Research Article Open Access
Isolation and Characterization of Relevant Algal and Bacterial Strains from Egyptian Environment for Potential Use in Photosynthetically Aerated Wastewater Treatment
| Keywords |
| Phenol; Pyridine; Wastewater; Photosynthesis; Microalgae; Biodegradation |
| Introduction |
| Organic pollutants represent a major environmental problem worldwide. Specifically, aromatic and hetero-aromatic compounds such as phenol and pyridine, which are massively used in the chemical, petrochemical and pharmaceutical industries and are therefore often found in many industrial wastewaters [1,2]. These compounds are highly toxic and have been listed by the US Environmental Protection Agency as priority pollutants [3]. Therefore, it is crucial to efficiently remove them from contaminated streams in order to prevent their entry in the environment. Several efficient methods were developed to treat these toxic pollutants such as chlorination, ozonation, adsorption, solvent extraction, membrane process, coagulation, flocculation and biological degradation [1,2,4]. However, these methods suffer from the drawbacks of either being costly and/or might generate secondary pollutants, which may be more toxic than the parent ones [5]. Accordingly, biological methods are generally preferred for being more economical and environmentally friendly [6,7]. |
| Eventually, among the most promising biological methods for the treatment of wastewater are those based on the use of algal-bacterial microcosms where algae provide the necessary oxygen for bacterial biodegradation. Indeed, algal-bacterial treatment strategy provided a safer and more economic alternative with tremendous advantages [8,9]. However, one of the major limitations to this promising strategy is the high fluctuation in influent toxicity [8-10]. In such cases, the use of acclimatized microorganisms especially adapted to metabolize and/ or tolerate the contaminants at high concentrations can provide an attractive solution to improve process performance [1,11,12]. |
| With this perspective, the present work reports the isolation and characterization of highly resistant bacterial and algal strains from the Egyptian environment. The biodegradation and/or tolerance capacities of the isolates were investigated. The most relevant isolates were further characterized for optimum biodegradation. Finally the impact of the relevant bacterial isolate on the growth of the corresponding algal isolate was investigated and vice versa. These isolates can be useful in the assembly of an algal-bacterial microcosm for the photosynthetically aerated biological treatment of industrial wastewater loaded with various organic pollutants. |
| Materials and Methods |
| All chemicals were reagent grade; phenol was obtained from Sigma–Aldrich (Steinheim, Germany). Unless otherwise specified, all experiments were conducted in triplicate at 28±2°C under sterile conditions. |
| Enrichment, isolation and maintenance of bacterial and algal strains |
| Screening, isolation and maintenance of all microbial cultures were conducted using a mineral salt medium (MSM) with a composition according to [1]. This medium was enriched with 500 mg phenol l-1 and/or 500 mg pyridine l-1 or 2000 mg NaHCO3 l-1 for cultivation and maintenance of bacterial or algal isolates respectively. |
| Bacterial strains were isolated according to [1] from several soil samples, collected from different locations in Giza and Cairo with incubation conditions of 28±2°C and agitation at 150 rpm. Algal strains were isolated from several water samples, collected from wastewater drainages at different locations in Giza and Cairo. Aliquots of 10 ml of the water samples were transferred to 100 ml of MSM enriched with 2000 mg NaHCO3 l-1 and incubated at 28±2°C in an incubator shaker with continuous illumination (5000 lux, Philips TLD 36W/840 fluorescent lamp) and agitation at 150 rpm for 72 h. A portion of 10 ml of the suspension was transferred into 100 ml of MSM supplemented with 500 mg ampicillin l-1, 80 mg garamycin l-1, 64 mg fluconazole l-1, 500 mg glucose l-1 and 2000 mg NaHCO3 l-1 and incubated under the same conditions. Aliquot of 0.1 ml of the concentrated algal suspension was streaked onto solid MSM supplemented with the same additives mentioned above and incubated under the same conditions. |
| Morphological and physiological characterization |
