Research Article Open Access
Molecular Detection of Helicobacter Species and Other Bacteria in Human Bile Samples of Patients with Biliary Diseases
Jamshid Vafaeimanesh1, Masoud Alebouyeh2, Mohammadreza Seyyedmajidi2*, Elahe Tajeddin2, Somayeh Jahani Sherafat2, Elahe Zanganeh2, Amirhoushang Mohammadalizadeh2 and Mohammadreza Zali2
1Department of Internal Medicine, Qom University of Medical Sciences, Qom, Iran
2Research Center for Gastroenterology and Liver Disease, Department of Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- *Corresponding Author:
- Mohammadreza Seyyedmajidi
Department of Gastroenterology and Liver Diseases
Shahid Beheshti University of Medical Sciences
Tehran, Iran
Tel/Fax: +98 21 22418885
E-mail: mrsmajidi55@yahoo.com
Received date: January 04, 2012; Accepted date: March 24, 2012; Published date: March 26, 2012
Citation: Vafaeimanesh J, Alebouyeh M, Seyyedmajidi M, Tajeddin E, Sherafat SJ, et al. (2012) Molecular Detection of Helicobacter Species and Other Bacteria in Human Bile Samples of Patients with Biliary Diseases. J Gastrointest Dig Syst 2:106. doi:10.4172/2161-069X.1000106
Copyright: © 2012 Vafaeimanesh J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Gastrointestinal & Digestive System
Abstract
Background & study aims: Bacterial infection is accepted as a precipitating factor in gallstone formation and recent studies have revealed the presence of Helicobacter species in the biliary system. The aim is to determine whether bacterial infections could be detected in bile obtained at ERCP and to evaluate the prevalence of these infections in patients with biliary diseases.
Patients & methods: 102 consecutive patients undergoing ERCP for various indications at Tehran Taleghani Hospital were asked to participate in this study. Bile juice was aspirated after selective cannulation of the common bile duct and stored at -20°C. Each of the patient samples had been tested by PCR on 16s rRNA region and RFLP-DGGE for bacterial infections.
Results: Helicobacter DNA was detected by PCR in bile samples 2 out of 74 with gallstone diseases, 1 out of 15 pancreatobiliary malignancies and 1 out of 13 other benign biliary diseases (p=0.582). Direct sequencing confirmed strains of H. pylori in all four bile samples. Bacteria were detected by the amplification of 16s rRNA 43.2% in gallstone diseases, 53.3% in pancreatobiliary malignancies and 53.8% in other benign biliary diseases (p=0.646).
Conclusion: E.coli, Enterococcus, Klebsiella, Pseudomonas, Acinetobacter and H. pylori were found in the biliary system, suggesting that these bacteria are of etiological importance in gallstone formation and other biliary diseases.
Keywords
Biliary disease; PCR; RFLP-DGGE; Bacterial infection; Helicobacter
Introduction
Detection of microbial agents is an important impress in diseases pathogenesis. Bacterial infections play a principal role in the formation of brown pigmented and pure cholesterol gallstones depends on cholesterol saturation and solubility. Consequently, bacterial infection is now accepted as a precipitating factor in the pathogenesis of mixed cholesterol gallstones. Attempts to culture potentially causative bacteria from gallstones have failed because the formation of gallstones takes a very long time, thus bacteria might be damaged or killed [1]. Swidsinski et al. [2] identified E. coli and Pseudomonas in cholesterol gallstones using PCR and these bacteria were suggested as the pathogens in cholesterol gallstone formation by Lee et al. [3].
Helicobacter pylori (H. pylori) is a Gram-negative, spiral-shaped, motile micro-organism that plays a causative role in the pathogenesis of chronic gastritis, peptic ulcer disease, development of gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma in humans. Although H. pylori is recognized as a pathogen associated with gastric lesions, recent studies have revealed the presence of several Helicobacter species in the hepatobiliary system [4-7]. In animals, Helicobacter species have been isolated from sites including the gastrointestinal tract, liver and biliary tree. H. hepaticus and H. bilis have been detected in liver tissue from mice with chronic active hepatitis and hepatocellular cancer [2,3]. Similarly, H. canis has been isolated from the liver of a dog with multifocal necrotizing hepatitis [8].
