Research Article Open Access
A Phenological Mid-Domain Analysis of Non-Native and Native Species Recruitment Richness
James F. Reinhardt1*, Richard W. Osman2 and Robert B. Whitlatch31Earth Resources Technologies, NOAA Restoration Center, 1315 East-West Highway, Silver Spring, MD 20910, USA
2Smithsonian Environmental Research Center, 647 Contees Wharf Road, Edgewater, MD 21037, USA
3Department of Marine Sciences, University of Connecticut, 1080 Shennecossett Road, Groton, CT 06340, USA
- *Corresponding Author:
- James F. Reinhardt
Earth Resources Technologies
NOAA Restoration Center
1315 East-West Highway
Silver Spring, MD 20910, USA
E-mail: JamesFReinhardt@hotmail.com
Received date: August 11, 2012; Accepted date: September 24, 2012; Published date: September 26, 2012
Citation: Reinhardt JF, Osman RW, Whitlatch RB (2012) A Phenological Mid-Domain Analysis of Non-Native and Native Species Recruitment Richness. J Ecosyst Ecogr 2:118. doi:10.4172/2157-7625.1000118
Copyright: © 2012 Reinhardt JF, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and and source are credited.
Visit for more related articles at Journal of Ecosystem & Ecography
Abstract
A mid-domain model was used to examine differences in patterns of species recruitment diversity between native and non-native species groups within shallow sub-tidal epifaunal communities of southern New England. A mid-domain model is a stochastic null model that predicts species richness patterns within a bounded domain. Deviations from the mid-domain model were examined to assess: 1) if each species group’s recruitment diversity differed from the mid-domain model; and 2) if there were differences in the patterns of deviation between groups. In general, the shape of mid-domain null was correlated to empirical patterns of recruitment diversity (i.e., mid-summer peaks in recruitment diversity) although among-year variation occurred in both the pattern of recruitment diversity and the level of fit. Akiake’s information criterion was used to determine if the mid-domain effect contributed to linear regression model predictions of phenological species recruitment patterns. We found distinct differences between the level of fit between native and non-native species groups. Native species did not significantly differ from the mid-domain null in five of seven years examined, while non-native species were significantly different in all years but two. Non-native species also showed a much weaker correlation to the mid-domain model and the seasonal water temperature signal. Collectively, this evidence supports the hypothesis that native and non-native species have different factors controlling the timing of their recruitment. Mid-domain nulls proved to be useful in the analysis of species richness patterns along temporal axes and provide a valuable baseline for further analysis.
Keywords
Native species; Non-native species; Mid-domain model; Species recruitment diversity; Water temperature signal; Phenological species
Introduction
Recruitment phenology, or the timing of seasonally recurring reproductive events, may influence intra- and inter-specific competition, food availability, predation rates, and survivorship of organisms [1-4] and ultimately the diversity of organisms present in a system. For marine benthic sessile species that permanently attach to hard substrates such as rocks, boulders, or reefs, the timing of recruitment may influence inter-specific competition for space [5] because these species often rely on preemptive colonization or priority to take and hold space. Changes to and differences in the timing of recruitment may also benefit non-native species over native species in habitats with significant levels of disturbance [6] if it enables them to recruit before native species.
Many studies have examined the temporal overlap of seasonally recurring events, particularly flowering times of plants [7], arrival and breeding times of birds [4] and the development and hatching of invertebrate larvae [1-4]. Organisms often exhibit seasonality because there is a limited period of time in that environmental conditions are optimal for certain life history events (e.g., time to maturity, spawning activity, recruitment). Relative changes in the phenology of organisms can lead to severe consequences, as exemplified by the match-mismatch hypothesis, when food available to predator organisms is reduced because of a shift in the relative timing between predators and prey [8]. Within a season, however, similar species also show differences in their phenologies, either because of differences in optimal conditions or from interspecific interactions such as competition or facilitation. Explanations for the difference in phenological timing between similar species of plants, invertebrates and birds include competition for food, pollinators, and seed dispersers [4,9-11]. These explanations most often invoke physical, genetic and ecological mechanisms to explain phenology patterns. Regardless, in all cases, an appropriate null model is necessary to adequately test patterns in phenology and to determine if patterns vary from random expectation.
A mid-domain model is a stochastic null model that predicts species richness patterns within a bounded domain. The concept of the mid-domain effect was originally developed to understand patterns and mechanisms of diversity over latitudinal gradients [13]. The model randomly assigns each species’ known or measured range a mid-point within the range. The characteristic hump-shaped pattern resulting from the null model has been termed the mid-domain effect [12]. The concept has been tested for species patterns in a broad variety of domains, including river and marine systems, time, and island to continental-scale latitudinal gradients [14-19]. However, the evaluation of the mid-domain effect along temporal domains remains relatively poorly studied see [17,20].
