A Deletion of Signal Peptide in Recombinant Murine Gamma-herpesvirus 68 M3 Protein Enhances its Binding Affinity to CCL5 Chemokine and Increases its Yield from Insect Cells
Received: 20-Dec-2017 / Accepted Date: 26-Dec-2017 / Published Date: 29-Dec-2017 DOI: 10.4172/2332-0877.1000347
Abstract
Objective: The M3 protein encoded by murine gamma-herpesvirus 68 (MHV-68) was the first secreted protein identified in herpesvirus. This protein is unique in its ability to bind a broad-spectrum of chemokines from all four subfamilies, thus it has been proposed to be a potential gene therapy candidate for controlling the overactive inflammatory responses of some human inflammatory diseases.
Methods: We prepared MHV-68 M3del protein with a deletion of its signal peptide (M3del) and full-length MHV-68 M3 protein (M3) using a baculovirus-mediated insect cell expression system and confirmed their specificity by Western blot using newly prepared mouse monoclonal anti-M3 antibody 1/27. Binding affinity of both proteins to human CCL5 and CXCL8 chemokine was examined by ELISA.
Results: M3del and M3 displayed affinity to both chemokines tested. M3 del concentration sufficient to bind 25% of CCL5 was two times smaller than that of M3 (IC25=3.5 nmol/l vs. 6.1 nmol/l). In contrary, the values of IC25 for CXCL8 for M3del and M3 were comparable (IC25=13.8 nmol/l vs. 13.3 nmol/l. We have found that the absence of signal peptide strongly affects the yield of recombinant M3 protein. The yields of M3del and M3 were in an unbalanced ratio of 67 to 1.
Conclusions: The results suggest that the absence of signal peptide in recombinant MHV-68 M3 protein allows increased binding activity of the protein to CCL5 but not to CXCL8, and even a very large increase in its yield from insect cells.
Keywords: Murine gamma-herpesvirus; M3 protein; Chemokines; Signal peptide; Insect cells
Highlights
The chemokine-binding properties of the recombinant MHV-68 M3 protein with deletion of its signal peptide.
Introduction
Chemokines are low molecular weight, chemoattractant cytokines (classified into four subfamilies-CC, CXC, CX3C and C) that regulate the trafficking and effector functions of leukocytes and immunocompetent cells and therefore play an important role in host defense against invading pathogens. The specific effects of chemokines are mediated by a subsequent intracellular cascade of signaling events leading to, among other events, the migration of immunocompetent cells to sites of inflammation in the appropriate tissues [1-3]. Given the central role that chemokine’s play in antiviral defenses, it is not surprising that many viruses have evolved strategies to alter host chemokine function to their benefit [4].
Large DNA viruses, particularly herpesviruses and poxviruses, encode viral chemokine binding proteins (vCKBPs) that effectively neutralize chemokines [5-10]. So far, five vCKBPs subfamilies have been described that differ in specificity as well as chemokine interaction mechanisms. However, only a few of them have been tested for their therapeutic potential in vivo even though neutralizing chemokine signaling is a very attractive therapeutic strategy for many diseases. Murine herpesvirus 68 (Murid herpesvirus 4 strain 68; MuHV-4; MHV-68) belongs to the genus Rhadinovirus, subfamily Gamma herpesvirinae [11]. This virus encodes a chemokine binding immunomodulatory M3 protein (also called vCKBP3) that has no sequence homology to any other known viral or human protein [12]. In addition to MHV-68, several other closely related murine herpesviruses were found in Apodemus spp. or Myodes spp. captured in Slovakia and Bohemia [13]. From these, the murine herpesviruses MHV-72 and MHV-4556 have been the most deeply studied with regards to their pathogenesis and molecular properties [14,15].
The first studies on the biological properties of the 406-residue MHV-68 M3 protein that is secreted into the media of virus-infected cells showed that it bound to a broad spectrum of chemokines from all four subfamilies, with the lowest binding affinities to members of the CXC subfamily [16,17]. M3 was the first example of a soluble inhibitor encoded by a herpesvirus. It specifically interacts with the N-terminal chemokine GPCR-binding domain, thereby blocking receptor recognition and inhibiting chemokine-mediated leukocyte migration [18-20]. Along with studies on its molecular properties, a variety of animal models have been developed to test the biological and pharmaceutical properties of the M3 protein, but they mainly relate to its potential use in gene therapy. Induction of M3 gene expression resulted in a 67% reduction in intimal area, suggesting that M3 may be effective in attenuating the intimal hyperplasia associated with arterial stenosis [21,22]. M3 expression overcame the cellular inflammatory responses in rat hepatocellular carcinoma lesions induced by a recombinant oncolytic vesicular stomatitis virus, prolonged the therapeutic effect of this virus, and improved animal survival [23]. Recombinant M3 also inhibited angiogenesis and neovascularization [24]. The experimental autoimmune encephalomyelitis animal model showed that M3 treatment significantly reduced the number of immune cells infiltrating neurons, indicating that M3 treatment might represent a novel therapeutic approach to neuroinflammatory disease [25]. Studies on double transgenic mice expressing both the M3 protein and different chemokines in pancreatic islets explored the role of chemokines and the effects of M3 on the development of diabetes mellitus type I [26-28]. Moreover, mice expressing M3 in the pancreas have also been shown to be resistant to induced diabetes [29].
In pioneering studies, Alexander et al. [19] prepared recombinant, full-length MHV-68 M3 in insect cells and determined the first three-dimensional crystal structure of M3, both alone and in complex with CCL2 (monocyte chemoattractant protein-1; MCP-1). Their model suggested how this viral protein is able to bind chemokines despite having no amino-acid homology to host chemokine receptors. Due to the complex structure of M3 and the broad spectrum of chemokines that it can potentially bind, the functions of its individual domains are only poorly understood. The binding capabilities of M3 to a variety of chemokines were mostly identified by crosslinking assays with radiolabeled chemokines, providing only semi-quantitative data on the binding of M3, which was either secreted into the media of virus-infected BHK-21 cells (baby hamster kidney-21) or prepared recombinantly in E. coli or insect cells, to human and mouse chemokines. In addition to the chemokines examined in this study, chemokines CCL3 (macrophage inflammatory protein; MIP-1α), CCL11 (eotaxin-1), CCL13 monocyte chemoattractant protein-4; MCP-4), CXCL6 (granulocyte chemotactic protein-2; GCP-2), CXCL13 (human B cell–attracting chemokine 1; BCA-1), and IL-18 (interleukin 18) [16,17] were also studied.