Kawaguchi et al. [9] demonstrated a microorganism resembling H. pylori in a resected gall-bladder specimen by H. pylori urease B gene and immunohistochemical staining and bile-resistant hepatic Helicobacter species such as H. bilis, H. pullorum, and F. rappini were extracted from gallbladder mucosa and bile juice of patients with chronic cholecystitis, suggesting that these agents may be key elements in the development of various biliary diseases, especially cancer [10]. H. pylori DNA in gallstone was detected by PCR in several reports [11-13]. The discovery of H. pylori in bile juice has led to the suggestion that Helicobacter species are etiological agents in gallstone formation [14-16]. However, one study in Germany reported no association of Helicobacter species with gallstone formation suggesting possible regional and ethical differences [17].
The16s rRNA is the most conserved (least variable) gene in all cells. Portions of the rDNA sequence from distantly-related organisms are remarkably similar. This means that sequences from distantly related organisms can be precisely aligned, making the true differences easy to measure. For this reason, genes that encode the rRNA (rDNA) have been used extensively to determine taxonomy, phylogeny and to estimate rates of species divergence among bacteria [18].
The aim of this study was to determine the frequency of bacterial infection using PCR and DNA sequencing in bile obtained from endoscopic retrograde cholangiopancreatography (ERCP) and the evaluation of the potential pathogenic role of Helicobacter species in biliary diseases in Iranian patients.
Patients and Methods
Clinical subjects
102 consecutive patients (≥18 years) undergoing ERCP for various indications at Tehran Taleghani Hospital were asked to participate in this study from March 2010 to September 2011. Subjects were excluded if they were taking antibiotics during the previous four weeks. Patients with history of recent pancreato-biliary infections (≤6 months), renal and hepatic impairment were not enrolled. All patients signed an informed consent form. This research was approved by the Ethical Committee of Research Center for Gastroenterology and Liver Disease in Shahid Beheshti University of Medical Sciences.
Patient demographics and diagnoses were recorded and bile was collected by aseptic aspiration after selective cannulation of the common bile duct. Bile samples stored at -20°C and each of the patient samples will be tested by 16s rRNA universal and specific PCR for bacterial identification. The frequencies of 16s rRNA will be confirmed by sequencing and identification of the bacterial genus will be determined by RFLP-DGGE (Restriction Fragment Length Polymorphism & Denaturing Gradient Gel Electrophoresis).
DNA extraction and amplification
1 mL of each bile samples was pelleted by centrifugation for 15 minutes at 14,000 rpm. Then DNA was extracted with phenolchloroform. To amplify DNA of eubacteria and Helicobacter species from the bile samples, the PCR method was used. To obtain higher amounts of DNA, the products were re-amplified with nested PCR primers (Tables 1, 2). Primer set 27F/1492R was used in separate reactions to amplify extracted rRNA genes. The PCR products were denatured at 94°C for 5 minutes, elongated at 30 cycles of 94°C for 30 seconds and 56°C for 30 seconds; and a final extension of 72°C for 10 minutes.
| Primers name | Primer sequence (5'to3') | Size (bps) |
|---|---|---|
| R | CCGTCAATTCTTTGAGTTT | ~1500 bp |
| GM5F-G-C-clamp | G-C-clamp-ACGGGAGGCAGCAG | ~1172 bp |
G-C-clamp:5’-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG- 3’
Table 1: Primers for Bacterial 16S rRNA.
| Bacterial Primers | Primer sequence (5'to3') | Genes | Size (bps) |
|---|---|---|---|
| Helicobacter species | F GGCTATGACGGGTATCCGGC R GCCGTGCAGCACCTGTTTTC |
16S rDNA | 764 |
| H.pylori | F GGATAAGCTTTTAGGGGTGTTAGGGG R GCTTACTTTCTAACACTAACGCGC |
ureC (glm) | 296 |
Table 2: Primers for Helicobacter species.