| Bacterial isolates were subjected to morphological characterization and motility tests using a light microscope (Olympus, USA). Gram stain reaction was done using Difco Gram stain set according to the standard protocol [13]. Further morphological examination was done using transmission scanning electron microscopy (JEOL Ltd., Tokyo, Japan) at 2 magnification scales (2 μm and 500 nm). Biochemical characterization was done using API 20 NE kit system (bioMérieux, France) according to the manufacturer’s instructions. The presence of oxidase was determined using a test strip (Microbiology Bactident Oxidase, Merck, Germany). Catalase activity was evaluated by transferring a loop of bacterial cells onto a microscope slide and adding a drop of 3% hydrogen peroxide solution [1]. |
| Micro algal isolates were morphologically characterized. This characterization was kindly carried out by Dr. Jiri Neustupa at the Department of Botany, Faculty of Science, Charles University of Prague, Czech Republic. |
| Partial 16S rRNA sequence analysis |
| The most relevant bacterial isolate was subjected to molecular identification using partial 16S rRNA sequence analysis according to [1] using 2 universal primers 28f 5’AGAGTTTGATCCTGGCTCAG-3’ (positions 8-28 in E. Coli numbering) and 1512R 5’ACGGCTACCTTGTTACGACT-3’ (positions 1512-1493 in E. Coli numbering). The GenBank database (NCBI, USA) was then used to search for 16S rRNA sequence similarities. |
| Determination of the maximum biodegradation capacities |
| For all bacterial isolates, a total of 10 ml of the bacterial culture suspension (107 cells ml-1) was inoculated into MSM supplemented with increasing pollutant concentrations (Table 1). Flasks were incubated for 3 d and up to 28 d at 28±2°C in an incubator shaker (New Brunswick, C25, USA) at 150 rpm. Samples were periodically withdrawn and tested for growth and pollutant removal. Controls were conducted using the same media but without bacterial inoculation. |
| Determination of the optimum biodegradation and biodegradation rate of phenol |
| Bacterial isolates with relevant biodegradation capacities (Table 1) were tested to determine the isolate with the highest biodegradation rate. This relevant isolate was then tested for optimum growth. In both tests, 90 ml of MSM supplemented with various phenol concentrations as mentioned (Table 1) were inoculated with 10 ml of the bacterial suspension (107 cells ml-1). Flasks were incubated for 48 h at 28 ±2°C in an incubator shaker (New Brunswick, C25, USA) at 150 rpm. Samples were periodically withdrawn and tested for growth and phenol concentration. |
| Toxicity of tested pollutants to the selected microalgae |
| Test tubes of 12 ml were filled with 10 ml MSM supplemented with 2000 mg NaHCO3 l-1 and containing phenol (50, 100, 200, 300, 400 and 500 mgl-1) or pyridine (100, 200, 500, 1000, 1500, 2000 and 3000 mgl-1) or a mixture of phenol and pyridine (50/100, 100/200, 200/400, 300/600, 400/800 and 500/1000) respectively. All tubes were inoculated with 6% v/v of C. vulgaris culture, flushed with N2 gas (whenever necessary to remove any atmospheric O2), sealed with plastic screw caps and incubated under continuous agitation (150 rpm) and illumination (5000 lux = 18 μW cm-2, Philips TLD 36W/840 lamp). After 72 h, 5 ml-samples were withdrawn and analyzed to measure the chlorophyll-A content. Blanks were conducted without adding any pollutants and algal inhibition (%) was calculated as the reduction of the average chlorophyll-A content in the test samples compared to that in the blanks. |
| Influence of bacterial metabolites on microalgal growth and vice versa |
| Both algal isolate A4 and bacterial isolate M4 were allowed to grow. Then a total of 20 ml of each growth suspension was centrifuged at 11300 g for 10 min (Denver instrument, USA). Ten milliliters of the supernatant of the centrifuged algal suspension was added into 40 ml MSM supplemented with 500 mg phenol l-1, inoculated with 6 % v/v of bacterial isolate M4 and incubated at 28±2°C with continuous agitation at 150 rpm for 48 h. Similarly, 10 ml of the supernatant of the centrifuged bacterial suspension was added into 40 ml MSM supplemented with 2000 mg NaHCO3 l-1, inoculated with 6% v/v of algal isolate A4 and incubated at 28±2°C with continuous agitation at 150 rpm and illumination (5000 lux = 18 μW cm-2, Philips TLD 36W/840 lamp) for 72 h. Samples were withdrawn at the end of the incubation period for analysis of phenol concentration, bacterial growth and chlorophyll-a content. Blanks were conducted without adding neither bacterial nor algal growth supernatant. Bacterial and algal inhibition % was calculated as the reduction in growth, phenol removal % or chlorophyll-a content compared to that in the blanks respectively. |