In an early study performed in our laboratory, Belvončíková et al. [30] evaluated the binding of MHV-68 M3 secreted into the media of infected BHK-21 cells to five human chemokines, CCL2, CCL3, CCL5 (regulated upon activation, normal T-cell expressed, and secreted; RANTES), CCL11 and CXCL8 (interleukin 8; IL-8), using an ELISA assay. The protein was compared to its counterpart from MHV-72 (clone h3.7), which possesses a unique Asp307Gly mutation near the C-terminus of the M3 CCL2-chemokine binding domain. Experimental infection of Balb/c mice with MHV-72 found that this virus elicited an attenuated immune response and exhibited weaker latency establishment and reactivation [31-33]. Belvončíková et al. [30] found that both M3 proteins had the weakest affinity for the CCL3 chemokine. Moreover, MHV-72 M3 could bind only 11% and 20% of the respective amounts of CCL5 and CXCL8 that MHV-68 M3 could. These results suggested that even a single mutation could affect the affinity of the M3 protein for individual chemokines.
The many advantages of the E. coli expression system continue to make it the first choice for high-level production of heterologous proteins [34]. E. coli cells have been well characterized, have relatively simple genetics and grow rapidly, making them the preferred hosts for recombinant protein production for both biochemical and structural studies [35-39]. Specific properties of the M3 protein, including its relatively small size, good solubility and absence of posttranslational modifications, allowed us to produce recombinant, full-length M3 protein from both MHV-68 and MHV-72 in E. coli cells [40]. Evaluation of the MHV-68 M3 protein affinity for CCL5, CXCL8 and CCL3 showed that binding to CCL5 is approximately 20 and 100-times stronger than its binding to CXCL8 and CCL3, respectively. An additional study on the MHV-72 M3 protein prepared in an E. coli system showed that its unique mutation reduces its binding to all chemokines tested, including CCL5, CXCL8 and CCL3, with the strongest impact on CCL3 binding [41]. Unfortunately, in both studies the majority of the expressed heterologous protein appeared in E. coli inclusion bodies, and approximately half of the recombinant protein produced formed insoluble aggregates.
In this study, we evaluated the effect of a deletion of the signal peptide (the first 24 residues) in recombinant MHV-68 M3 protein on its chemokine binding properties. To prepare recombinant proteins with and without signal peptide, we used a baculovirus-mediated insect cell expression system and compared both M3del and M3, focusing on their binding to CCL5 and CXCL8.
Materials and Methods
Virus and cells
The twice-plaque purified clone f2.6 of MHV-68, obtained from an isolate of the bank vole Myodes glareolus, was used to prepare pure virion DNA as previously described [12,15,42]. The Spodoptera frugiperda 9 (Sf9) insect ovary cell line (ATCC® CRL-1711) was maintained as an adherent culture in Sf-900TM II SFM media (Gibco) supplemented with 2.5-10% (v/v) fetal bovine serum (FBS, Sigma-Aldrich) and 50 µg/ml of gentamicin (Sandoz) at 27.5°C. The baby hamster kidney (BHK-21) cell line was cultivated in DMEM (Lonza) at 37°C in 5% CO2. Escherichia coli cells (JM109) were used according to the manufacturer’s instructions (Promega).
Preparation of anti-M3 monoclonal antibody
To prepare an antibody against the M3, two 6- to 8-week-old inbred Balb/c mice were immunized with purified full-length recombinant MHV-68 M3 protein produced in E. coli cells according Pančík et al. [40]. The mice were inoculated subcutaneously in the groin with different antigen doses (50 µg and 100 µg) diluted in PBS and emulsified with 100 µl of complete Freund’s adjuvant. Another mouse, inoculated with PBS and managed as the immunized mice, served as a negative control. Two weeks later, a second immunization was performed in the same manner, but using incomplete Freund’s adjuvant. To achieve as high a titer of anti-M3 antibodies as possible, mice were subcutaneously boosted by injecting 25 µg of antigen in 100 µl PBS. The titer of anti-M3 polyclonal antibodies in the serum of immunized mice was screened by an indirect ELISA. The serum of immunized mice No 1 (poly anti-M3-Ab/1) with the highest titer of anti-M3 polyclonal antibodies was used in following experiments.
Three days following the last inoculation, the mice spleens were removed into serum-free DMEM medium (SFM) and homogenized. Spleen cells were then used for hybridization and fusion with NS0 murine myeloma cells using polyethylene glycol as a fusing agent, following conventional methods for the generation of monoclonal antibodies [43], slightly modified for our requests. Antibody-secreting fused cells, growing in selective HAT medium (DMEM with 20% fetal calf serum FCS with HAT supplement (50x), Hybri-Max (Sigma, Germany)), were screened by indirect ELISA. Positive clones were selected and expanded by additional culture in HT selective medium (DMEM with 10-20% FCS with HT supplement (50x), Hybri-Max) and finally in complete medium (DMEM with 10% FCS). The specificity of a set of monoclonal antibodies was continuously determined by testing hybridoma culture supernatants by indirect ELISA with recombinant MHV-68 M3 protein mentioned above. Cells from positive clones were frozen and then cloned after limiting dilution; afterwards, they were expanded as above, and their cultivation medium was screened with an indirect ELISA. To prepare an anti-M3 monoclonal antibody suitable for Western blot analyses, 1 × 107 hybridoma cells were first cultivated in DMEM with 10% FCS and then again in SFM for three and two days, respectively. Prior to use, the presence of anti-M3 activity of each monoclonal antibody in aliquots of SFM was tested by indirect ELISA. Data shown represents the mean ± SEM of triplicate experiments.