Mid-domain models can serve as an excellent starting point for examining patterns of species richness, because they can establish a random expectation for richness patterns and are able to consider multiple species or groups simultaneously. Species richness patterns are one of the most commonly studied patterns in ecology. These patterns are often used to characterize habitats along spatial or temporal (e.g., succession) gradients. Although, mid-domain models have instigated considerable debate over the underlying mechanism (or lack thereof) contributing to species richness patterns, it is important and often instructive to understand under what circumstances communities deviate from the mid-domain expectations [12]. Differences in deviations from a null model between sets (or groups) of organisms may indicate underlying mechanisms that create patterns [16,17,21]. Alternatively, one can utilize mid-domain models to predict the expected contribution of geometric constraints on range location to species richness curves as one factor among many [22]. For example, factors such as energy or productivity (along latitudinal or temporal gradients) may contribute to species richness in that greater productivity will allow for more individuals, larger populations, perhaps allowing lower extinction rates, and therefore, more species [23-26]. The inclusion of geometric constraints (i.e., mid-domain effect) is useful when it improves model predictions of species richness patterns in conjunction with other predictors.
In the recent past, increases in species invasions have altered the distribution of species and their ranges. In some cases, species introductions have led to increased species diversity [27]; while in others they have caused native species extinction [28]. A general concern is that an increased rate of species introductions is causing a global homogenization of flora and fauna [29]. The consequences of species invasions are not fully understood but can often be disastrous [30]. In southern New England, new invaders to the shallow epifaunal community can rapidly dominate habitat and exclude native organism from primary substrate [31]. Many introductions into the hard substrate epifaunal community have led to large scale changes in trophic structure [32] and have resulted in whole-sale shifts in community structure [33]. One of the most fundamental questions ecologists pose is what makes a species a successful invader.
To better understand patterns of species recruitment and differences in recruitment between native and non-native species, we examined the explanatory potential of the mid-domain effect for inter-annual patterns of recruitment for marine benthic species using a 7-year data set. The effects of geometric constraints were assessed for recruitment of all species and then for the subsets of both non-native and native species for each year. The deviations from the mid-domain expectations were examined and compared to assess: 1) if each group’s recruitment diversity differed from the mid-domain expectations; 2) if there were differences in patterns of deviation between species groups; and 3) if the mid-domain effect makes a significant contribution to predictions of species richness patterns using model selection guided by Akaike’s information corrected criterion (AICc). By examining differences in recruitment patterns insight was gained regarding the possible reasons for invasion success, while serving as a starting point for examining the utility of defining a truncated temporal domain for the hard substrate assemblages.
Methods
Data collection
Recruitment data from 2002-2008 were collected in Avery Point, CT, USA at the University of Connecticut Marine Science docks. Four 10x10cm roughened PVC panels were suspended facing the seafloor from a floating dock (~1 m below the surface). Panels were collected and replaced by new ones each week. All animals attached to each panel were identified to lowest level of certainty and counted using a dissecting microscope. Species that recruited included 43 species of bryozoans, ascidians, cnidarians, barnacles, polychaetes, and sponges. The species include a number of species of bryozoans and ascidians that have invaded eastern Long Island Sound over the past 35 years (See Appendix 1). The “recruitment period” of each species was defined as the time between the first and last observance of a recruit of that species for each year, within the predetermined domain. We define the domain in this study as the “recruitment season”. The beginning of the recruitment season was (based on the average of 7 years) Julian day, so that the water reached 20 growing degrees days (base 10°C). This time coincided with the rapid increase in recruitment activity of all species (data not shown). The last day of the recruitment season was defined as the average day (of all 7 years) on which growing degree days no longer accumulated. We use recruitment season to refer to the recruitment of all species while a recruitment period refers to the recruitment of a single species. Growing degree days (GDD) were calculated as the average of the daily temperature minimum and daily temperature maximum minus the base temperature. Negative values are excluded and GDD are cumulatively summed throughout a season. The domain used in this study (defined by GDD) corresponded to greater than 90% of the recruitment. This rule was used in order to avoid being completely arbitrary in defining the domain boundaries, but also to limit the domain to a biologically active period of the year. We used the average from all years to minimize correlation between the domain length and annual temperature patterns.
The domain was thus, defined as the recruitment season (from Julian Day 140 to Julian Day 301 [May 19 to October 27]), slightly adjusted for weekly sampling times. Thus, the domain was actually from the first sampling day before day 140 until the last sampling day before day 301. Although, some species recruited throughout the year (albeit at very low densities; <5 recruits 100 cm-2 wk-1) the domain was truncated to 21-23 weeks (depending upon the year) as 90% of the recruitment of all species occurred during this period.
