Research Article Open Access
CMEIAS Quadrat Maker: A Digital Software Tool to Optimize Grid Dimensions and Produce Quadrat Images for Landscape Ecology Spatial Analysis
Frank B. Dazzo* and Colin GrossDepartment of Microbiology and Molecular Genetics, Michigan State University, East Lansing, USA
- *Corresponding Author:
- Frank B Dazzo
Department of Microbiology and Molecular Genetics
Michigan State University, East Lansing
Michigan 48824 USA
Tel: 517-884-5394
E-mail: dazzo@msu.edu
Received date: July 15, 2013; Accepted date: September 10, 2013; Published date: September 12, 2013
Citation: Dazzo FB, Gross C (2013) CMEIAS Quadrat Maker: A Digital Software Tool to Optimize Grid Dimensions and Produce Quadrat Images for Landscape Ecology Spatial Analysis. J Ecosys Ecograph 3:136. doi:10.4172/2157-7625.1000136
Copyright: © 2013 Dazzo FB, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and and source are credited.
Visit for more related articles at Journal of Ecosystem & Ecography
Abstract
This paper describes CMEIAS Quadrat Maker, a new digital computing tool designed to alleviate the nontrivial problem of optimizing the grid-lattice dimensions and automating the production of size-optimized quadrat images for plot-based spatial pattern analysis in landscape ecology. The program is written for 32-bit and 64-bit Window’s operating systems and handles both 8-bit grayscale and 24-bit color input images. Following a brief user interaction, the software application transforms a copy of the input landscape image into an annotated, color index image with optimized grid overlay and column-row labeling of individual quadrats, cuts a copy of the landscape image into quadrats defined by the optimized grid raster, and then saves each individual quadrat image with a file name indicating its unique location within the landscape domain, now ready for stack building and automated image analysis. Version 1.0 of this computing technology is implemented into a software package containing the executable file, user manual and tutorial images that will be freely available at http://cme.msu.edu/cmeias/. This new computing technology will facilitate quadrat-based analyses of how spatial patterns vary with the scale at which they are measured, and will also strengthen microscopy-based approaches for understanding the spatial ecology of microbial biofilm communities.
Keywords
Biofilm; CMEIAS; Microbial Ecology; Landscape Ecology; Quadrat; Plot-Based Spatial Pattern Analysis
Introduction
Quadrats are standard sampling units of relatively small, uniformly defined areas that divide a habitat of interest into subregions of equal size. They are used in ecography for 2-dimensional sampling of plants, slowmoving animals, and (more recently) microorganisms to (i) estimate population sizes when it is not feasible to measure their abundance in entirety, (ii) define the patterns of their spatial distribution, and (iii) compute species-area and distance-decay curves that define how spatial patterns of diversity and other categories of observations differ as a function of scale. All of these uses have important implications in biogeography, the conservation of biodiversity, and ecophysiological adaptations. In addition, defining these patterns contributes to our understanding of community structure and function within a landscape.
Passive quadrat sampling is done by examining the organisms on site, or more commonly, by acquiring photographs of the landscape domain with sufficient resolution and detail to reveal the organisms of interest for spatial analysis back in the laboratory. Although typically having a square or rectangular shape, quadrat-sampling units can theoretically be any shape and size so long as their area is known and they are standardized to a single, common-based unit area without location bias. The most definitive way to avoid location bias is to analyze all contiguous quadrats in a grid that covers the entire landscape domain under investigation.
The results of quadrat-based spatial analyses are heavily dependent on quadrat size. This is because its size will significantly influence the extent of edge effects that can cause counting errors, the recognition of spatially discrete clusters of organisms, and the resolution of void areas that separate them. Thus, a major non-trivial challenge in the quadratbased analysis of spatial patterns is how to optimize quadrat dimensions for the particular study in order to maximize the precision, accuracy and usefulness of the extracted data and the conclusions derived from them. One recommendation is to use at least two quadrat sizes, e.g., one smaller and the other larger than the largest patch where an event is occurring, and interpret the results according to that appropriate spatial scale. Two experimental methods to optimize quadrat size for estimating population size are described by Krebs [1], based on the operational rule of minimizing the cumulative product of relative variability in the object density and the cost to sample. Both methods may be appropriate for ecological studies of plants and animals, but are less than ideal for comprehensive studies of microorganisms attached to surfaces, a common lifestyle in natural and managed habitats known as biofilms.