Final nested PCR products were electrophoretically separated in a 1% agarose gel, then the DNA band from the agarose gel was cut by Sma I (from Serratia marcescens S) enzyme (Roche, Germany) and stained with ethidium bromide. Resulting sequences were compared to databases accessed through the NCBI (National Center for Biotechnology Information) server.
Statistical analysis
Statistical analysis was performed with Chi-square test as well as Fisher’s exact test, and one-way analysis of variance (ANOVA) test. P values of 0.05 or less were considered statistically significant. All the data were analyzed using SPSS 16 for Windows (SPSS Inc., Chicago, IL, USA) and the values were expressed as mean ± standard deviation (SD) for continuous variables and percentages for categorical variables.
Results
One hundred and two patients were included in this study. Fiftytwo patients (50.98%) were female, and the age ranged from 18 to 92 years (mean, 59.5 ± 16.6 years). The diagnoses of these patients are listed in Table 3. We divided the patients in 3 groups based on diagnosis; Group A: 74 patients (72.6%) with stones including CBD (Common Bile Duct) stones only, Gallbladder and CBD stones and microlithiasis. Group B: 15 patients (14.7%) with malignancy including cholangiocarcinoma, ampullary carcinoma, pancreatic carcinoma and duodenal adenocarcinoma. Group C: 13 patients (12.7%) with other benign diseases including SOD (Sphincter of Oddi Dysfunction) and PSC (Primary Sclerosing Cholangitis). There were no statistical differences among the study groups in age, gender and body mass index (BMI) (Table 3).
| Diagnosis | n | Female (%) | Age(years) | BMI(kg/m2) |
|---|---|---|---|---|
| Group A: Stones CBD only Gall-bladder & CBD stones CBD microlithiasis |
74 19 44 11 |
41 (55.4%) 14 (73.6%) 23 (52.2%) 4 (36.3%) |
60.8 ± 16.7 57.1 ± 17.3 62.8 ± 16.3 58.9 ± 17.7 |
23.38 ± 2.49 23.67 ± 2.93 23.40 ± 2.35 22.84 ± 2.33 |
| Group B: Malignancy Cholangiocarcinoma Pancreatic carcinoma Duodenal adenocarcinoma Ampullary carcinoma |
15 6 6 1 2 |
4 (26.7%) 2 (33.3%) 1 (16.7%) 0 1 (50%) |
60.8 ± 14.5 62.8 ± 9.2 63.5 ± 16.3 75.0 40.0 ± 8.4 |
22.21 ±1.92 21.75 ± 1.85 21.92 ± 2.19 23.91 23.57 ± 1.55 |
| Group C: Other Benign Dis. SOD PSC |
13 8 5 |
7 (53.8%) 6 (75%) 1 (20%) |
50.4 ± 16.7 56.5 ± 18.1 40.8 ± 8.4 |
23.90 ± 1.74 24.32 ± 0.98 23.22 ± 2.55 |
| p. value | - | 0.124 | 0.111 | 0.270 |
| Total | 102 | 52 (50.9%) | 59.5 ± 16.6 | 23.28 ± 2.36 |
Table 3: Diagnoses and characteristics of study subjects in different groups.