Model and statistical analysis
A variety of mid-domain models have been proposed and used to analyze species richness patterns. The mid-domain model used for this study has previously been applied to flowering phenologies of alpine annual plants [17] and assumes a continuous recruitment period. In this model, the mid-point of a species’ recruitment period was randomly placed within the domain, while the length of the recruitment period remained fixed at its observed value. For example, if a species’ recruitment period extended the entire length of the recruitment season (or domain) there was only one possible mid-point for that species. If a species’ recruitment period was only one week then its mid-point could be anywhere in the domain. The possible number of mid-points is, therefore, an inverse function of the length of each species’ specific recruitment period. This random placement of a species recruitment range was done for each species and for all years, to generate random species richness curves. The process of assigning random mid-points was done 1000 times to generate the species recruitment curve expected under this model, with 95% confidence bounds around the curves.
To test if recruitment species richness was significantly different from the pattern expected under the mid-domain null model, we utilized a random re-sampling technique that tested if the observed richness fit within the 95% confidence limits of the model [34]. This method is useful to test whether empirical patterns show departures from the null model. This procedure was performed for each year for each of the three groups of species (all, native, and non-native). In a second analysis, observed species richness patterns were correlated with both the species richness pattern produced by the model and seawater temperatures (7-day moving average), to obtain a measure of collinearity between the mid-domain model and temperature. Temperature data were collected using HOBO TidBits© temperature loggers located adjacent to the recruitment panels. As there were some collection gaps caused by equipment loss, the gaps were filled in by first creating a seasonal water temperature signal by averaging all available water temperature data at Avery Point (AP) by date (from 2002 to 2008). Missing water temperature data were interpolated using multiple regression with the seasonal signal and daily mean air temperature from the Groton Airport (>1 km from the field site) as the independent variables (r2 = 0.97). Air temperature is highly influential on AP temperature data because temperature was collected in the top 1 m of the water column and better reflects the daily variability in surface sea water temperature than sea water temperature taken at distant sites.
Multiple linear regressions, using the Newey-West variancecovariance estimation [35] to correct for auto-correlation in residuals, were used to assess the contribution of geometric constraints (as represented by the MDE null model) to predictions of recruitment species richness patterns. All possible combinations of temperature, the sum of all individual recruits per week, and the mid-domain null were used as predictors of recruitment species richness. We included the weekly sum of all individuals recruiting as an additional parameter because population sizes can influence species richness [23]. Residuals from the regressions were then used to calculate the AICc and ΔAICc (the difference in AICc between a parameter and the parameter with the lowest AICc value). We then calculated the number of times a specific combination of regressors had the lowest ΔAICc over all species groups and years. If the mid-domain model was frequently included in the best linear regression model, this would support the inference that geometric constraints on range location influence patterns of recruitment species richness [22].
Paired t-tests were used to compare the timing of peak species richness between non-native species and native species groups paired by year (years being the replicate). Lastly, correlation coefficients were generated for the residuals of each year and species group to better understand patterns and to look for generalization in species richness across years and species groups. All statistical and null model analysis was done in Mat lab 2009a.