We are developing software applications designed to strengthen quantitative, microscopy-based approaches for understanding microbial ecology at single-cell resolution and spatial scales relevant to the individual microbes and their ecological niches in situ. The software suite is called CMEIAS (Center for Microbial Ecology Image Analysis System), and as components become fully developed and documented, they are released for free download at the project website, (available online: http://cme.msu.edu/cmeias/). The CMEIAS upgrade currently being developed [2] includes a spatial ecology module, based on motivation to create quantitative computing tools designed to help users obtain better, statistically defendable answers to spatially explicit questions of relevant microbial ecophysiology during biofilm colonization of (non)biological surfaces.
In recent studies [3-6], we used image analysis to evaluate the ability of many point-pattern, quadrat-lattice, and geostatistical autocorrelation/ kriging methods to scrutinize the highly structured spatial patterns of microbial diversity and abundance in biofilm communities. Our interest in these methods has been to produce predictive models that reveal the intensity of ecophysiological processes driving their in situ colonization behavior within biofilm habitats. Those studies revealed that each method has its strengths and trade-offs, but when combined, they harmonize into a powerful, multivariate data-mining approach to understand how spatial scale affects cell-cell interactions that cooperatively or competitively affect microbial colonization behavior [3-6]. For example, a major benefit of the quadrat –based approach is that it forces the analysis to take into account possible scales within which statistically significant inhomogeneous events may be occurring [7]. Its major limitation of ignoring the location of each individual foreground object is nicely complemented when combined with other point pattern and geostatistical methods [3-6].
Here, we describe a new, digital software computing tool (called CMEIAS Quadrat Maker) that is uniquely designed to alleviate the important but cumbersome, time-consuming tasks of optimizing the grid dimensions of the raster that divides digital landscapes into contiguous quadrats, followed by preparing individual image files of each quadrat sample saved with column-row location for plot-based spatial pattern analysis of the landscape domain.
Materials and Methods
CMEIAS Quadrat Maker was coded to run in 32-bit and 64-bit Window’s operating systems. Tiff images for documentation were acquired using a Zeiss Photomicroscope equipped with an Insight digital camera (Diagnostic Instruments), Jing screen capture software (Techsmith) and Photoshop (Adobe Systems). The Quadrat Maker application was used to optimize grid dimensions and produce quadrat images. To facilitate working with landscape images containing elongated foreground objects, their object centroid X|Y Cartesian coordinates were extracted using CMEIAS-ImageTool [8], then used to make georeferenced 2-dimensional dot maps in Microsoft™ Excel, converted to tiff images using Photoshop, and then processed using Quadrat Maker. Quadrat image stacks were built and analyzed using CMEIAS-ImageTool and their frequencies of object counts were used to compute the various commonly used indices of spatial dispersion [1].
Results and Discussion
The CMEIAS Quadrat Maker application is designed to: 1) interactively evaluate the landscape image and efficiently produce a grid-lattice raster overlay with optimized quadrat dimensions that provide the spatial resolution of foreground objects by avoiding their over- or under-sampling of points; 2) automatically produce and save an indexed image of the original landscape overlaid with the optimized grid-lattice that is annotation to indicate each quadrat’s location; and 3) automatically cut a copy of the input landscape image into the smaller, equal-sized, individual quadrat images that are saved with column-row numbered annotation of their location defined by the optimized grid.