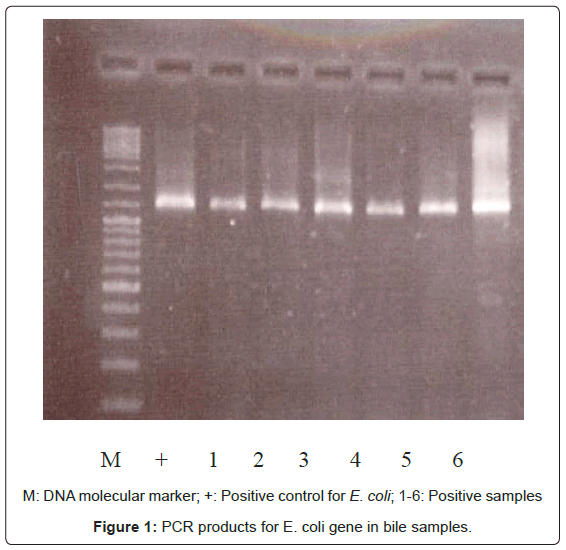
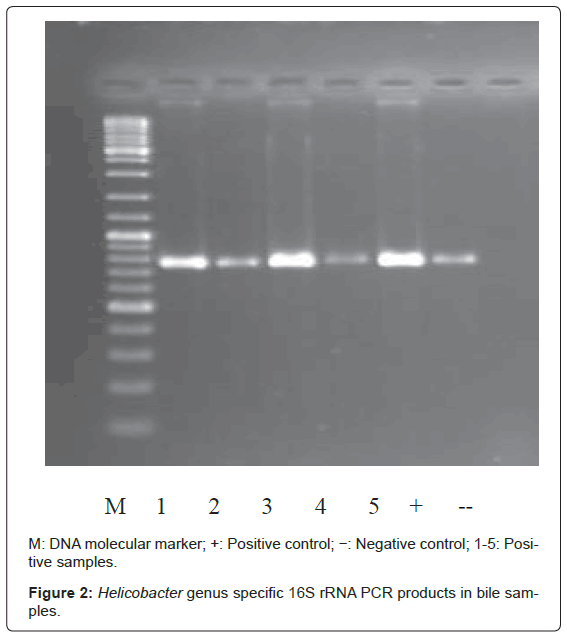
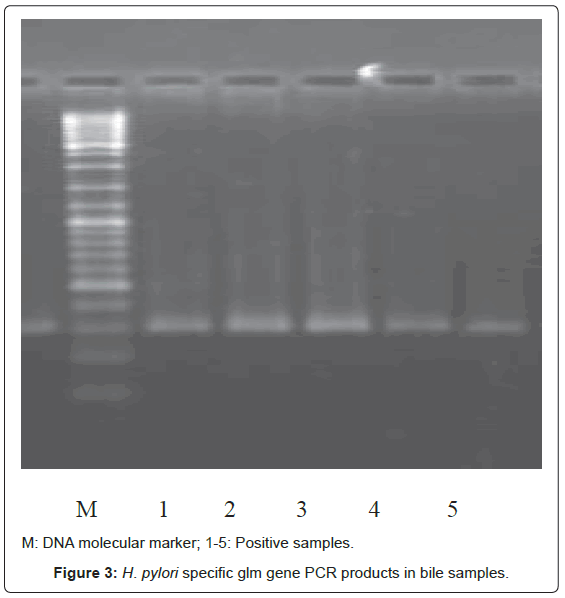
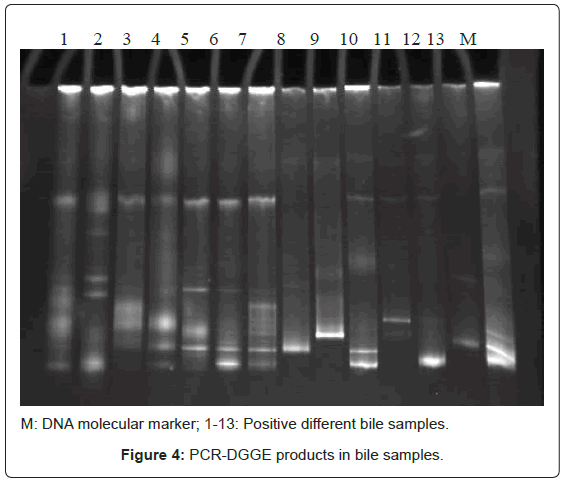
Bacterial DNA was detected in 47 (46.1%) of 102 samples of bile juice by PCR assay (Figure 1). Additional PCR assays were done on the 47 positive bacterial DNA bile samples using Helicobacter genusspecific primers. Four samples had positive PCR for Helicobacter species (Figure 2). Helicobacter pylori-specific primer pairs HPF and HPR were used to identify H. pylori by PCR, and generate amplicons of approximately 296 bases for H. pylori ureC genes (glm region) in 4 samples positive for Helicobacter 16S rRNA (Figure 3). All of them (4 of 4) were positive for H. pylori ureC genes (glm). For detection of different bacterial isolates, molecular typing with RFLP-DGGE method was done in 47 positive bile samples for bacterial DNA (Figure 4) and 51 bacterial isolates were found in 47 positive samples (Table 4), including E.coli (20.6%), Enterococcus (11.7), Klebsiella (9.8%), Pseudomonas (9.8%), Acinetobacter (3.9%) and H. Pylori(3.9%). There was no difference in the identification of bacterial DNA and Helicobacter species among the groups based on diagnoses (Table 4).