Determination of the specificity of the mouse monoclonal anti-M3 antibody 1/27
From a set of newly prepared monoclonal antibodies we chose anti-M3 antibody 1/27 and tested its specificity as follows. BHK-21 cells were seeded onto 6-well plates (5 × 105 cells/well) and cultivated in DMEM supplemented with 5% (v/v) FBS, penicillin-streptomycin (100 U-100 μg/ml), and amphotericin B (2.5 μg/ml) at 37°C in 5% CO2 overnight. Then, BHK-21 cells were infected with MHV-68 at an MOI of 2, 1 or 0.1. The cells were washed with physiological saline (pH 7.2) 45 min post-infection and incubated in 2 ml of fresh DMEM supplemented with 1% (v/v) FBS at 37°C in 5% CO2. All samples were tested in duplicate. The medium and cells were harvested at 24 h post infection. The cells were lysed in 300 µl of RIPA lysis buffer supplemented with a protease inhibitor cocktail (Sigma Fast Protease Inhibitor Tablets; Sigma-Aldrich). Then, the protein concentrations of the lysates of BHK-21 cells infected with MHV-68 as well as the medium were determined using a bi-cinchoninic acid assay (Pierce) according to the manufacturer’s instructions. Protein samples (22 µg) were separated on a 12.5% SDS-PAGE gel, transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore) and probed with either the primary mouse monoclonal anti-M3 antibody 1/27 or poly anti-M3-Ab/1 (both diluted in 5% (w/v) nonfat dry milk in TBST buffer (10 mmol/l Tris-HCl, 150 mmol/l NaCl, and 0.05% (v/v) Tween 20; pH 7.5)) at a 1:100 ratio at 21°C for 2 h followed by incubation with the secondary goat anti-mouse IgG-horseradish peroxidase (HRP) conjugate antibody (Novagen) (diluted at a 1:5000 ratio) for 1 h. The membrane was washed five times with TBST for 15 min after incubation with each antibody. Finally, DAB (3,3'-diaminobenzidine, Santa Cruz Biotechnology) was used as substrate for HRP to observe a specific signal for the M3 protein.
Detection of anti-M3 antibodies in mouse serum and cultivation media by indirect ELISA
The binding of the anti-M3 antibodies present in mouse sera and cell media was examined in triplicate. Ninety-six-well plates were first coated with different amounts of antigen (500, 250, 50 and 5 ng of recombinant MHV-68 M3 protein) [40] in a 50 µl/well volume and incubated overnight. The plates were then washed three times with 0.05% Tween 20 (MP Biomedicals) in PBS and blocked with 1% BSA (Sigma) in PBS with 0.05% Tween 20 for 90 min. The mouse serum (diluted 102 to 105 times) or medium being tested was added and incubated for 2 h. HRP conjugated to a swine anti-mouse antibody diluted in a 1:1000 ratio (Lachema) was used as a secondary antibody; ortho-phenylenediamine (OPD, Sigma) diluted in a pH 6 phosphate buffer served as the substrate. The reaction was stopped with 2 M H2SO4. All incubations were performed at room temperature. The binding of anti-M3 antibodies to the recombinant M3 protein present in the samples was evaluated by determining the optical density at 492 nm (OD492) using an ELISA reader (ELx808, BioTek).
OD492 values were normalized to values of serum of uninfected mouse or uninfected Sf9 cell medium used as a negative control.
Construction of a baculovirusexpression vector
MHV-68 DNA was used as a template to amplify a fragment of the full-length M3 gene (nt 6058–7206) or M3 gene without its signal peptide sequence encoding M3del (aa 25–406) (GenBank accession. No AAF19271.1). A proofreading Takara ExTaq DNA polymerase (Clontech) was used in the PCR mixture with the primers either pAcGP-F3M3Bam, 5´ CCGGGATCCATGGCCTTCCTATCCACATCTGTGC-3´ or pAcGP-M3BamF, 5′-CCGGGATCCCCTTACTCTAGGTTTGGCAC-3′ and pAcGP-M3EcoRHis, 5′-CCGGAATTCTCAATGGTGATGGTGATGATGATCCCCAAAATAC-3′. An uncleavable penta-histidine tag was introduced, and the appropriate restriction sites were added to 3´ end of this fragment encoding the M3 or M3del fused with His5. The PCR procedure was performed in a 50-µl volume with 35 cycles of 94°C for 60 s, 67.2°C for 60 s and 72°C for 90 s, followed by an extension at 72°C for 3 min in a thermal Labcycler (SensoQuest). The PCR product with a length of 1251 bp or 1183 bp was purified using a QIAquick PCR Purification Kit (Qiagen), digested with BamHI and EcoRI (Termo Scientific) and cloned into the baculovirus transfer vector pAcGP67A (BD Biosciences Pharmingen) downstream of the polyhedrin promoter and gp67 signal sequence of the vector. The construction of recombinant baculovirus transfer vector pAcGP67A/M3 or pAcGP67A/ΔssM3 was verified by restriction fragment length polymorphism (RFLP) analysis and sequencing (BITCET, Slovakia). This vector was then propagated in E. coli JM109 competent cells according to the manufacturer’s instructions.