Results
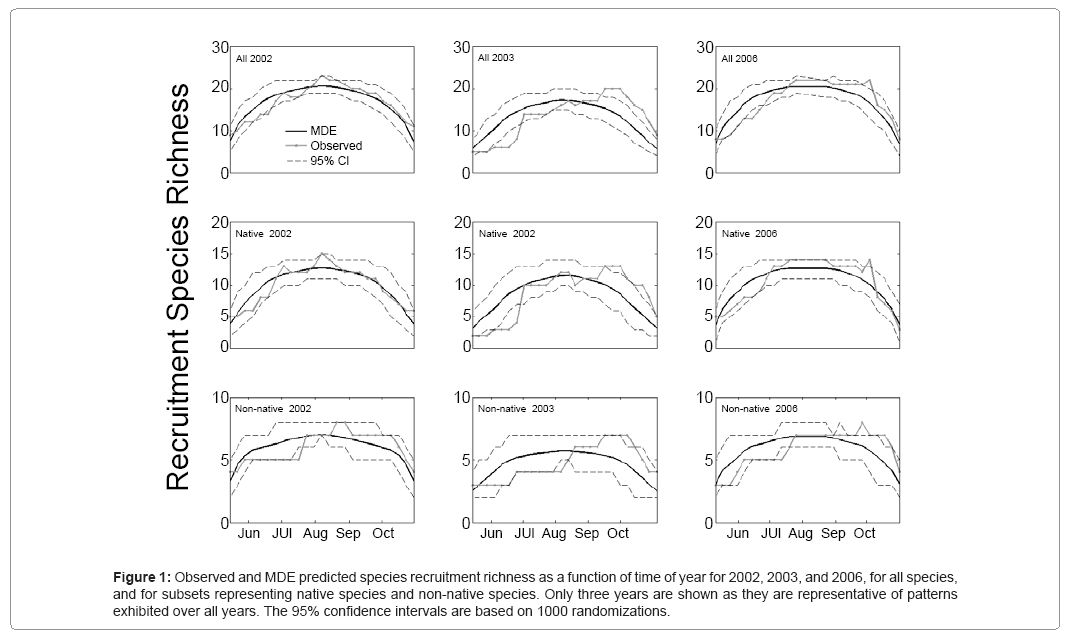
General patterns of species recruitment richness were similar among all species, native and non native species groups. Like parabolic patterns of species richness (Figure1), there was an initial period of increasing recruitment richness, a period of sustained high richness, followed by a period of decreasing recruitment richness. In general, the mid-domain null model was highly correlated to the observed pattern of recruitment richness of all species (averaged across years r = 0.83, S.D. = 0.13). Native species recruitment richness showed an even stronger correlation to the null model (averaged across years r = 0.88, S.D. = 0.12). Non-native species group showed the weakest average correlation between species recruitment diversity and the null model (averaged across years r = 0.60, S.D. = 0.19).
Figure 1: Observed and MDE predicted species recruitment richness as a function of time of year for 2002, 2003, and 2006, for all species, and for subsets representing native species and non-native species. Only three years are shown as they are representative of patterns exhibited over all years. The 95% confidence intervals are based on 1000 randomizations.
On average, species recruitment richness was highest from Julian day 233 to 257 (Aug. 20 to Sep. 13). Native species had their highest richness from Julian day 222 to 243 (Aug. 9 to Aug. 30), while nonnative species showed their highest richness from Julian day 240 to 268 (Aug. 27 to Sep. 24). There were clear differences in the timing of maximum species recruitment richness between native and non-native species groups. Native species recruitment richness tended to reach maximum richness earlier in the season (although not significantly [paired t-test, p = 0.18]) and to end peak diversity periods earlier (paired t-test; p = 0.033).
Although there was generally high correlation between patterns of species recruitment richness and those predicted by the model, there were equally high correlations between the observed recruitment richness and the seawater temperature (Table 1). Similar to correlations with the null model, seawater temperature showed the lowest correlation to species recruitment richness for the non-native species group (compared to non-natives and the all species groups). There was high degree of correlation between temperature and the mid-domain model (Pearson correlation coefficient averaged over years for all, native, and non-native species, respectively; r = 0.94, S.D. = 0.03; r = 0.95, S.D. = 0.03; r = 0.93, S.D. = 0.03).
| Year | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | Mean | S.D. | |
|---|---|---|---|---|---|---|---|---|---|---|
| Annual Mean Temp | 12.86 | 11.4 | 11.91 | 12.57 | 12.77 | 12.27 | 12.32 | 12.3 | 0.51 | |
| All Species | ||||||||||
| r model-observed | 0.93 | 0.54 | 0.82 | 0.91 | 0.83 | 0.92 | 0.85 | 0.83 | 0.13 | |
| r model-temp | 0.97 | 0.93 | 0.93 | 0.98 | 0.97 | 0.89 | 0.94 | 0.94 | 0.03 | |
| r temp-observed | 0.96 | 0.76 | 0.93 | 0.84 | 0.75 | 0.94 | 0.77 | 0.85 | 0.09 | |
| P-value* | 0.28 | 0 | 0.03 | 0.10 | 0.02 | 0.11 | 0.01 | |||
| Native Species | ||||||||||
| r model-observed | 0.95 | 0.62 | 0.89 | 0.94 | 0.92 | 0.95 | 0.86 | 0.88 | 0.12 | |
| r model-temp | 0.98 | 0.94 | 0.93 | 0.98 | 0.98 | 0.89 | 0.95 | 0.95 | 0.03 | |
| r temp-observed | 0.95 | 0.82 | 0.91 | 0.87 | 0.86 | 0.87 | 0.82 | 0.87 | 0.05 | |
| P-value | 0.45 | 0.01 | 0.37 | 0.72 | 0.3 | 0.58 | 0.01 | |||
| Non-native Species | ||||||||||
| r model-observed | 0.59 | 0.31 | 0.47 | 0.75 | 0.51 | 0.82 | 0.77 | 0.60 | 0.19 | |
| r model-temp | 0.92 | 0.91 | 0.93 | 0.99 | 0.96 | 0.89 | 0.91 | 0.93 | 0.03 | |
| r temp-observed | 0.59 | 0.54 | 0.7 | 0.66 | 0.38 | 0.93 | 0.55 | 0.62 | 0.17 | |
| P-value | 0.02 | 0 | 0 | 0.05 | 0 | 0.08 | 0.06 | |||
* P-values calculated by [34]. Significant (α = 0.05) values in bold.