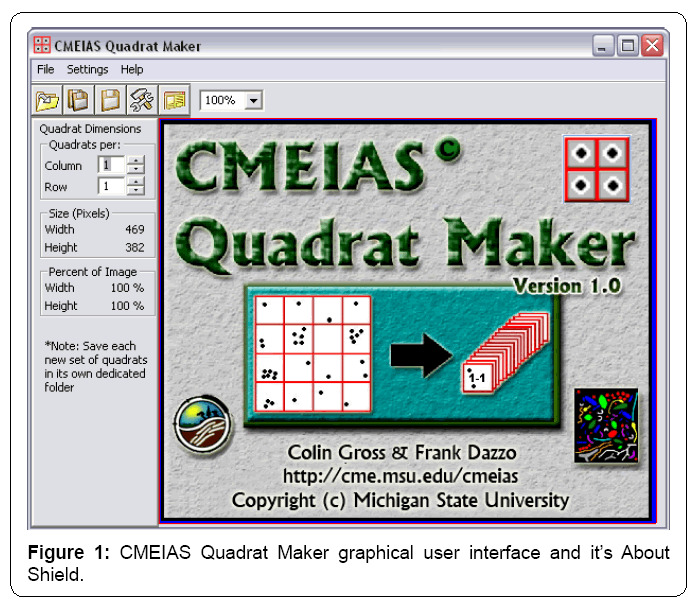
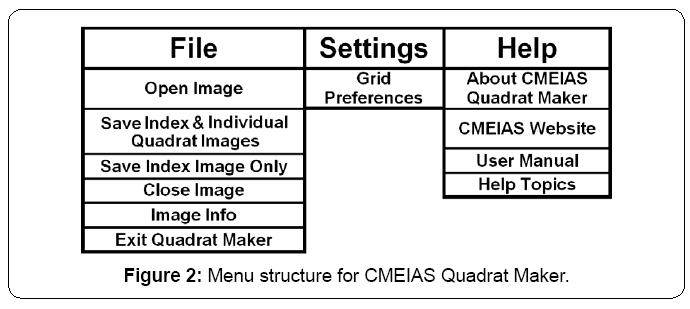
The application has a simple Graphical User Interface (GUI) containing a shortcut toolbar of main workflow steps, workspace to display the entire landscape image with grid-lattice overlay, and easily accessed interactive features to adjust the image zoom setting and grid dimensions (Figure 1). Input images can be 8-bit grayscale or 24-bit RGB color in tiff, bmp, jpg or png format. Grid specifications, quadrat size in pixels, and percentage of the landscape area represented by each quadrat are automatically displayed in the GUI as the grid optimization procedure is interactively performed. The diagram in the About Shield (Figure 1) displayed briefly when the application is launched shows the key functions of optimizing the grid-lattice and producing the stack of annotated quadrat images derived from the input landscape image. The full menu structure is indicated in Figure 2.
In a typical run, the user 1) opens the input, segmented landscape image; 2) adjusts the GUI workspace size and zoom settings to display the entire image; 3) selects the size and display color of grid lines and quadrat labels; 4) runs the grid optimization routine; 5) activates the image cutting routine that automatically divides a copy of the input landscape image into multiple quadrat images defined by the gridlattice; 6) saves output image files of the annotated indexed image and the multiple 8-bit grayscale quadrat images at a user-specified drive location; and 7) closes the input landscape image. What may take hours to perform this entire sequence when done manually on a large landscape image can be done within a few minutes using the software application.
Ideally, the landscape input image should have an object-free border on all four sides and have width and height dimensions that are multiples of the number of columns and rows in the optimized grid. If not possible, the extra object-free pixels will be excluded on the right and bottom borders of the image, respectively.
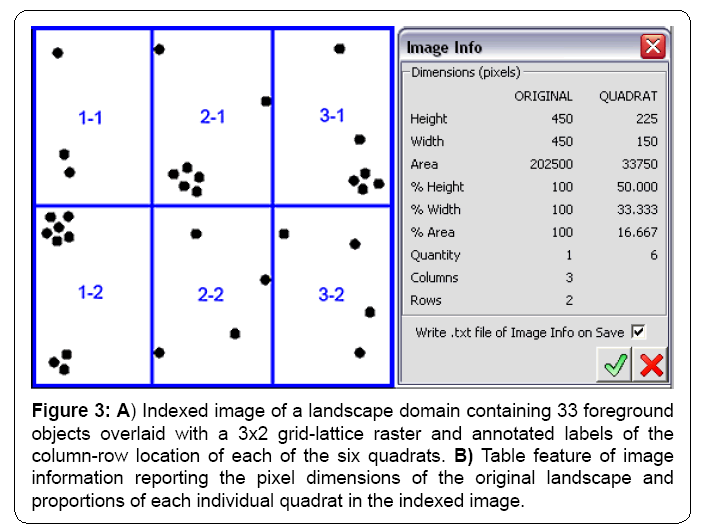
By default, all output images are saved in the same folder where the original segmented landscape image is located, but an alternate location can be designated. The indexed image displays the user-selected colors of the grid lines and column-row labels of each quadrat in the optimized grid-lattice raster overlay on the parent landscape image. It is automatically saved as a 24-bit RGB jpg image when the quadrat images are made or it can be produced after grid optimization and saved independently (Figures 2 and 3a). The individual, output quadrat images are saved as flattened 8-bit grayscale in Tiff format with the same pixel resolution as the parent landscape image and are ready for incorporation into an image stack for automated CMEIAS image analysis. Another optional feature automatically saves a *.txt file of tabled information on the input landscape and individual output quadrat images when the image cutting routine is run (Figure 3b).