| Analysis |
| Analysis of phenol and pyridine was conducted by HPLC-UV (Schimadzu 10A VP, USA), equipped with a Supelco LC-18 column. Samples were first centrifuged at 11300 g for 10 min (Denver, USA) and portions of the supernatant were injected into HPLC. The samples were eluted with a mobile phase which consisted of acetonitrile : water (300 : 700), 0.11 g heptane sulphonic acid, 0.29 g anhydrous sodium acetate, and 2.5 ml glacial acetic acid. The detection was conducted at 280 nm for phenol and 254 nm for pyridine. External standards allowed for quantification and the detection limit was below 1 mg l-1. |
| Chlorophyll-A content was measured according to [14]. Microbial density was estimated by measuring the absorbency of the culture at 600 nm (OD600) using a UV/visible spectrophotometer (Schimadzu 1650, USA). |
| Results and Discussion |
| Identification and characterization of the bacterial isolates |
| Nine bacterial cultures were isolated (M1-M9). Preliminary examination showed that 8 out of these cultures were single bacterial strains with negative Gram reaction and rod shaped. While isolated culture M1 was a mixture of Gram positive oval shaped microorganism and Gram negative rod shaped bacterium. Therefore, this culture was excluded from further identification scheme. Similarly isolates M8 and M9 were excluded due to their recorded relative low biodegradation capacities (Table 2). Further identification of the selected isolates showed that all were motile and had positive catalase reaction. Only 2 isolates (M2 and M4) had positive oxidase reaction while 4 isolates gave negative reactions (Table 2). |
| Strain M4 was identified biochemically and molecularly to be a member of Pseudomonas sp. whiles the other 6 identified bacterial isolates were classified (using API 20 NE kit system) as members of Burkholderia sp., Sphingomonas sp. and Chryseomonas sp. (Table 2). However, all these species were formerly classified as part of the Pseudomonads [15]. These isolates showed a remarkable ability to tolerate and/or degrade phenol. Members of Pseudomonas sp. and related species are well known for their ability to biodegrade aromatic compounds and especially phenol [16-18]. |
| All of the 9 bacterial isolates were able to degrade phenol where 6 of these isolates were able to degrade up to 1700 mg l-1 (Table 2). The other 3 isolates (M6, M8 and M9) showed lower capacities of 1300, 1000 and 1000 mg phenol l-1, respectively (Table 2). Interestingly, among these 9 isolates only 4 isolates were additionally able to biodegrade 2000 - 3000 mg pyridine l-1 (Table 2). Previous studies reported the ability of the isolated genera to degrade phenol and/or pyridine [17-19] and even [2] have reported the combined biodegradation of phenol and pyridine by members of Pseudomonas sp. where the authors have also reported that phenol was more toxic than pyridine, which is in agreement with the recorded data in the present study. |
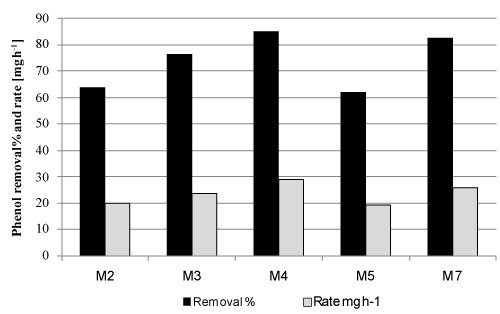
| The biodegradation rate of 1000 mg phenol l-1 was determined for the 5 isolates with the highest biodegradation capacities (Table 2). All isolates were able to achieve complete biodegradation after 48 h (data not shown). However, the maximum phenol removal rates for all tested isolates were recorded after 32 h of incubation (Figure 1). Although isolates M4 and M7 showed the highest biodegradation efficiency (86 and 82 %) and rate (29 and 26 mgh-1) respectively (Figure 1), M7 was not able to grow or degrade pyridine (Table 2). Therefore, M4 was selected and subjected for further identification and characterization. |
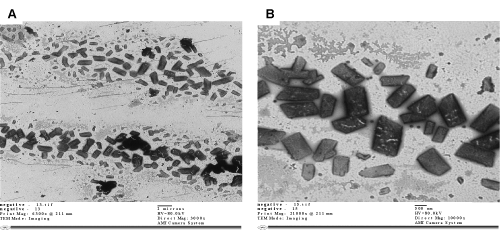
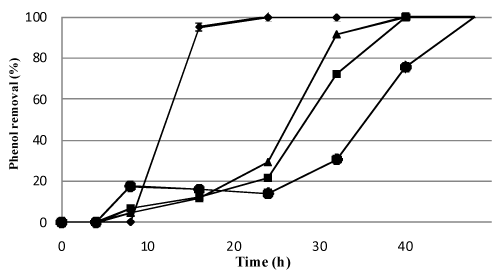
| Transmission electron microscope examination showed that isolate M4 had rod (rectangular) shape with 0.5-1 μm length, with neither visible flagella, nor abnormal extracellular structures and was arranged singly (Figure 2). The partial 16S rRNA sequencing of strain M4 (Genbank accession number JQ178342) showed 99% identity to the sequence of members of Pseudomonas sp., therefore it was named Pseudomonas MT1. This strain was preferably grown on phenol rather than pyridine (data not shown) at 28±2°C, pH 7, with agitation at 150 rpm and phenol concentration of 1000-1250 mg l-1. Lower concentrations resulted in lower biodegradation rates and higher concentrations resulted in prolonged lag phase (Figure 3). |