| Bacterial isolates | Group A Stones n=74 |
Group B Malignancy n=15 |
Group C Other Benign Dis. n=13 |
p. value |
|---|---|---|---|---|
| Positive PCR | 32 (%43.2) | 8 (%53.3) | 7 (%53.8) | 0.646 |
| E. coli | 16 (%21.6) | 2 (%13.3) | 3 (%23.1) | 0.748 |
| Enterococcus spp. | 10 (%13.5) | 1 (%6.6) | 1 (%7.6) | 0.801 |
| Klebsiella Pneumonia | 7 (%9.4) | 2 (%13.3) | 1 (%7.6) | 0.728 |
| Pseudomonas | 7 (%9.4) | 2 (%13.3) | 1 (%7.6) | 0.866 |
| Acinetobacter spp. | 3 (%4.1) | 0 (%0) | 1 (%7.6) | 0.575 |
| H. Pylori | 2 (%2.7) | 1 (%6.6) | 1 (%7.6) | 0.582 |
Table 4: Bacterial isolates among the groups based on diagnoses.
Seventy-four gallstones were classified by their gross appearance and composition of cholesterol, resulting in 63.5% cholesterol gallstones, 31.1% black pigmented stones and 5.4% brown pigmented stones. There was no difference in the isolation of bacterial DNA and Helicobacter species among the groups based on gallstone composition (Table 5).
| Bacterial isolates | Cholesterol gallstones n=47 |
Black pigmented stones n=23 |
Brown pigmented stones n=4 |
p. value |
|---|---|---|---|---|
| Positive PCR | 19 (%40.4) | 12 (%52.1) | 1 (%25) | 0.486 |
| E. coli | 9 (%19.1) | 6 (%26.1) | 1 (%25) | 0.792 |
| Enterococcus spp. | 5 (%10.6) | 4 (%17.3) | 1 (%25) | 0.583 |
| Klebsiella Pneumonia | 5 (%10.6) | 2 (%8.6) | 0 (%0) | 0.775 |
| Pseudomonas | 3 (%6.3) | 3 (%13.1) | 1 (%25) | 0.369 |
| Acinetobacter spp. | 1 (%2.1) | 2 (%8.6) | 0 (%0) | 0.388 |
| H. Pylori | 2 (%4.2) | 0 (%0) | 0 (%0) | 0.554 |
Table 5: Bacterial isolates among the groups based on gallstone composition.
Discussion
The biliary system is thought to be sterile but this sterility could be broken under certain conditions. Two major routes of infection are ascending through the sphincter of Oddi and descending through the portal system [19,20]. For example, bacterial infection was found in bile (20%) and the liver (17%) on post-operative individuals without any hepato-biliary abnormalities. Also, patients underwent non-biliary abdominal surgery had a 32% positive rate of bacterial infection in their portal blood [21,22]. Leung et al. [23] found bacteria in 84% of the inner cut surface of the pigmented gallstones using electron microscopy but not cholesterol stones.
Lee et al. [1] extracted bacterial DNA from the specimens of gallstones, bile juice and Gall-bladder (GB) mucosa at cholecystectomy. Bacterial DNA was positive in 69.4% of the mixed cholesterol stones compared with 10% of pure cholesterol stones. Helicobacter DNA was detected in 4 out of 58 gallstones, 6 out of 48 bile samples and 5 out of 46 gallbladder specimens. Almost all mixed-cholesterol gallstones appear to harbor bacterial DNA, predominantly E. coli and identified the DNA of Helicobacter species in 27.7% of the GB mucosa, 25% of the bile juice, and 11.4% of the gallstones.