Figure 3: A) Indexed image of a landscape domain containing 33 foreground objects overlaid with a 3x2 grid-lattice raster and annotated labels of the column-row location of each of the six quadrats. B) Table feature of image information reporting the pixel dimensions of the original landscape and proportions of each individual quadrat in the indexed image.
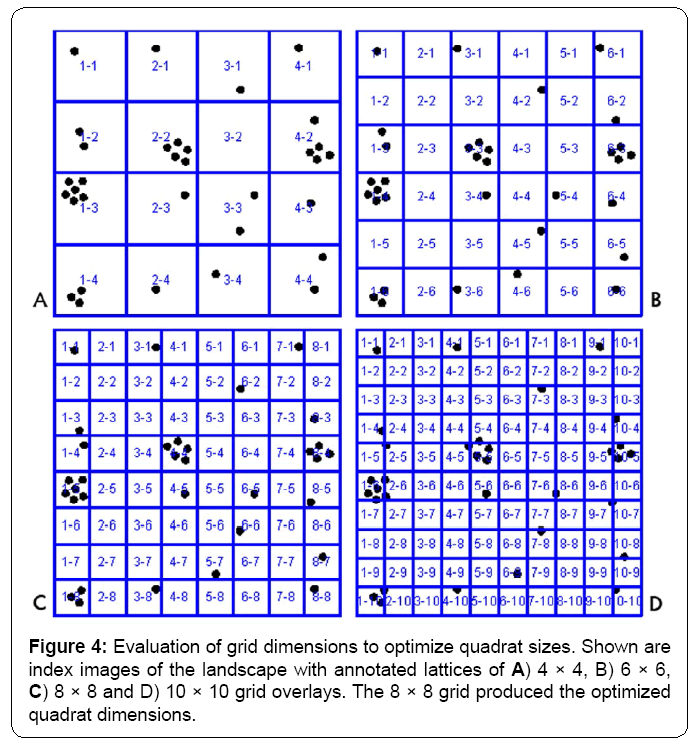
Commonly, optimization of quadrat size requires examination of multiple grid-lattice dimensions. This important process of determining which quadrat size works best can be greatly facilitated by the software application since it takes only a few seconds to produce and save multiple indexed images with different grid dimensions on the landscape that can then be visually inspected to reduce the number of candidates for subsequent image analysis. For example, consider the four different annotated, indexed images containing 4 × 4, 6 × 6, 8 × 8 and 10 × 10 grid-lattice overlays of the demo landscape image containing 33 foreground objects (Figure 4A-4D).
Visual inspection of the point distribution within the quadrat samples ranked the level of optimization for the 8x8 grid as high for the resolution of full clusters. The 4 × 4 and 10 × 10 grids are less than optimal because they oversample and undersample clustered points, respectively. The 6 × 6 grid produces an acceptable grid but with slightly less resolution between clusters compared to the 8 × 8 grid. Quantitative image analysis of the point distribution within the quadrats followed by computation of various indices of spatial dispersion agreed with this visual ranking, and definitively identified the 8 × 8 grid dimension as optimal (Table 1). Statistical analysis of the quadrat data from the optimized 8 × 8 grid indicated that the objects have a significantly clustered spatial pattern, with the probability of their pattern being due to random chance (hence representing complete spatial randomness) as less than 1 in 100,000. The variation in Chi-square and associated p values computed for the variance/mean index of dispersion derived from the four different landscape grids clearly emphasizes the significant importance of optimizing the quadrat dimensions for this type of spatial pattern analysis.