| Recently, many studies have reported the isolation of bacterial strains with relevant phenol tolerance and biodegradation capacity [1]. However, isolated bacterial strains in the present study, particularly strain M4 showed one of the highest degradation capacities and rate among reported wild bacterial isolates [20]. Strain M4 was able to biodegrade up to 1700 mg phenol l-1 with a biodegradation rate of 29 mg h-1. Indeed, such high resistance and fast biodegradation rate is a prerequisite to rapidly bring down phenol concentration at satisfactory levels and to ensure efficient and stable phenol removal in continuous and especially batch treatment of real industrial wastewaters [1,8]. |
| Identification and characterization of algal isolates |
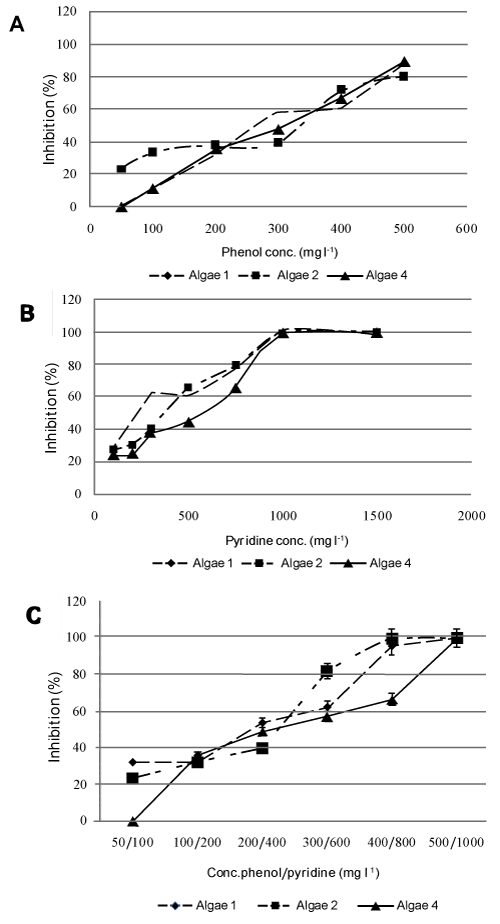
| Four algal strains were isolated (A1-A4) and morphologically characterized as members of the Chlorella genus. However, only 3 isolates showed remarkable tolerance to phenol and/or pyridine (Figure 4). Interestingly, all of the 3 algal isolates had almost similar tolerance pattern to phenol (Figure 4A) while A4 was more resistant against pyridine (Figure 4B). When a mixture of both pollutants was used at low concentrations, almost all algal isolates showed the same order of magnitude of inhibition; however, at a higher concentration (400/800), A4 had the lowest inhibition % (Figure 4C). All algal isolates were completely inhibited when incubated with a mixture of 500/1000 mg l-1 phenol/pyridine respectively (Figure 4C). |
| Although some studies [12] have reported the isolation of phenol biodegrading algal strains and even polycyclic aromatic hydrocarbons, none of the isolates algal strains in the present study showed a proof to degrade any of the tested pollutants. Similarly, many other studies reported that algal isolates lack the degradation capacities [21,22]. Hence algae commonly participate in the algal bacterial treatment of wastewater by providing the photosynthetic oxygen to the bacterial partner [8,22]. |
| Interestingly, phenol was more toxic to algae than pyridine especially at high concentrations where complete inhibition of algal growth was observed at 500 mg phenol l-1 compared to 1000 mg pyridine l-1. This agrees with previous studies reporting a phenol 96h- (Effective Concentration) EC50 of 370 mg l-1 to C. vulgaris [21], compared to a pyridine EC50 of 628 mg l-1 to algae and aquatic plants [23]. |
| Correlation between algal and bacterial isolates |
| The symbiotic algal–bacterial relationship is often beneficial when microalgae provide the O2 necessary for aerobic bacteria to biodegrade organic pollutants, consuming in turn the CO2 released from bacterial respiration. However, interaction between algae and bacteria is not limited to a simple CO2/O2 exchange where algae can have a detrimental effect on bacterial activity and vice versa [8,24]. However, in the present study neither bacterial isolate M4 nor algal isolate A4 had detrimental effect on each other (data not shown). Therefore, they can be used for the construction of an algal-bacterial microcosm for biological wastewater treatment. |
| Conclusion |
| Highly biodegrading and/or tolerant bacterial and algal strains were isolated. Both bacterial and algal isolates with the highest degradation and/or tolerance capacities had neither inhibitory nor beneficial impact on the growth of each other. Hence, these isolates could be assembled into a microcosm for photocatalytic -biological treatment of toxic effluents in algal–bacterial bioreactors. |
| Acknowledgements |
| This project was supported by funds from Cairo University, administrated by the Biotechnology Centre, Faculty of Pharmacy, Cairo University. |
| References |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
A number of environmental samples were collected from different locations in Egypt. Briefly, 9 bacterial strains and 4 algal strains were isolated and characterized. All isolated bacterial strains showed a remarkable ability to tolerate and/or biodegrade phenol (aromatic pollutant) and pyridine (hetero-aromatic pollutant). Phenol showed higher toxicity than pyridine to both bacterial and algal isolates. The bacterial isolates were identified as members of Pseudomonas, Chryseomonas, Sphingomonas and Burkholderiae species. The highest biodegradation rate and capacity were reported to bacterial isolate M4, identified as Pseudomonas MT1. This strain was able to degrade up to 1700 and 3000 mg l-1 of phenol and pyridine respectively. Pseudomonas