Table 1: Annual mean temperature (°C), and correlation coefficients for 1) predicted and empirical recruitment richness, 2) predicted richness and temperature, and 3) empirical recruitment richness and temperature.
In four out of seven years, there was a significant deviation from the null model when examining all species pooled together (Table 1). The non-native species group showed significant difference from the null in five out of seven years, while the native species group showed significant difference in only two out of seven years (Table 1).
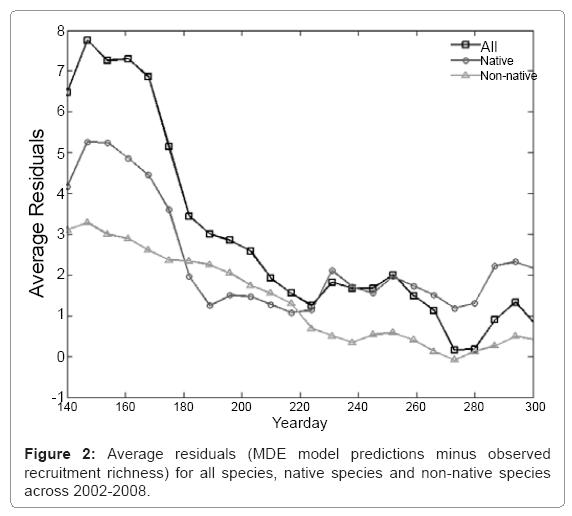
For all species, there was a clear pattern in residuals averaged across years (Figure 2). The all species group had lower richness than predicted by the mid-domain model at the beginning of the season (Figure 1). By the end of the season, for both all species and non-native species groups, residuals trended toward zero, while the native species group often displayed a decrease in recruitment richness that was not predicted by the null model.
Multiple regressions and AICc showed that of 21 multiple linear regression models the mid-domain parameter was included in 13 of the best models; temperature was included in 12 of the best models, and the sum of individuals was included in 7 of the best models (Table 2). Temperature and the MDE were both included in the best model 5 times. When the linear regression models are broken down by species group, the mid-domain parameter is included in 5/7, 3/7 and 5/7 of the best models of all, non-native and native species groups, respectively.
| Year | Treatment | T1 | T M | T M S | M2 | M S | S3 | S T |
|---|---|---|---|---|---|---|---|---|
| 2002 | All | 10.4 | 0 | 0.5 | 27.8 | 27.8 | 49.5 | 11.7 |
| 2003 | All | 10.1 | 0 | 1.8 | 18.9 | 20.7 | 22.3 | 11.4 |
| 2004 | All | 0 | 2 | 3.9 | 3.1 | 5.1 | 24.3 | 1.9 |
| 2005 | All | 9.1 | 10.6 | 1.2 | 9.7 | 0 | 6.6 | 2.9 |
| 2006 | All | 4.9 | 6.3 | 2 | 7.2 | 0 | 13 | 2.6 |
| 2007 | All | 0 | 1.4 | 3.1 | 21.7 | 22.9 | 47.8 | 1.8 |
| 2008 | All | 44.3 | 1 | 2.8 | 0 | 2 | 38.2 | 39.7 |
| Times in the best model | 2 | 2 | 0 | 1 | 2 | 0 | 0 | |
| 2002 | Native | 0.3 | 0 | 0.6 | 8.3 | 8.7 | 34.9 | 1.2 |
| 2003 | Native | 11.1 | 0 | 2 | 20.8 | 21.4 | 23.9 | 13.1 |
| 2004 | Native | 18.4 | 9.4 | 1.8 | 9.1 | 0 | 58.8 | 7.3 |
| 2005 | Native | 2.8 | 1.5 | 2.7 | 0 | 0.7 | 24.9 | 4.7 |
| 2006 | Native | 0 | 0.5 | 2.5 | 2.2 | 3.7 | 36 | 1.9 |
| 2007 | Native | 1.5 | 0 | 1.9 | 5.6 | 7.4 | 46.9 | 3.2 |
| 2008 | Native | 45.3 | 1.8 | 1.4 | 0 | 0.2 | 37.4 | 38.1 |
| Times in the best model | 1 | 2 | 0 | 2 | 1 | 0 | 0 | |
| 2002 | Non-native | 4.3 | 6.2 | 1.6 | 7.1 | 1.6 | 23.1 | 0 |
| 2003 | Non-native | 4.5 | 3.4 | 0.5 | 7.6 | 4.6 | 11.3 | 0 |
| 2004 | Non-native | 0.1 | 1 | 1.5 | 1.6 | 1.7 | 8.1 | 0 |
| 2004 | Non-native | 0.1 | 1 | 1.5 | 1.6 | 1.7 | 8.1 | 0 |
| 2005 | Non-native | 1.3 | 2 | 3.8 | 0 | 1.8 | 6.5 | 3.2 |
| 2006 | Non-native | 0 | 2 | 3.6 | 0.7 | 2.7 | 7.9 | 1.6 |
| 2007 | Non-native | 4.2 | 0 | 0.3 | 21.2 | 23.1 | 38.4 | 4.4 |
| 2008 | Non-native | 27.2 | 2.1 | 1.1 | 1.6 | 0 | 29.2 | 29.2 |
| Times in the best model | 1 | 1 | 0 | 1 | 1 | 0 | 3 | |
| Σ times in the best model | 4 | 5 | 0 | 4 | 4 | 0 | 3 | |
1. Temperature, 2. Mid-domain model, 3. Sum of all individuals 100cm-2 wk-1
Table 2: The ΔAICc for each combination of model parameters.