Preparation of a recombinant baculovirus
Healthy and dividing Sf9 insect cells were seeded on a T-25 (25 cm2) flask (4 × 105 cells/ml) and cultivated in serum-free Sf-900TM II media without antibiotics. To transfect cells, the culture medium was removed and a transfection mixture containing 3 µg of the baculovirus transfer vector, 0.2 µg of ProGreen™ Baculovirus DNA (carrying a green fluorescent protein (GFP) reporter gene) (AB Vector) and 20 µl of Cellfectin reagent in 1 ml of cultivation medium was added dropwise. This was followed by incubation in the dark at 21°C for 4 h with hourly back-and-forth agitation. A mixture with only 20 µl of Cellfectin reagent and medium was used as a negative control. Following cultivation, the transfection mixture was replaced with 5 ml of fresh media containing 10% (v/v) fetal bovine serum (FBS, Sigma-Aldrich) and 50 µg/ml of gentamicin (Sandoz), and cells were cultivated at 27.5°C for 7 days. The cultivation medium containing the released recombinant baculovirus was purified from cell debris (1000 × g for 10 min) and then filtered through a 0.45 µm filter; the titer of recombinant baculovirus was determined using an end-point dilution assay. Afterwards, the first and subsequent viral stocks were prepared by infecting 2 × 106 cells (seeded on a T-25 flask with 5 ml medium) and 1.4 × 107 cells (seeded on a T-125 flask with 50 ml of medium) with a multiplicity of infection below 1 (MOI<1 PFU/cell) followed by 7 days of cultivation, respectively. To prepare a stock with a high viral titer, the cells were infected with an MOI of 1 [44]. All viral stocks were purified as described above and stored at 4°C in the dark.
The expression of M3del and M3 in insect cells
First, Sf9 cells were sequentially adapted for growth in low serum medium (2.5% v/v) in a six-well culture plate (2 × 106 cells/ml) and were then infected with the baculovirus (at MOI=3). After 1 h of culture, fresh medium was replaced and cells were cultivated at 27.5°C for 9 days. The production of recombinant protein in infected cells was followed by fluorescence microscopy (Axiovert 40 CFL, Zeiss) and screened by harvesting cell and medium aliquots 1, 3, 6 and 9 days post infection (d.p.i.). Uninfected Sf9 cells grown in parallel were used as a negative control. Nested RT-PCR was used to follow M3 gene transcription in virus-infected cells. Briefly, total cellular RNA was purified using an EZ-10 Spin Column Total RNA Mini-Preps Super Kit (Bio Basic) and treated with 1 U of RNase free DNase I (Invitrogen). Then, 750 ng of this RNA was used to prepare cDNA using a Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Scientific) and oligo dT16-18 primers. Ten nanograms of this cDNA was used as template in a 25-µl reaction mixture for the first nested RT-PCR round with the following outer primers: M3PP1F, 5′-ACTCCAGCCTGTACTGTTGC-3′ (nt 6075–6094), and M3PP1R, 5′-TCTGCCCCACAACCAAGTTT-3′ (nt 6575–6594); this reaction amplified a 520-bp long fragment of the M3 gene. This PCR procedure was performed with 35 cycles of 94°C for 40 s, 59.3°C for 30 s and 72°C for 45 s, followed by an extension at 72°C for 3 min in a SensoQuest Lab Cycler. The second nested RT-PCR reaction mixture contained 1 µl of the products from the first-round PCR as template and the following inner primers to amplify a 241-base pair fragment: M3PP1FN, 5′-ACTGGCCCTCAACCAGTCTA-3′ (nt 6171–6190), and M3PP1RN, 5′-TACAAGTACAGCGTGAGCCC-3′ (nt 6392–6411). This PCR procedure was performed as described above, except the annealing temperature was 59.8°C, the polymerization step was 20 s, and 40 cycles were run. The nested RT-PCR product was evaluated on a 1.5% agarose gel using the GoodViewTM Nucleic Acid Stain (Beijing SBS, Genetech).
The production of recombinant M3del or M3 by infected Sf9 cells was followed by Western blotting using the mouse anti-M3 monoclonal antibody prepared in this study. The cell medium was harvested at the time intervals noted above and screened for proteins. Samples were separated on a 12.5% (w/v) SDS-PAGE gel, transferred onto a nitrocellulose membrane (Sigma-Aldrich) and probed with the primary mouse anti-M3 monoclonal antibody (diluted in 5% (w/v) nonfat dry milk in TBST buffer (10 mmol/l Tris-HCl, 150 mmol/l NaCl, and 0.05% (v/v) Tween 20; pH 7.5)) at a 1:100 ratio at 21°C for 2 h followed by incubation with the secondary goat anti-mouse IgG-HRP conjugate antibody (Novagen) (diluted at a 1:5000 ratio) for 1 h. After incubation with both primary and secondary antibodies, the membrane was washed five times with TBST for 15 min. Finally, HRP color was developed using the DAB substrate (3,3′-diaminobenzidine, Santa Cruz Biotechnology), allowing the specific signal of the M3 protein to be observed in the samples.
Purification of M3del and M3 by affinity chromatography
The manufacturer’s recommended batch procedure was used to purify recombinant M3 proteins from the cultivation medium after 6 days of culturing Sf9 cells infected with baculovirus. The medium was first collected and purified as described above. Next, the purified medium was diluted with an equivalent volume of buffer A (10 mmol/l Tris-HCl and 10 mmol/l imidazole; pH 8) and gently mixed with HIS-Select Cobalt Affinity Gel (Sigma) or Ni-NTA agarose (Qiagen) that had been equilibrated in buffer A. After 1 h of incubation at 4°C with gentle shaking, the mixture was loaded onto a chromatography column and subsequently washed with 5 ml of buffer A to remove non-specifically bound proteins. To elute the purified recombinant protein, the column was washed six times with 150 µl of elution buffer B (10 mmol/l Tris-HCl and 100 mmol/l imidazole; pH 8). To prevent protein degradation, the pooled elution fractions were dialyzed against buffer C (20 mmol/l Tris-HCl, 150 mmol/l NaCl, and 20% (v/v) glycerol; pH 8.0) at 4°C on an Amicon Ultracel 30 K (Merck Millipore) filter. The amount of protein in the sample was determined using a NanoDrop 2000 Spectrophotometer (280 nm) (Thermo Scientific) and was stored at -20°C.