| Spatial Analysis Metrica | Quadrat Grid Matrix | |||
|---|---|---|---|---|
| 4X4 | 6X6 | 8X8 | 10X10 | |
| Quadrat Unit Size | 6.25 | 2.78 | 1.56 | 1.00 |
| Taylor’s Density Variance/Mean ratio | 1.517 | 2.268 | 2.667 | 1.582 |
| Chi-Square for V/M ratio | 22.76 | 79.36 | 168.00 | 126.54 |
| V/M ratio χ2 Significance (p)< | 0.08908 (random) |
0.00003 (clustered) |
0.00000 (very clustered) |
0.00072 (clustered) |
| Morista Dispersion Index | 1.242 | 2.386 | 4.387 | 2.455 |
| Standadized Morista Coefficient | 0.3106 | 0.5119 | 0.521 | 0.5039 |
| Index of Cluster Size | 0.517 | 1.268 | 1.667 | 0.582 |
aIndices of dispersion as defined in [1]
Table 1: Influence of grid dimensions on quadrat-based indices of dispersiona for spatial distribution analysis of the landscape image in Figure 4.
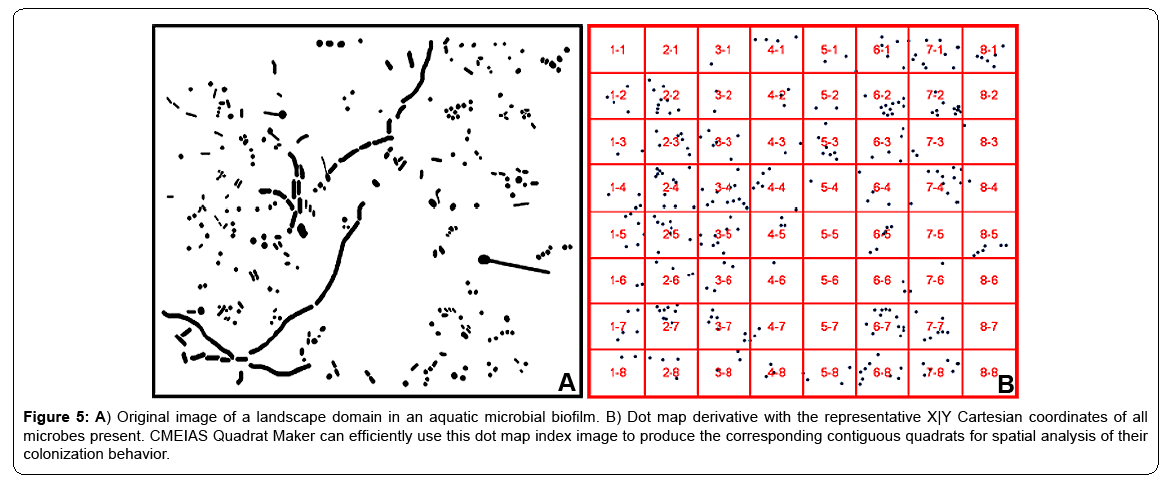
A significant problem arises when the best possible grid-lattice on the landscape index image still significantly overlaps foreground objects, causing serious edge effects. This problem often occurs in landscape images of natural biofilm communities containing microbes that vary widely in morphology (see example in Figure 5A). This nontrivial problem can be alleviated by producing a dot map derivative of the original landscape image. Dot map images are prepared by first extracting the X|Y Cartesian coordinates of object centroids (along with other optional features of body size and morphotype classification) of each individual cell in the segmented landscape image using CMEIAS image analysis, followed by constructing 2-dimensional scatterplots of the object X|Y centroid coordinates using symbols of small filled circles within Microsoft Excel, then adjusting the size and pixel resolution of the cropped dot map image to match the original landscape image and saving it in Adobe Photoshop. Figure 5B shows the index image of the dot map representation derived from the original biofilm landscape image with the optimized grid raster overlay having significantly reduced edge effects, now ready for preparation of individual quadrat images using the new software application.