Discussion
While mid-domain models have been able to indicate significant geometric constraints on patterns of species richness [22], in some cases they have not been able account for such patterns [34]. Studies examining mid-domain effects along temporal domains are less common [17] but are theoretically analogous to studies of geographical mid-domain effects. This study adds to the ongoing discussion of the usefulness of mid-domain models and specifically addresses differences in recruitment patterns between native and non-native species of sessile benthic assemblages found in southern New England coastal waters.
The mid-domain model used in this study was often highly correlated with annual patterns of species recruitment richness with the exception of 2003 when a much weaker correlation was found. This year was notable as it was the coldest year within the entire data set. In general, the mid-domain model was effective in capturing overall seasonal species recruitment trends but did not capture overall intraseasonal patterns as judged by the varying success of the MDE model among years.
We tested whether predictions of species recruitment richness would be enhanced with the inclusion of the mid-domain effect in multiple regressions. More often than not (65% of the time), the middomain parameter was included in the best multiple linear model of recruitment richness (Table 2). In fact, the best regression model included the mid-domain effect more often than temperature or the sum of individual parameters. This result, however, does not discount the overall importance of temperature or the number of individuals in influencing species richness patterns.
Native species and non-native species groups showed strong differences in their agreement with their respective null models. The native species had only two years when the empirical pattern differed significantly from the null model (2003 and 2008), while the nonnative species group differed significantly in five years. The native species group showed a high correlation between the empirical species richness and the null model. The non-native species group was less well correlated. Although, differences in agreement with the null model were more often significant for the non-native species group, both groups showed similar deviations from the null, as indicated by plots of average residuals (Figure 2).
One of the major criticisms of mid-domain models is that nearly all null predictions strongly correlate with other environmental parameters [36]. This study is no exception; the mid-domain prediction was highly correlated with seawater temperature. Colwell et al. [22] countered this criticism by stating that the mid-domain effect should not be treated as an “all or nothing” approach. Rather, the mid-domain effect should be considered as a contributing factor to overall patterns of species richness. Here we show that the mid-domain effect often improved the model performance as indicated by Akaike’s information criterion (AIC). The mid-domain can be a valuable contributor to empirical species richness patterns.
It is interesting to note that in this study native species recruitment diversity correlated well with both the mid-domain prediction and seawater temperature, while the non-native species group did not (except for 2007). This may indicate different mechanisms contributing to patterns of recruitment richness between native and non-native species. The time of maximum native species richness preceded maximum richness of non-native species in all years except for one. Further, the inter-annual average placed the peak in native species recruitment richness 18 days before the peak in non-native species recruitment richness (Table 1). Similarly, the period of maximum native species richness ended before the end of the maximum period for non-native species.
Previous work on the recruitment patterns of non-native species at our study site has shown that onset of recruitment can be regulated by late winter/early spring water temperatures [3]. In contrast, many native species did not show as strong a relationship between recruitment onset and water temperature [3], although the timing of native species recruitment can be predicted by temperature parameters (Reinhardt per observation). Despite this finding, our analysis showed that non-native species recruitment richness is less correlated with patterns of annual water temperature than native species recruitment.