Similarly, in our study, we extracted bacterial DNA from the specimens of bile juice samples %43.2 in stone group, %53.3 in malignant group and %53.8 in other benign group including E.coli (20.6%), Enterococcus (11.7%), Klebsiella (9.8%), Pseudomonas (9.8%), Acinetobacter (3.9%) and H. Pylori (3.9%) without any difference in the identification of bacterial DNA and Helicobacter species among the groups based on diagnoses. The exact mechanism by which bacterial infections contribute to cholesterol gallstones is not known, bacterial biofilm is suggested to play a role as a nucleation factor in cholesterol gallstone formation, like a mucin. Other causative factors are changes of bile composition and bacterial phospholipase, excessive mucin production of Gall-bladder epithelial cells triggered by bacterial lipopolysaccharides [1].
More than 25 species of Helicobacter have been found and these microorganisms have been caused various diseases of the stomach, intestine and the liver in mammals. Hepatitis and hepatomas are caused by H. hepaticus in mice [24-26]. H. pulorum, Flexispira rappini, and H.canis are isolated in diarrheal patients, showing the possibility of zoonosis [27]. H. pylori is sensitive to bile acid and it is difficult to grow in bile juice but the extraction of bile-resistant Helicobacter species from animals bile showed that F. rappini, H. hepaticus, H. bilis, H. canis, H. cholecystus, and H. pullorum may be able to grow and survive in bile juice [21].
Lin et al. investigated the presence of urease A gene in bile sampled by the percutaneous transhepatic route, 3 out of 7 cases showed positive findings, suggesting that H. pylori might be the cause of subclinical cholangitis [14]. Figura et al. [28] implicated H.pylori as a precipitating factor in gallstone formation by identifying H. pylori antibodies in the bile juice of gallstone patients. de Martel et al. [13] conducted a literature review of the epidemiological evidence linking the presence of Helicobacter species in bile or biliary tract (BT). In 4 of 9 studies Helicobacter species were detected in patients with BT cancer significantly more frequently than in controls. In two studies, no Helicobacter species were detected in either cases or controls. Helicobacter species were also often detected in benign BT diseases such as gallstone disease or chronic cholecystitis [13].
Hamada et al. [29] collected 126 bile samples from patients with cholelithiasis, cholecystitis, gallbladder polyp and other non-biliary diseases. H. hepaticus was detected in bile samples with nested PCR whereas H. bilis was not. IgG antibodies to H. hepaticus were detected by Western blotting. Helicobacter hepaticus was detected in 32% of total samples. Patients with cholelithiasis (41%) and cholecystitis with gastric cancer (36%) had higher prevalence of H. hepaticus infection than samples from patients with other diseases [29]. Kobayashi et al. [30] examined 57 bile samples from 30 patients with benign biliary diseases, 6 malignant biliary diseases and 21 non-biliary diseases. Helicobacter genus DNA (shorter amplicons, 400 bp) was statistically frequently detected in bile samples from 53% (16/30) and 86% (5/6) of benign and malignant biliary diseases, compared with 9% (2/21) of nonbiliary diseases, but longer amplicons (1200 bp) were not detectable in any samples. The H. pylori urease A gene (nested amplicon) was also frequently found in bile, whether benign, malignant, or control, though neither H. pylori 16S rRNA nor the 26K protein gene was detectable in any bile samples. H. bilis-16S rRNA genes were detectable in only two cases. H. hepaticus was not detectable in any samples [30].
In study of Fallone et al. [7] bile juice was collected from 125 patients with various hepato-biliary diseases (75 with biliary stones, 15 with pancreatico-biliary malignancies and 4 with primary sclerosing cholangitis) undergoing ERCP. All bile samples of 122 patients with hepato-biliary diseases were negative for Helicobacter DNA. In a similar German study no Helicobacter species were found in bile juice, suggesting that there may be racial and demographic differences [17].