Figure 5: A) Original image of a landscape domain in an aquatic microbial biofilm. B) Dot map derivative with the representative X|Y Cartesian coordinates of all microbes present. CMEIAS Quadrat Maker can efficiently use this dot map index image to produce the corresponding contiguous quadrats for spatial analysis of their colonization behavior.
CMEIAS Quadrat Maker is intended for research and educational applications, and serves as a useful computing tool for anyone who needs to evaluate the grid-lattice and produce the size-optimized quadrat dimensions for plot-based spatial analysis of digital images in landscape ecology. We use it in our spatial ecology studies of microbial biofilms developed in freshwater aquatic ecosystems [3-5] and on plant surfaces [6]. Version 1.0 of this computing technology is being implemented into a software package containing the executable file, user manual and tutorial images that will be freely available at http://cme.msu.edu/cmeias/. This application adds to our CMEIAS© suite of integrated software whose combined mission is to strengthen microscopy-based approaches for advancing a greater understanding of microbial ecology at single-cell resolution and spatial scales relevant to the organisms themselves.
Acknowledgements
We thank Lei Gao for his contribution to early development of Quadrat Maker, and Prabhat Jha, Kyle Card and Gulnihal Ozbay for their helpful comments on the paper. Previous support for development of CMEIAS software has been provided by the Center for Microbial Ecology and Long-Term Ecological Research programs at Michigan State University, and by the US-Egypt Science and Technology Joint Program.
References
- Krebs CJ (1999) Ecological methodology. Addison-Wesley Educational Publishers Inc, Menlo Park, CA, USA.
- Dazzo FB, Liu J, Gross C, Reddy C, Monosmith C, et al (2013) CMEIAS v3.10: advanced computational tools of image analysis software designed to strengthen microscopy-based approaches for understanding microbial ecology. 2013 Proceedings of the All-Scientist Meeting for Kellogg Biological Station Long Term Ecological Research.
- Dazzo FB (2010) CMEIAS digital microscopy and quantitative image analysis of microorganisms. Microscopy: science, technology, applications and education, Microscopy Book Series #4, Formatex Research Center, Badajoz, Spain 1083-1090.
- Dazzo FB (2012) CMEIAS-aided microscopy of the spatial ecology of individual bacterial interactions involving cell-to-cell communication within biofilms. Sensors 12: 7047-7062.
- Dazzo FB, Klemmer K, Chandler R, Yanni Y (2013) In situ ecophysiology of microbial biofilm communities analyzed by CMEIAS computer-assisted microscopy at single-cell resolution. Diversity 5:426-460.
- Dazzo FB, Yanni YG (2013) CMEIAS: an improved computing technology for quantitative image analysis of root colonization by rhizobacteria in situ at single-cell resolution. Molecular microbial ecology of the rhizosphere, J Wiley & Sons, New York, USA 733-742.
- Wikipedia (2012) Point pattern analysis.
- Liu J, Dazzo FB, Glagoleva O, Yu B, Jain AK (2001) CMEIAS: a computer-aided system for the image analysis of bacterial morphotypes in microbial communities. Microbial Ecology 41: 173-194.
Relevant Topics
- Aquatic Ecosystems
- Biodiversity
- Conservation Biology
- Coral Reef Ecology
- Distribution Aggregation
- Ecology and Migration of Animal
- Ecosystem Service
- Ecosystem-Level Measuring
- Endangered Species
- Environmental Tourism
- Forest Biome
- Lake Circulation
- Leaf Morphology
- Marine Conservation
- Marine Ecosystems
- Phytoplankton Abundance
- Population Dyanamics
- Semiarid Ecosystem Soil Properties
- Spatial Distribution
- Species Composition
- Species Rarity
- Sustainability Dynamics
- Sustainable Forest Management
- Tropical Aquaculture
- Tropical Ecosystems
Recommended Journals
Article Tools
Article Usage
- Total views: 18299
- [From(publication date):
December-2013 - Nov 14, 2025] - Breakdown by view type
- HTML page views : 13420
- PDF downloads : 4879