroff J (1998) Increased apolipoprotein E mRNA in the hippocampus in Alzheimer disease and in rats after entorhinal cortex lesioning. ExpNeurol 149: 79-86.
- Matsui T, Ingelsson M, Fukumoto H, Ramasamy K, Kowa H, et al. (2007) Expression of APP pathway mRNAs and proteins in Alzheimer's disease. Brain Res 1161: 116-123.
- Akram A, Schmeidler J, Katsel P, Hof PR, Haroutunian V (2012) Association of ApoE and LRP mRNA levels with dementia and AD neuropathology. Neurobiol Aging 33: 628.
- Tambini MD, Pera M, Kanter E, Yang H, Guardia-Laguarta C, et al. (2016) ApoE4 upregulates the activity of mitochondria-associated ER membranes. EMBO Rep 17: 27-36.
- Area-Gomez E, Del Carmen Lara Castillo M, Tambini MD, Guardia-Laguarta C, de Groof AJ, et al. (2012) Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J 31: 4106-4123.
- Schon EA, Area-Gomez E (2013) Mitochondria-associated ER membranes in Alzheimer disease. Mol Cell Neurosci 55: 26-36.
- Schmechel DE, Saunders AM, Strittmatter WJ, Crain BJ, Hulette CM, et al. (1993) Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. ProcNatlAcadSci U S A 90: 9649-9653.
- Wisniewski T, Castaño EM, Golabek A, Vogel T, Frangione B (1994) Acceleration of Alzheimer's fibril formation by apolipoprotein E in vitro. Am J Pathol 145: 1030-1035.
- Hatters DM, Zhong N, Rutenber E, Weisgraber KH (2006) Amino-terminal Domain Stability Mediates Apolipoprotein E Aggregation into Neurotoxic Fibrils. J MolBiol 361: 932-944.
- Harris FM, Brecht WJ, Xu Q, Tesseur I, Kekonius L, et al. (2003) Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer's disease-like neurodegeneration and behavioral deficits in transgenic mice. ProcNatlAcadSci U S A 100: 10966-10971.
- Huang Y, Liu XQ, Wyss-Coray T, Brecht WJ, Sanan DA, et al. (2001) Apolipoprotein E fragments present in Alzheimer's disease brains induce neurofibrillary tangle-like intracellular inclusions in neurons. ProcNatlAcadSci U S A 98: 8838-8843.
- Chang S, ran Ma T, Miranda RD, Balestra ME, Mahley RW, et al. (2005) Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. ProcNatlAcadSci U S A 102: 18694-18699.
- Morrow JA, Segall ML, Lund-Katz S, Phillips MC, Knapp M, et al. (2000) Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry 39: 11657-11666.
- Ji ZS, Miranda RD, Newhouse YM, Weisgraber KH, Huang Y, et al. (2002) Apolipoprotein E4 potentiates amyloid beta peptide-induced lysosomal leakage and apoptosis in neuronal cells. J BiolChem 277: 21821-21828.
- Sullivan PM, Mace BE, Maeda N, Schmechel DE (2004) Marked regional differences of brain human apolipoprotein E expression in targeted replacement mice. Neuroscience 124: 725-733.
- Sullivan PM, Mezdour H, Aratani Y, Knouff C, Najib J, et al. (1997) Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J BiolChem 272: 17972-17980.
- Bekris LM, Galloway NM, Montine TJ, Schellenberg GD, Yu CE (2010) APOE mRNA and protein expression in postmortem brain are modulated by an extended haplotype structure. Am J Med Genet B Neuropsychiatr Genet 153B: 409-417.
- Bekris LM, Lutz F, Yu CE (2012) Functional analysis of APOE locus genetic variation implicates regional enhancers in the regulation of both TOMM40 and APOE. J Hum Genet 57: 18-25.
- Bekris LM, Millard SP, Galloway NM, Vuletic S, Albers JJ, et al. (2008) Multiple SNPs within and surrounding the apolipoprotein E gene influence cerebrospinal fluid apolipoprotein E protein levels. J Alzheimers Dis 13: 255-266.
- Yu CE, Cudaback E, Foraker J, Thomson Z, Leong L, et al. (2013) Epigenetic signature and enhancer activity of the human APOE gene. Hum Mol Genet 22: 5036-5047.
- Roses AD (2010) An inherited variable poly-T repeat genotype in TOMM40 in Alzheimer disease. Arch Neurol 67: 536-541.
- Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, et al. (2010) A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease. Pharmacogenomics J 10: 375-384.
- Bruno D, Nierenberg JJ, Ritchie JC, Lutz MW, Pomara N (2012) Cerebrospinal fluid cortisol concentrations in healthy elderly are affected by both APOE and TOMM40 variants. Psychoneuroendocrinology 37: 366-371.
- Bruno D, Pomara N, Nierenberg J, Ritchie JC, Lutz MW, et al. (2012) Levels of cerebrospinal fluid neurofilament light protein in healthy elderly vary as a function of TOMM40 variants. ExpGerontol 47: 347-352.
- Hayden KM, McEvoy JM, Linnertz C, Attix D, Kuchibhatla M, et al. (2012) Ahomopolymer polymorphism in the TOMM40 gene contributes to cognitive performance in aging. Alzheimers Dement 8: 381-388.
- Johnson SC (2011) The effect of TOMM40 poly-T length on gray matter volume and cognition in middle-aged persons with APOEvarepsilon3/varepsilon3 genotype. Alzheimers Dement 7: 456-65.
- Payton A, Sindrewicz P, Pessoa V, Platt H, Horan M, et al. (2015) A TOMM40 poly-T variant modulates gene expression and is associated with vocabulary ability and decline in nonpathologic aging. Neurobiol Aging.
Relevant Topics
- Advanced Parkinson Treatment
- Advances in Alzheimers Therapy
- Alzheimers Medicine
- Alzheimers Products & Market Analysis
- Alzheimers Symptoms
- Degenerative Disorders
- Diagnostic Alzheimer
- Parkinson
- Parkinsonism Diagnosis
- Parkinsonism Gene Therapy
- Parkinsonism Stages and Treatment
- Stem cell Treatment Parkinson
Recommended Journals
Article Tools
Article Usage
- Total views: 12228
- [From(publication date):
March-2016 - Aug 29, 2025] - Breakdown by view type
- HTML page views : 11266
- PDF downloads : 962