Nilsson et al. [31] have also studied the relationship between H. pylori and primary biliary cirrhosis and primary biliary sclerosing cholangitis. H. pylori was found in the liver tissue but H. bilis, H. pullorum and H. hepaticus was not [31]. In our study, we found the H. pylori from only one of five samples in the patients with PSC.
In summary, RFLP-DGGE is very accurate method but there are many limitations to identify all the organisms in cases of infection with multiple Helicobacter subtypes. Although disadvantage of lack of comparative healthy control group, we could identify bacteria including E.coli, Enterococcus, Klebsiella, Pseudomonas, Acinetobacter and H. pylori in bile juice samples of Iranian patients with biliary diseases. It may be a just innocent bystander, but bacterial infection is of etiological importance in benign and malignant biliary diseases. In future, researches will be needed to show the route of H. pylori infection in biliary diseases and the effect of bacterial eradication on the development of these diseases.
Acknowledgements
The study was supported by the Shahid Beheshti University of Medical Sciences. We wish to thank all the researchers who took part in this research project.
References
- Lee JW, Lee DH, Lee JI, Jeong S, Kwon KS, et al. (2010) Identification of Helicobacter pylori in Gallstone, Bile, and Other Hepatobiliary Tissues of Patients with Cholecystitis. Gut Liver 1: 60-67.
- Swidsinski A, Ludwig W, Pahlig H, Priem F (1995) Molecular genetic evidence of bacterial colonization of cholesterol gallstones. Gastroenterology 108: 860-864.
- Lee DK, Tarr PI, Haigh WG, Lee SP (1999) Bacterial DNA in mixed cholesterol gallstones. Am J Gastroenterol 94: 3502-3506.
- Graham DY (1991) Helicobacter pylori: its epidemiology and its role in duodenal ulcer disease. J Gastroenterol Hepatol 6: 105-113.
- Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, et al. (1991) Helicobacter pyloriinfection and the risk of gastric carcinoma. N Engl J Med 325: 1127-1131.
- Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, et al. (2001) Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 345: 784-789.
- Fallone CA, Tran S, Semret M, Discepola F, Behr M, et al. (2003) Helicobacter DNA in bile: correlation with hepato-biliary diseases. Aliment Pharmacol Ther 17: 453-458.
- Fox JG, Yan LL, Dewhirst FE, Paster BJ, Shames B, et al. (1995) Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol 33: 445-454.
- Kawaguchi M, Saito T, Ohno H, Midorikawa S, Sanji T, et al. (1996) Bacteria closely resembling Helicobacter pylori detected immunohistologically and genetically in resected gallbladder mucosa. J Gastroenterol 31: 294-298.
- Fox JG, Dewhirst FE, Shen Z, Feng Y, Taylor NS, et al. (1998) Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology 114: 755-763.
- Monstein HJ, Jonsson Y, Zdolsek J, Svanvik J (2002) Identification of Helicobacter pylori DNA in human cholesterol gallstones. Scand J Gastroenterol 37: 112-119.
- Abayli B, Colakoglu S, Serin M, Erdogan S, Isiksal YF, et al. (2005) Helicobacter pylori in the etiology of cholesterol gallstones. J Clin Gastroenterol 39: 134-137.
- de Martel C, Plummer M, Parsonnet J, van Doorn LJ, Franceschi S (2009) Helicobacter species in cancers of the gallbladder and extrahepatic biliary tract. Br J Cancer 100: 194-199.
- Lin TT, Yeh CT, Wu CS, Liaw YF (1995) Detection and partial sequence analysis of Helicobacter pylori DNA in the bile samples. Dig Dis Sci 40: 2214-2219.
- Monti J, Fay M, Banchio C, Améndola R, Farías R, et al. (1999) [Detection of Helicobacterpyloriby polymerase reaction in bile samples from gallbladder and bile stones]. Acta Gastroenterol Latinoam 29: 251-253.
- Neri V, Margiotta M, de Francesco V, Ambrosi A, Valle ND, et al. (2005) DNA sequences and proteic antigens of H. pyloriin cholecystic bile and tissue of patients with gallstones. Aliment Pharmacol Ther 22: 715-720.