Why do non-native species more often differ from the mid-domain model prediction and native species? Why is there a difference in the timing of peak recruitment richness between native and non-native species? Presumably, native and non-native species have evolved in different regions with different selective pressures. Therefore, factors that cause peaks in non-native species recruitment may be different from those influencing recruitment dynamics of native species. Nonnatives species tend to have peaks of recruitment later in the season. Non-native species may be taking advantage of periods when there is less recruitment by native species in order to avoid interspecific competition for space: a common limiting-resource for most sessile epifaunal species [36]. Recruiting later in the season may also take advantage of natural- and/or human-mediated disturbance regimes, as Altman and Whit latch [6] have shown that non-native species appear to response positively to disturbances while native species do not. Simply, we may see more non-native species recruiting at the end of the season because those are the non-native species that have survived. Their survival may be a result of recruiting later in the season.
It is particularly challenging to define the domain of a temporal middomain model. We used ‘soft’ boundaries based on water temperature. This is different than geographical mid-domain studies that utilize a hard boundary such as the margins of an island [22]. It is often more difficult to objectively define a soft boundary, such as those needed in studies of a temporal axis [37]. To avoid being overly arbitrary, and designing the model to fit the data, our domain was determined a priori based on the average time of the year when growing degreedays accumulated and corresponded to the vast majority (e.g., greater than 90%) of recruitment in the system (Reinhardt per observation). Although it is likely that all species recruitment could potentially fit another mid-domain model better, it would not change the fact that there is persistent disagreement between native and non-native species recruitment richness.
Conclusion
Both temperature and mid-domain models have the potential to match empirical patterns of species recruitment diversity. The middomain effect more often than not was included in the best model of species recruitment richness temperature was included almost as often. Native and non-native species show distinct differences in their agreement with the respective mid-domain models possibly indicating underlying differences between the two species groups. The frequent inclusion of the MDE parameter in linear regressions support the notion that geometric constraints are important to consider in understanding the recruitment phenology in hard substrate epifaunal communities. Mid-domain models can be useful in highlighting differences or similarities between species subsets within a single community, or perhaps between multiple communities as well. When comparing subsets of species, especially on a temporal axis, it may prove useful to determine a priori rules for establishing domain boundaries to avoid some common criticisms of mid-domain models. Utilizing middomain models is useful in the characterization of species diversity patterns along temporal axes by providing a strong null baseline to make comparisons.
Acknowledgements
We would like to thank the numerous interns and graduate students who helped collect the data and Rob Colwell for his helpful comments.
References
- Tokeshi M (1986) Population dynamics, life histories and species richness in an epiphytic chironomid community. Freshw Biol 16: 431-441.
- Hurd LE, Eisenberg RM (1990) Experimentally synchronized phenology and interspecific competition in mantids. American Midland Naturalist 124: 390-394.
- Stachowicz JJ, Terwin JR, Whitlatch RB, Osman RW (2002) Linking climate change and biological invasions: Ocean warming facilitates non indigenous species invasions. PNAS 99: 15497-15500.
- Visser ME, Both C (2005) Shifts in phenology due to global climate change: The need for a yardstick. Proc R Soc B 272: 2561-2569.
- Stachowicz JJ, Byrnes JE (2006) Species diversity, invasion success, and ecosystem functioning: disentangling the influence of resource competition, facilitation, and extrinsic factors. Mar Ecol Prog Ser 311: 251-262.
- Altman S, Whitlatch RB (2007) Effects of small-scale disturbance on invasion success in marine communities. J Exp Mar Bio Ecol 342: 15-29.
- Gotelli NJ, Graves GR (1996) Null Models in Ecology. Smithsonian Institution Press, Washington D.C.
- Stenseth N, Mysterud A (2002) Climate, changing phenology, and other life history traits: nonlinearity and match–mismatch to the environment. PNAS 99: 13379-13381.
- Waser NM (1978) Competition for hummingbird pollination and sequential flowering in two Colorado wildflowers. Ecology 59: 934-944.
- Rathcke B, Lacey EP (1985) Phenological patterns of terrestrial plants. Annual Review of Ecology and Systematics 16: 179-214.
- Rathcke B (1988) Flowering phenologies in a shrub community: competition and constraints. J Ecology 76: 975-994.
- Colwell RK, Lees DC (2000) The mid-domain effect: geometric constraints on the geography of species richness. Trends Ecol Evol 15: 70-76.
- Colwell RK, Hurtt GC (1994) Nonbiological gradients in species richness and a spurious rapoport effect. American Naturalist 144: 570-595.
- Willig MR, Lyons SK (1998) An analytical model of latitudinal gradients of species richness with an empirical test for marsupials and bats in the New World. Oikos 81: 93-98.
- Jetz W, Rahbek C (2001) Geometric constraints explain much of the species richness pattern in African birds. PNAS 98: 5661-5666.