MT1 showed the highest phenol biodegradation rate of 29 mg h-1 and lag phase approximately of 8 h and was optimally grown on 1000 -1250 mg phenol l-1. All algal isolates were morphologically identified as members of the Chlorella genus. None of the isolated algal strains showed biodegradation ability of any of the tested organic pollutants. However, isolates A1, A2 and A4 showed remarkable tolerance to both phenol and/or pyridine. The highest tolerance capacity was reported to algal isolate A4, identified as Chlorella vulgaris MM1 with a toxicity cut off of 500 and 1000 mg l-1 of phenol and pyridine respectively. Both Pseudomonas MT1 and Chlorella vulgaris MM1 had no inhibitory effect on each other. Therefore, they represented potential candidates for the construction of algal bacterial microcosm used for the photosynthetically aerated biological degradation of effluents loaded with various organic pollutants.
| Keywords |
| Phenol; Pyridine; Wastewater; Photosynthesis; Microalgae; Biodegradation |
| Introduction |
| Organic pollutants represent a major environmental problem worldwide. Specifically, aromatic and hetero-aromatic compounds such as phenol and pyridine, which are massively used in the chemical, petrochemical and pharmaceutical industries and are therefore often found in many industrial wastewaters [1,2]. These compounds are highly toxic and have been listed by the US Environmental Protection Agency as priority pollutants [3]. Therefore, it is crucial to efficiently remove them from contaminated streams in order to prevent their entry in the environment. Several efficient methods were developed to treat these toxic pollutants such as chlorination, ozonation, adsorption, solvent extraction, membrane process, coagulation, flocculation and biological degradation [1,2,4]. However, these methods suffer from the drawbacks of either being costly and/or might generate secondary pollutants, which may be more toxic than the parent ones [5]. Accordingly, biological methods are generally preferred for being more economical and environmentally friendly [6,7]. |
| Eventually, among the most promising biological methods for the treatment of wastewater are those based on the use of algal-bacterial microcosms where algae provide the necessary oxygen for bacterial biodegradation. Indeed, algal-bacterial treatment strategy provided a safer and more economic alternative with tremendous advantages [8,9]. However, one of the major limitations to this promising strategy is the high fluctuation in influent toxicity [8-10]. In such cases, the use of acclimatized microorganisms especially adapted to metabolize and/ or tolerate the contaminants at high concentrations can provide an attractive solution to improve process performance [1,11,12]. |
| With this perspective, the present work reports the isolation and characterization of highly resistant bacterial and algal strains from the Egyptian environment. The biodegradation and/or tolerance capacities of the isolates were investigated. The most relevant isolates were further characterized for optimum biodegradation. Finally the impact of the relevant bacterial isolate on the growth of the corresponding algal isolate was investigated and vice versa. These isolates can be useful in the assembly of an algal-bacterial microcosm for the photosynthetically aerated biological treatment of industrial wastewater loaded with various organic pollutants. |
| Materials and Methods |
| All chemicals were reagent grade; phenol was obtained from Sigma–Aldrich (Steinheim, Germany). Unless otherwise specified, all experiments were conducted in triplicate at 28±2°C under sterile conditions. |
| Enrichment, isolation and maintenance of bacterial and algal strains |
| Screening, isolation and maintenance of all microbial cultures were conducted using a mineral salt medium (MSM) with a composition according to [1]. This medium was enriched with 500 mg phenol l-1 and/or 500 mg pyridine l-1 or 2000 mg NaHCO3 l-1 for cultivation and maintenance of bacterial or algal isolates respectively. |