- Rudi J, Rudy A, Maiwald M, Stremmel W (1999) Helicobactersp. are not detectable in bile from German patients with biliary disease. Gastroenterology 116: 1016-1017.
- Boye K, Høgdall E, Borre M (1999) Identification of bacteria using two degenerate 16s rDNA sequencing primers. Microbiol Res 154: 23-26.
- Csendes A, Fernandez M, Uribe P (1975) Bacteriology of the gallbladder bile in normal subjects. Am J Surg 129: 629-631.
- Csendes A, Becerra M, Burdiles P, Demian I, Bancalari K, et al. (1994) Bacteriological studies of bile from the gallbladder in patients with carcinoma of the gallbladder, cholelithiasis, common bile duct stones and no gallstones disease. Eur J Surg 160: 363-367.
- Dye M, MacDonald A, Smith G (1978) The bacterial flora of the biliary tract and liver in man. Br J Surg 65: 285-287.
- Schatten WE, Desprez JD, Holden WD (1955) A bacteriologic study of portal-vein blood in man. AMA Arch Surg 71: 404-409.
- Leung JW, Sung JY, Costerton JW (1989) Bacteriological and electron microscopy examination of brown pigment stones. J Clin Microbiol 27: 915-921.
- Fox JG, Dewhirst FE, Tully JG, Paster BJ, Yan L, et al. (1994) Helicobacter hepaticussp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol 32: 1238-1245.
- Parsonnet J, Hansen S, Rodriguez L, Gelb AB, Warnke RA, et al. (1994) Helicobacter pyloriinfection and gastric lymphoma. N Engl J Med 330: 1267-1271.
- Ward JM, Anver MR, Haines DC, Benveniste RE (1994) Chronic active hepatitis in mice caused by Helicobacter hepaticus. Am J Pathol 145: 959-968.
- Stanley J, Linton D, Burnens AP, Dewhirst FE, On SL, et al. (1994) Helicobacter pullorumsp. nov.-genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology 140: 3441-3449.
- Figura N, Cetta F, Angelico M, Montalto G, Cetta D, et al. (1998) Most Helicobacter pylori-infected patients have specific antibodies, and some also have H. pyloriantigens and genomic material in bile: is it a risk factor for gallstone formation? Dig Dis Sci 43: 854-862.
- Hamada T, Yokota K, Ayada K, Hirai K, Kamada T, et al. (2009) Detection of Helicobacter hepaticus in Human Bile Samples of Patients with Biliary Disease. Helicobacter 14: 545-551.
- Kobayashi T, Harada K, Miwa K, Nakanuma Y (2005) Helicobacter Genus DNA Fragments Are Commonly Detectable in Bile from Patients with Extrahepatic Biliary Diseases and Associated with Their Pathogenesis. Dig Dis Sci 50: 862-867.
- Nilsson HO, Taneera J, Castedal M, Glatz E, Olsson R, et al. (2000) Identification of Helicobacter pylori and other Helicobacter species by PCR, hybridization, and partial DNA sequencing in human liver samples from patients with primary sclerosing cholangitis or primary biliary cirrhosis. J Clin Microbiol 38: 1072-1076.
Relevant Topics
- Constipation
- Digestive Enzymes
- Endoscopy
- Epigastric Pain
- Gall Bladder
- Gastric Cancer
- Gastrointestinal Bleeding
- Gastrointestinal Hormones
- Gastrointestinal Infections
- Gastrointestinal Inflammation
- Gastrointestinal Pathology
- Gastrointestinal Pharmacology
- Gastrointestinal Radiology
- Gastrointestinal Surgery
- Gastrointestinal Tuberculosis
- GIST Sarcoma
- Intestinal Blockage
- Pancreas
- Salivary Glands
- Stomach Bloating
- Stomach Cramps
- Stomach Disorders
- Stomach Ulcer
Recommended Journals
Article Tools
Article Usage
- Total views: 14275
- [From(publication date):
May-2012 - Sep 25, 2024] - Breakdown by view type
- HTML page views : 9829
- PDF downloads : 4446