Determination of the chemokine binding properties of M3del and M3
The binding of purified recombinant M3 proteins to the chemokines CCL5 and CXCL8 was determined using the Human CCL5/RANTES and Human CXCL8/IL-8 DuoSet ELISA kits (R&D Systems). In each assay, 50 pg of recombinant chemokine were mixed with 2-400 ng of protein in a total volume of 110 µl. Each mixture was incubated at 21°C for 1.5 h with gentle shaking and then applied to the ELISA plates (100 µl/well; Greiner Bio-one) following the manufacturer’s instructions except that OPD was used as the substrate. A reduction in the sample’s OD492 relative to that of 50 pg of commercial recombinant chemokine was interpreted as evidence of M3 binding. Each assay for each chemokine was measured in triplicate. The elution buffer B served as a negative control. The strength of both M3 proteins binding to each chemokine was evaluated by determining the amount of protein that bound to 25% of the chemokine and data for both M3 proteins were expressed as inhibitory concentration (IC25) of protein sufficient for 25% inhibition of the chemokines.
Results
Preparation of anti-M3 antibodies
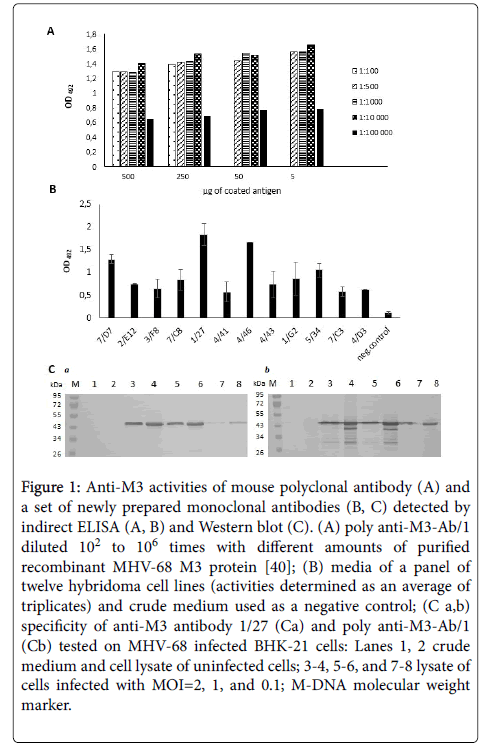
We induced the production of specific anti-M3 polyclonal antibodies in inbred Balb/c mice by immunizing them with recombinant MHV-68 M3 protein produced by E. coli cells [40]. In the ELISA assays, serum from immunized mice, but not the non-immunized mouse, bound the M3 protein (data not shown). We found that using poly anti-M3-Ab/1 diluted from 102 to 104 provided the most reproducible results with 5 ng of coated antigen (Figure 1A). By fusing mouse splenocytes and NS0 cells, we prepared twelve hybridoma clones, which produced anti-M3 monoclonal antibodies. We confirmed the specificity of this panel of monoclonal antibodies by indirect ELISA (Figure 1B). In addition, we obtained two stable hybridoma cell lines, 7/D7 and 1/27, which produced high levels of anti-M3 monoclonal antibodies which bound well recombinant M3 protein prepared in E. coli cells [40]. As shown in Figure 1C, the specificity of anti-M3 antibody 1/27 is in compliance with that of mouse poly anti-M3-Ab/1. Three additional stable hybridoma cell lines, 4/41, 4/43 and 1/G2 also produced antibodies, but only about half the amount of the 7/D7 and 1/27 cell lines. Some hybridoma cell lines, 2/E12, 3/F8, 7/C8, 4/46, 5/34, 7/C3 and 4/D3, stopped multiplying during the expansion procedure and lost the ability to produce antibodies (data not shown). An anti-M3 antibody 1/27 was used in the following experiments to identify the recombinant MHV-68 M3 protein produced by insect cells infected with recombinant baculovirus.
Figure 1: Anti-M3 activities of mouse polyclonal antibody (A) and a set of newly prepared monoclonal antibodies (B, C) detected by indirect ELISA (A, B) and Western blot (C). (A) poly anti-M3-Ab/1 diluted 102 to 106 times with different amounts of purified recombinant MHV-68 M3 protein [40]; (B) media of a panel of twelve hybridoma cell lines (activities determined as an average of triplicates) and crude medium used as a negative control; (C a,b) specificity of anti-M3 antibody 1/27 (Ca) and poly anti-M3-Ab/1 (Cb) tested on MHV-68 infected BHK-21 cells: Lanes 1, 2 crude medium and cell lysate of uninfected cells; 3-4, 5-6, and 7-8 lysate of cells infected with MOI=2, 1, and 0.1; M-DNA molecular weight marker.