| Bacterial strains were isolated according to [1] from several soil samples, collected from different locations in Giza and Cairo with incubation conditions of 28±2°C and agitation at 150 rpm. Algal strains were isolated from several water samples, collected from wastewater drainages at different locations in Giza and Cairo. Aliquots of 10 ml of the water samples were transferred to 100 ml of MSM enriched with 2000 mg NaHCO3 l-1 and incubated at 28±2°C in an incubator shaker with continuous illumination (5000 lux, Philips TLD 36W/840 fluorescent lamp) and agitation at 150 rpm for 72 h. A portion of 10 ml of the suspension was transferred into 100 ml of MSM supplemented with 500 mg ampicillin l-1, 80 mg garamycin l-1, 64 mg fluconazole l-1, 500 mg glucose l-1 and 2000 mg NaHCO3 l-1 and incubated under the same conditions. Aliquot of 0.1 ml of the concentrated algal suspension was streaked onto solid MSM supplemented with the same additives mentioned above and incubated under the same conditions. |
| Morphological and physiological characterization |
| Bacterial isolates were subjected to morphological characterization and motility tests using a light microscope (Olympus, USA). Gram stain reaction was done using Difco Gram stain set according to the standard protocol [13]. Further morphological examination was done using transmission scanning electron microscopy (JEOL Ltd., Tokyo, Japan) at 2 magnification scales (2 μm and 500 nm). Biochemical characterization was done using API 20 NE kit system (bioMérieux, France) according to the manufacturer’s instructions. The presence of oxidase was determined using a test strip (Microbiology Bactident Oxidase, Merck, Germany). Catalase activity was evaluated by transferring a loop of bacterial cells onto a microscope slide and adding a drop of 3% hydrogen peroxide solution [1]. |
| Micro algal isolates were morphologically characterized. This characterization was kindly carried out by Dr. Jiri Neustupa at the Department of Botany, Faculty of Science, Charles University of Prague, Czech Republic. |
| Partial 16S rRNA sequence analysis |
| The most relevant bacterial isolate was subjected to molecular identification using partial 16S rRNA sequence analysis according to [1] using 2 universal primers 28f 5’AGAGTTTGATCCTGGCTCAG-3’ (positions 8-28 in E. Coli numbering) and 1512R 5’ACGGCTACCTTGTTACGACT-3’ (positions 1512-1493 in E. Coli numbering). The GenBank database (NCBI, USA) was then used to search for 16S rRNA sequence similarities. |
| Determination of the maximum biodegradation capacities |
| For all bacterial isolates, a total of 10 ml of the bacterial culture suspension (107 cells ml-1) was inoculated into MSM supplemented with increasing pollutant concentrations (Table 1). Flasks were incubated for 3 d and up to 28 d at 28±2°C in an incubator shaker (New Brunswick, C25, USA) at 150 rpm. Samples were periodically withdrawn and tested for growth and pollutant removal. Controls were conducted using the same media but without bacterial inoculation. |
| Determination of the optimum biodegradation and biodegradation rate of phenol |
| Bacterial isolates with relevant biodegradation capacities (Table 1) were tested to determine the isolate with the highest biodegradation rate. This relevant isolate was then tested for optimum growth. In both tests, 90 ml of MSM supplemented with various phenol concentrations as mentioned (Table 1) were inoculated with 10 ml of the bacterial suspension (107 cells ml-1). Flasks were incubated for 48 h at 28 ±2°C in an incubator shaker (New Brunswick, C25, USA) at 150 rpm. Samples were periodically withdrawn and tested for growth and phenol concentration. |
| Toxicity of tested pollutants to the selected microalgae |
| Test tubes of 12 ml were filled with 10 ml MSM supplemented with 2000 mg NaHCO3 l-1 and containing phenol (50, 100, 200, 300, 400 and 500 mgl-1) or pyridine (100, 200, 500, 1000, 1500, 2000 and 3000 mgl-1) or a mixture of phenol and pyridine (50/100, 100/200, 200/400, 300/600, 400/800 and 500/1000) respectively. All tubes were inoculated with 6% v/v of C. vulgaris culture, flushed with N2 gas (whenever necessary to remove any atmospheric O2), sealed with plastic screw caps and incubated under continuous agitation (150 rpm) and illumination (5000 lux = 18 μW cm-2, Philips TLD 36W/840 lamp). After 72 h, 5 ml-samples were withdrawn and analyzed to measure the chlorophyll-A content. Blanks were conducted without adding any pollutants and algal inhibition (%) was calculated as the reduction of the average chlorophyll-A content in the test samples compared to that in the blanks. |
| Influence of bacterial metabolites on microalgal growth and vice versa |