- Connolly SR, Bellwood DR, Hughes TP (2003) Indo-Pacific Biodiversity of Coral Reefs: Deviations from a Mid-Domain Model. Ecology 84: 2178-2190.
- Morales MA, Dodge GJ, Inouye DW (2005) A phenological mid-domain effect in flowering diversity. Oecologia 142: 83-89.
- Dunn RR, Colwell RK, Nilsson C (2006) The river domain: Why are there more species halfway up the river. Ecography 29: 251-259.
- Carranza A, Colwell RK, Rangel TFVB (2008) Distribution of megabenthic gastropods along environmental gradients: the mid-domain effect and beyond. Mar Ecol Prog Ser 367: 193-202.
- Tiwari M, Bjorndal KA, Bolten AB, Bolker BM (2005) Intraspecific application of the mid-domain effect model: spatial and temporal nest distributions of green turtles, Chelonia mydas, at Tortuguero, Costa Rica. Ecology Letters 8: 918-924.
- Jetz W, Rahbek C (2002) Geographic range size and determinants of avian species richness. Science 297:1548-1551.
- Colwell RK, Rahbek C, Gotelli NJ (2004) The mid-domain effect and species richness patterns: what have we learned so far. American Naturalist 163: E1-E23.
- Connell JH, Orias E (1964) The ecological regulation of species diversity. American Naturalist 98: 399-414.
- Pianka ER (1966) Latitudinal diversity in species diversity: A review of concepts. American Naturalist 100: 33-46.
- Waide R, Willig M, Steiner C, Mittelbach G, Gough L, et al. (1999) The relationship between productivity and species richness. Annu Rev Ecol Syst 30: 257-300.
- Currie DJ, Mittelbach H. Cornell, Field R, Guegan J, Hawkins B, et al. (2004) Predictions and tests of climate-based hypotheses of broad-scale variation in taxonomic richness. Ecology Letters 7: 1121-1134.
- Gurevitch J, Padilla DK (2004) Are non-native species a major cause of extinctions. Trends Ecol Evol 19:470-474.
- Fritts TH, Rodda GH (1998) The role of introduced species in the degradation of island ecosystems: A case history of Guam. Annual Review of Ecology and Systematics 29: 113-140.
- Ricciardi A (2007) Are modern biological invasions an unprecedented form of global change. Conservation Biology 21: 329-336.
- Pimental D, Zuniga R, Morrison D (2005) Update on the environmental and economic costs associated with alien-non-native species in the United States. Ecological Economics 52:273-288.
- Reinhardt JF, Stefaniak L, Hudson D, Mangiafico J, Glagych R, et al. (2010) First record of the non-native light bulb tunicate Clavelina lepadiformis (Müller , 1776 ) in the northwest Atlantic. Aquat Invasions 5: 185-190.
- Byrnes JE, Reynolds PL, Stachowicz JJ (2007) Invasions and extinctions reshape coastal marine food webs PLoS ONE 2: e295.
- Dijkstra JA, Harris LG (2009) Maintenance of diversity altered by a shift in dominant species: Implications for species coexistence. Mar Ecol Prog Ser 387: 71-80.
- Veech JA (2000) A null model for detecting nonrandom patterns of species richness along spatial gradients. Ecology 81: 1143-1149.
- Newey WK, West KD (1987) A simple, positive semi-definite, heteroskedasticity and autocorrelation consistent covariance matrix. Econometrica 55: 703-708.
- Currie DJ, Kerr JT (2008) Tests of the mid-domain hypothesis: A review of the evidence. Ecol Monogr 78:3-18.
- Jackson JB (1977) Competition on Marine Hard Substrata: The adaptive significance of solitary and colonial strategies. American Naturalist 111: 743-767.
- Zapata FA, Gaston KJ, Chown SL (2005) The mid-domain effect revisited. American Naturalist 166: E144-148.
Relevant Topics
- Aquatic Ecosystems
- Biodiversity
- Conservation Biology
- Coral Reef Ecology
- Distribution Aggregation
- Ecology and Migration of Animal
- Ecosystem Service
- Ecosystem-Level Measuring
- Endangered Species
- Environmental Tourism
- Forest Biome
- Lake Circulation
- Leaf Morphology
- Marine Conservation
- Marine Ecosystems
- Phytoplankton Abundance
- Population Dyanamics
- Semiarid Ecosystem Soil Properties
- Spatial Distribution
- Species Composition
- Species Rarity
- Sustainability Dynamics
- Sustainable Forest Management
- Tropical Aquaculture
- Tropical Ecosystems
Recommended Journals
Article Tools
Article Usage
- Total views: 15670
- [From(publication date):
September-2012 - Nov 17, 2025] - Breakdown by view type
- HTML page views : 10942
- PDF downloads : 4728