Expression in insect cells and purification of the MHV-68 M3del and full-length M3 protein
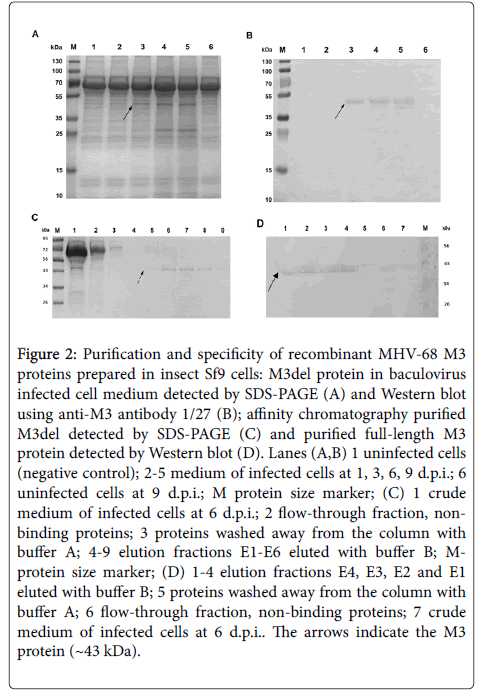
The recombinant baculovirus transfer vector carrying the M3 gene or M3 gene which lacks its signal peptide sequence was first confirmed to be constructed correctly via RFLP analysis by digestion with BamHI+EcoRI, PvuII and DraI (data not shown); both transfer vectors were also confirmed by sequencing (data not shown). Then, Sf9 cells were infected with recombinant baculovirus at an MOI of 3 PFU/cells. The expression of M3del or M3 or was screened after 1, 3, 6 and 9 days of infection using light and fluorescence microscopy to follow the cytopathic effects and the emission of GFP, respectively (data not shown). The earliest over expression of the GFP reporter protein was found 3 days after infection. To identify the best time for production of the recombinant protein, the expression of transcript of both M3 proteins was investigated using RT-PCR after 1, 3, 6 and 9 days of infection. We found comparable M3 transcript expression levels from 1 to 6 d.p.i. (data not shown), but declining levels after 6 d.p.i. To find the optimal infection time for secretion of each M3 protein into the medium, its amount in the media was investigated at the same time intervals on a 12.5% SDS-PAGE. As shown on Figure 2A, M3del was secreted from producing cells 3 to 9 d.p.i., with the highest amount appearing in the two last intervals tested. We gained the same data for M3 (data not shown). Based on these results, we purified both M3 proteins from the baculovirus-infected insect cell media after six days of cultivation at 27.5°C by affinity chromatography. SDS-PAGE and Western blot analyses of M3del in cell media (Figure 2B) and purified M3del (Figure 2C) as well Western blot analysis of purified M3 (Figure 2D), confirmed the identity of proteins. It should be mentioned that 14 times more media of baculovirus-infected insect cells should be used to obtain M3 in quality comparable with M3del. As shown in Figure 2C (lanes 5–9) and Figure 2D (lanes 1–4), the elution fractions E2–E6 and E4–E1 contained the majority of the M3del and M3. However, the evaluation of effectiveness of protein production shows that the yield of M3 was about 67 times smaller compared to M3del yield. However, both M3 proteins were then used for subsequent analysis of their biological properties.
Figure 2: Purification and specificity of recombinant MHV-68 M3 proteins prepared in insect Sf9 cells: M3del protein in baculovirus infected cell medium detected by SDS-PAGE (A) and Western blot using anti-M3 antibody 1/27 (B); affinity chromatography purified M3del detected by SDS-PAGE (C) and purified full-length M3 protein detected by Western blot (D). Lanes (A,B) 1 uninfected cells (negative control); 2-5 medium of infected cells at 1, 3, 6, 9 d.p.i.; 6 uninfected cells at 9 d.p.i.; M protein size marker; (C) 1 crude medium of infected cells at 6 d.p.i.; 2 flow-through fraction, non-binding proteins; 3 proteins washed away from the column with buffer A; 4-9 elution fractions E1-E6 eluted with buffer B; M-protein size marker; (D) 1-4 elution fractions E4, E3, E2 and E1 eluted with buffer B; 5 proteins washed away from the column with buffer A; 6 flow-through fraction, non-binding proteins; 7 crude medium of infected cells at 6 d.p.i.. The arrows indicate the M3 protein (~43 kDa).
Binding of the MHV-68 M3del and M3 to chemokines
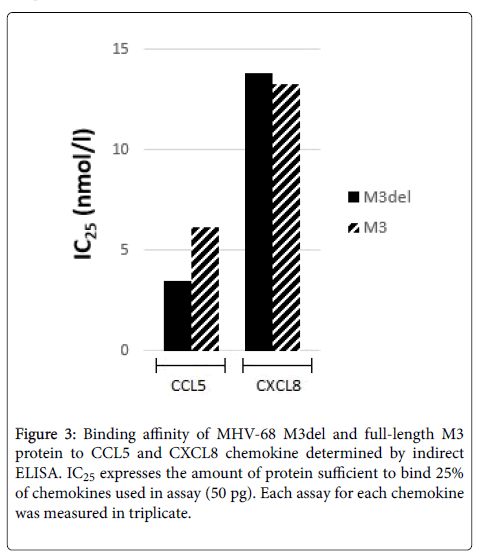
An ELISA assay designed to discern the chemokine-binding properties of M3del and M3 demonstrated that they both had binding affinities to CCL5 and CXCL8. Although we obtained a very low yield of M3, it was about 67 times smaller compared to M3del yield, we could compare proteins by determining their IC25 values for CCL5 and CXCL8 (Figure 3). We found that the concentration sufficient to bind 25% of CCL5 of M3del was 3.51 nmol/l, while about two times more M3 was required to bind the same amount of CCL5 (IC25=6.17 nmol/l). The values of IC25 for CXCL8 for M3del and M3 were comparable (IC25=13.8 nmol/l vs. 13.3 nmol/l).
Discussion
Gamma-herpesviruses are predominantly host-specific, so the virus vs. host immune system interaction plays a crucial role in viral infection, particularly in latency establishment and reactivation from latency [45]. It is very important for proper function of the host immune system to effectively deliver immunocompetent cells to the site of virus replication. Chemokines trigger many signaling cascades involved in innate immunity by binding to their receptors on the surface of immunocompetent cells. Many viruses have developed strategies to overcome this signal transduction, which clearly illustrates the significance of its role in the battle against viral infection [6]. Murid and human gamma-herpesviruses share several common pathogenic characteristics as a consequence of their genome homology.
MHV-68 infection of its host results in an acute productive infection of lung alveolar epithelial cells and in the latent infection of several cell types including B lymphocytes, macrophages and epithelia. The viral infection induces an inflammatory infiltrate in the lungs and enlargement of the lymph nodes and spleen, caused by a strong chemokine response. In spite of the high expression of chemokines in the lungs and spleen of experimentally infected laboratory mice, the significant benefit of this inflammatory response to virus replication or pathogenicity has not been successfully proven [46]. The first secreted herpesvirus protein, vCKBP-3 encoded by MHV-68, was initially characterized as a unique, soluble viral decoy receptor that bound a broad spectrum of chemokines with high affinity and destabilized the chemokine network both in vitro and in vivo [16,17,26]. The binding affinities of M3 for a broad-spectrum of chemokines in all four subfamilies have been well characterized [16,19,30,40,41], but data on its binding to individual chemokines are rather rare.