| Both algal isolate A4 and bacterial isolate M4 were allowed to grow. Then a total of 20 ml of each growth suspension was centrifuged at 11300 g for 10 min (Denver instrument, USA). Ten milliliters of the supernatant of the centrifuged algal suspension was added into 40 ml MSM supplemented with 500 mg phenol l-1, inoculated with 6 % v/v of bacterial isolate M4 and incubated at 28±2°C with continuous agitation at 150 rpm for 48 h. Similarly, 10 ml of the supernatant of the centrifuged bacterial suspension was added into 40 ml MSM supplemented with 2000 mg NaHCO3 l-1, inoculated with 6% v/v of algal isolate A4 and incubated at 28±2°C with continuous agitation at 150 rpm and illumination (5000 lux = 18 μW cm-2, Philips TLD 36W/840 lamp) for 72 h. Samples were withdrawn at the end of the incubation period for analysis of phenol concentration, bacterial growth and chlorophyll-a content. Blanks were conducted without adding neither bacterial nor algal growth supernatant. Bacterial and algal inhibition % was calculated as the reduction in growth, phenol removal % or chlorophyll-a content compared to that in the blanks respectively. |
| Analysis |
| Analysis of phenol and pyridine was conducted by HPLC-UV (Schimadzu 10A VP, USA), equipped with a Supelco LC-18 column. Samples were first centrifuged at 11300 g for 10 min (Denver, USA) and portions of the supernatant were injected into HPLC. The samples were eluted with a mobile phase which consisted of acetonitrile : water (300 : 700), 0.11 g heptane sulphonic acid, 0.29 g anhydrous sodium acetate, and 2.5 ml glacial acetic acid. The detection was conducted at 280 nm for phenol and 254 nm for pyridine. External standards allowed for quantification and the detection limit was below 1 mg l-1. |
| Chlorophyll-A content was measured according to [14]. Microbial density was estimated by measuring the absorbency of the culture at 600 nm (OD600) using a UV/visible spectrophotometer (Schimadzu 1650, USA). |
| Results and Discussion |
| Identification and characterization of the bacterial isolates |
| Nine bacterial cultures were isolated (M1-M9). Preliminary examination showed that 8 out of these cultures were single bacterial strains with negative Gram reaction and rod shaped. While isolated culture M1 was a mixture of Gram positive oval shaped microorganism and Gram negative rod shaped bacterium. Therefore, this culture was excluded from further identification scheme. Similarly isolates M8 and M9 were excluded due to their recorded relative low biodegradation capacities (Table 2). Further identification of the selected isolates showed that all were motile and had positive catalase reaction. Only 2 isolates (M2 and M4) had positive oxidase reaction while 4 isolates gave negative reactions (Table 2). |
| Strain M4 was identified biochemically and molecularly to be a member of Pseudomonas sp. whiles the other 6 identified bacterial isolates were classified (using API 20 NE kit system) as members of Burkholderia sp., Sphingomonas sp. and Chryseomonas sp. (Table 2). However, all these species were formerly classified as part of the Pseudomonads [15]. These isolates showed a remarkable ability to tolerate and/or degrade phenol. Members of Pseudomonas sp. and related species are well known for their ability to biodegrade aromatic compounds and especially phenol [16-18]. |
| All of the 9 bacterial isolates were able to degrade phenol where 6 of these isolates were able to degrade up to 1700 mg l-1 (Table 2). The other 3 isolates (M6, M8 and M9) showed lower capacities of 1300, 1000 and 1000 mg phenol l-1, respectively (Table 2). Interestingly, among these 9 isolates only 4 isolates were additionally able to biodegrade 2000 - 3000 mg pyridine l-1 (Table 2). Previous studies reported the ability of the isolated genera to degrade phenol and/or pyridine [17-19] and even [2] have reported the combined biodegradation of phenol and pyridine by members of Pseudomonas sp. where the authors have also reported that phenol was more toxic than pyridine, which is in agreement with the recorded data in the present study. |
| The biodegradation rate of 1000 mg phenol l-1 was determined for the 5 isolates with the highest biodegradation capacities (Table 2). All isolates were able to achieve complete biodegradation after 48 h (data not shown). However, the maximum phenol removal rates for all tested isolates were recorded after 32 h of incubation (Figure 1). Although isolates M4 and M7 showed the highest biodegradation efficiency (86 and 82 %) and rate (29 and 26 mgh-1) respectively (Figure 1), M7 was not able to grow or degrade pyridine (Table 2). Therefore, M4 was selected and subjected for further identification and characterization. |
| Transmission electron microscope examination showed that isolate M4 had rod (rectangular) shape with 0.5-1 μm length, with neither visible flagella, nor abnormal extracellular structures and was arranged singly (Figure 2). The partial 16S rRNA sequencing of strain M4 (Genbank accession number JQ178342) showed 99% identity to the sequence of members of Pseudomonas sp., therefore it was named Pseudomonas MT1. This strain was preferably grown on phenol rather than pyridine (data not shown) at 28±2°C, pH 7, with agitation at 150 rpm and phenol concentration of 1000-1250 mg l-1. Lower concentrations resulted in lower biodegradation rates and higher concentrations resulted in prolonged lag phase (Figure 3). |
| Recently, many studies have reported the isolation of bacterial strains with relevant phenol tolerance and biodegradation capacity [1]. However, isolated bacterial strains in the present study, particularly strain M4 showed one of the highest degradation capacities and rate among reported wild bacterial isolates [20]. Strain M4 was able to biodegrade up to 1700 mg phenol l-1 with a biodegradation rate of 29 mg h-1. Indeed, such high resistance and fast biodegradation rate is a prerequisite to rapidly bring down phenol concentration at satisfactory levels and to ensure efficient and stable phenol removal in continuous and especially batch treatment of real industrial wastewaters [1,8]. |
| Identification and characterization of algal isolates |
| Four algal strains were isolated (A1-A4) and morphologically characterized as members of the Chlorella genus. However, only 3 isolates showed remarkable tolerance to phenol and/or pyridine (Figure 4). Interestingly, all of the 3 algal isolates had almost similar tolerance pattern to phenol (Figure 4A) while A4 was more resistant against pyridine (Figure 4B). When a mixture of both pollutants was used at low concentrations, almost all algal isolates showed the same order of magnitude of inhibition; however, at a higher concentration (400/800), A4 had the lowest inhibition % (Figure 4C). All algal isolates were completely inhibited when incubated with a mixture of 500/1000 mg l-1 phenol/pyridine respectively (Figure 4C). |
| Although some studies [12] have reported the isolation of phenol biodegrading algal strains and even polycyclic aromatic hydrocarbons, none of the isolates algal strains in the present study showed a proof to degrade any of the tested pollutants. Similarly, many other studies reported that algal isolates lack the degradation capacities [21,22]. Hence algae commonly participate in the algal bacterial treatment of wastewater by providing the photosynthetic oxygen to the bacterial partner [8,22]. |
| Interestingly, phenol was more toxic to algae than pyridine especially at high concentrations where complete inhibition of algal growth was observed at 500 mg phenol l-1 compared to 1000 mg pyridine l-1. This agrees with previous studies reporting a phenol 96h- (Effective Concentration) EC50 of 370 mg l-1 to C. vulgaris [21], compared to a pyridine EC50 of 628 mg l-1 to algae and aquatic plants [23]. |
| Correlation between algal and bacterial isolates |
| The symbiotic algal–bacterial relationship is often beneficial when microalgae provide the O2 necessary for aerobic bacteria to biodegrade organic pollutants, consuming in turn the CO2 released from bacterial respiration. However, interaction between algae and bacteria is not limited to a simple CO2/O2 exchange where algae can have a detrimental effect on bacterial activity and vice versa [8,24]. However, in the present study neither bacterial isolate M4 nor algal isolate A4 had detrimental effect on each other (data not shown). Therefore, they can be used for the construction of an algal-bacterial microcosm for biological wastewater treatment. |
| Conclusion |
| Highly biodegrading and/or tolerant bacterial and algal strains were isolated. Both bacterial and algal isolates with the highest degradation and/or tolerance capacities had neither inhibitory nor beneficial impact on the growth of each other. Hence, these isolates could be assembled into a microcosm for photocatalytic -biological treatment of toxic effluents in algal–bacterial bioreactors. |
| Acknowledgements |
| This project was supported by funds from Cairo University, administrated by the Biotechnology Centre, Faculty of Pharmacy, Cairo University. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 |
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 14995
- [From(publication date):
June-2012 - Dec 23, 2025] - Breakdown by view type
- HTML page views : 10258
- PDF downloads : 4737