The unique properties of MHV-68 M3 protein have kept it a topic of interest beyond the pioneering work of Alexander et al. [19]. In this work, we studied a functional form of M3 without the first 24 residues that function as a signal peptide, which is cleaved in the mature protein [19,47]. Understanding the importance of this signal peptide sequence for the binding of M3 protein to variety of chemokines would expand our knowledge of its biology and increase the prospects of using its unique properties in gene therapy. To examine how the deletion of signal sequence could change the overall chemokine-binding properties of MHV-68 M3 protein we compared M3del and M3 prepared in insect cells that were successfully used to prepare a recombinant form of the human cytomegalovirus immunomodulatory protein UL141 of similar size [44]. To test the chemokine binding properties of recombinant M3 proteins proven to be specific by newly prepared monoclonal anti-M3 antibody we chose two human chemokines. Though they come from different subfamilies, CCL5 and CXCL8, any recombinant antagonist would be an interesting candidate for gene therapy. The inflammatory chemokine CCL5 plays a critical role in T-lymphocyte, macrophage, eosinophil and basophil activation, proliferation and recruitment. Although the exact function of CCL5 in tumorigenesis remains unclear, it may be involved in melanoma and has also been implicated in tumor growth and progression [48]. The second chemokine, CXCL8, is highly involved in wound healing and triggers the infiltration of both macrophages and neutrophils in cystic fibrosis [49]. In addition, CXCL8 is associated with a highly metastatic phenotype [50].
We found that deletion of the N-terminal signal peptide in M3del does not hamper its binding to both the CCL5 and CXCL8, but this binding is different for these chemokines. Comparing the values of IC25 for CCL5 and CXCL8 has shown that approximately 3.9 times more M3del is needed to bind CXCL8 than the same amount of CCL5. Our results correlate with the results of previous study on MHV-68 M3del protein prepared in E.coli cells which exhibited different abilities to bind chemokines as approximately 5 times more M3del protein was needed to bind CXCL8 than the same amount of CCL5 [41]. An important outcome of this study is the finding that the absence of signal peptide strongly affects the yield of recombinant M3 protein being produced in insect cells. We found that from the same amount of cells (managed by the same manner) it is possible to obtain up to 70 times less full-length M3 protein than the protein with deleted signal peptide.
Finding that the concentration of M3del sufficient to bind 25% of CCL5 was two times smaller than that of M3 (IC25=3.51 nmol/l vs. 6.17 nmol/l) suggest that the cleavage of signal peptide might play an important role also in the proper protein folding required for effective binding of M3 protein to chemokines, particularly to CCL5. However, this conclusion does not seem to concern the second chemokine, CXCL8, when the values of IC25 for M3del and M3 were comparable. Contrary to MHV-68 M3del produced in E. coli cells displaying approximately twenty times decreased binding to both CCL5 and CXCL8 compared to its full length counterpart [41], MHV-68 M3del produced in insect cells had higher binding affinity to CCL5 and unchanged binding affinity to CXCL8. It is significant that comparable affinities for CCL5 and CXCL8 were demonstrated for the M3 protein secreted by virus-infected BHK-21 cells [30], while affinity of full-length M3 protein produced in E. coli cells to CXCL8 was found to be at least orderly reduced compared to CCL5. The results of all studies using different cells show the importance of the cells in which protein is being produced should be considered. It can be concluded that the production of recombinant M3 proteins in insect cells was the best choice for following studies on the possibilities to increase and control chemokine binding of MHV-68 M3 protein. Taking in account that MHV-68 M3 protein is able to bind a broad spectrum of chemokines, detecting such huge increase of the yield we found for M3del protein produced in insect cells is a quite surprising but very good chance of assessing the possibility to modulate the binding of M3 protein to desirable chemokines. Following experiments are in the progress to identify an impact of selected mutations introduced to M3 protein sequence on its chemokine binding properties.
Conclusions
The existence of a large number of chemokines involved in different human diseases suggests that the therapeutic use of a broad-spectrum chemokine antagonist may be effective. Examination of viral chemokine inhibitors that have been actively interacting with the chemokine system for millions of years may help to control an over-reactive inflammatory response in a number of human inflammatory diseases in the near future. Virus-encoded vCKBPs with different binding specificities, such as the M3 protein of MHV-68, offer a valuable and unique source to further develop and design chemokine inhibitors.
Results conclude that the MHV-68 M3del protein lacking its N-terminal signal peptide sequence prepared in insect cells is a fully functional protein with selective affinity to CCL5 and CXCL8 chemokines. The absence of signal peptide in M3 protein prepared in insect cells was clearly demonstrated to increase protein binding to CCL5 and to have no effect on its binding to CXCL8. It also significantly increases the yield of recombinant M3 protein from insect cells. Our study has revealed new information about the biological properties of the MHV-68 M3 protein, which is a potentially useful candidate for regulating the immune response in future therapeutic strategies.
Acknowledgements
The authors are grateful to Dr. Jacob Bauer for his helpful remarks and discussion. This work was supported by grants from the Slovak Research and Development Agency [#APVV-0621-12, APVV-15-0474] and the Scientific Grant Agency of Slovak Republic [#VEGA 2/0087/17].
Ethics Statement
Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the Institute of Virology, BMC SAS. The animals were treated according to the European Union standards, and fundamental ethical principles, including animal welfare requirements, were respected. The experiments were evaluated and approved by the State Veterinary and Food Administration of the Slovak Republic, Permit Number: 292/16-221c.
Compliance with Ethical Standards Conflict of Interest
The authors declare that they have no conflicts of interest regarding the publication of this paper.
References
- Baggiolini M (1998) Chemokines and leukocyte traffic. Nature 392: 565-568.
- Fernandez EJ, Lolis E (2002) Structure, function, and inhibition of chemokines. Annu Rev Pharmacol Toxicol 42: 469-499
- Sallusto F, Baggiolini M (2008) Chemokines and leukocyte traffic. Nat Immunol 9: 949-952.
- Alcami A, Koszionowski UH (2000) Viral mechanisms of immune evasion. Immunol Today 21: 447-455.
- Lalani AS, Barrett JW, McFadden G (2000) Modulating chemokines: more lessons from viruses. Immunol Today 21: 100-106.
- Alcami A (2003) Structural basis of the hespesvirus M3-chemokine interaction. Trends Microbiol 11: 191-192.
- Seet BT, Barrett J, Robichaud J, Shilton B, McFadden G (2001) Glycosaminoglycan binding properties of the myxoma virus CC-chemokine inhibitor, M-T1. J Biol Chem 276: 30504-30513.
- Bryant NA, Davis-Poynter N, Vanderplasschen A, Alcami A (2003) Glycoprotein G isoforms form some alphaherpesviruses function as broad-spectrum chemokine binding proteins. EMBO J 22: 833-846.
- Costes B, Ruiz-Argüello MB, Bryant N, Alcami A, Vanderplasschen A (2005) Both soluble and membrane-anchored forms of Felid herpesvirus 1 glycoprotein G function as a broad-spectrum chemokine-binding protein. J Gen Virol 86: 3209-3214.
- Van de Walle GR, May ML, Sukhumavasi W, vonEinem J, Osterrieder N (2007) Herpesvirus chemokine-binding glycoprotein G (gG) efficiently inhibits neutrophil chemotaxis in vitro and in vivo. J Immunol 179: 4161-4169
- van Regenmortel MHV, Fauquet CM, Bishop DHL (2000) Herpesvirus family; in: Virus Taxonomy: Classification and Nomenclature of Viruses, 7th ICTV Report Academic Press 220-226
- Blaskovic D, Stanceková M, Svobodová J, MistrÃková J (1980) Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol 24: 468.
- MistrÃková J, RaÅ¡lová H, Mrmusová M, Kúdelová M (2000) A murine gammaherpesvirus. Acta Virol 44: 211-226.
- Halásová Z, ValoviÄová M, MaÄáková K, PanÄÃk P, BelvonÄÃková P, et al. (2011) Partial genome sequence of murine gammaherpesvirus 72 and its analysis. Acta Virol 55: 317-325.
- Kúdelová M, Halásová Z, BelvonÄÃková P, PanÄÃk P, Režuchová I, et al. (2012) Partial genome analysis of murine gammaherpesvirus 4556. Acta Virol 56: 177-186.
- Parry CM, Simas JP, Smith VP, Stewart CA, Minson AC, et al. (2000) A broad spectrum secreted chemokine-binding protein encoded by a herpesvirus. J Exp Med 191: 573-578.
- van Berkel V, Barrett J, Tiffany HL, Fremont DH, Murphy PM, et al. (2000) Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J Virol 74: 6741-6747.
- Webb LMC, Clark-Lewis I, Alcami A (2003) The gammaherpesvirus chemokine binding protein binds to the N-terminus of CXCL8. J Virol 77: 8588-8592
- Alexander JM, Nelson Ch A, van Berkel V, Lau EK, Studts JM, et al. (2002) Structural basis of chemokine sequestration by a herpesvirus decoy receptor. Cell 111: 343-356
- Sarawar SR, Lee BJ, Anderson M, Teng YC, Zuberi R, et al. (2002) Chemokine induction and leukocyte trafficking to the lungs during murine gammaherpesvirus 68 (MHV-68) infection. Virology 293: 54-62.
- Gossen M, Bujard H (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A 89: 5547-5551.
- Pyo R, Jensen KK, Wiekowski MT, Manfra D, Alcami A, et al. (2004) Inhibition of hyperplasia in transgenic mice conditionally expressing the chemokine-binding protein M3. Am J Pathol 164: 2289-2297
- Wu L, Huang T, Meseck M, Altomonte J, Ebert O, et al. (2008) rVSV (M∆51)-M3 is an effective and safe oncolytic virus for cancer therapy. Hum Gene Ther 19: 635-647
- Andrés G, Leali D, Milota S, Coltrini D, Camozzi M, et al. (2009) A pro-inflammatory signature mediates FGF-2 induced angiogenesis. J Cell Mol Med 13: 2083-2108.
- Millward JM, Holst PJ, Høgh-Petersen M, Thomsen AR, Christensen JP, et al. (2010) The murine gammaherpesvirus-68 chemokine-binding protein M3 inhibits experimental autoimmune encephalomyelitis. J Neuroimmunol 224: 45-50.
- Jensen KK, Chen S, Hipkin RW, Wiekowski MT, Schwarz MA, et al. (2003) Disruption of CCL21-induced chemotaxis in vitro and in vivo by M3, chemokine-binding protein encoded by Murine gammaherpesvirus 68. J Virol 77: 624-630.
- Martin AP, Canasto-Chibuque C, Shang L, Rollins BJ, Lira SA (2006) The chemokine decoy receptor M3 blocks CC chemokine ligand 2 and CXC chemokine ligand 13 function in vivo. J Immunol 177: 7296-7302.
- Lira SA, Viejo-Borbolla A, Shang L, Martin AP (2009) The chemokine-binding protein M3 as a tool to understand the chemokine network in vivo. Methods Enzymol 460: 193-207
- Martin AP, Alexander-Brett JM, Canasto-Chibuque C, Garin A, Bromberg JS, et al. (2007) The chemokine binding protein M3 prevents diabetes induced by multiple low doses of streptozotocin. J Immunol 178: 4623-4631.
- BelvonÄÃková P, Kráľová A, Kúdelová M, Hajnická V, Režuchová I, et al. (2008) Chemokine-binding activities of M3 protein encoded by Murine gammaherpesvirus 72. Acta Virol 52: 91-97.
- Halásová Z, ValoviÄová M, MaÄáková K, PanÄÃk P, BelvonÄÃková P, et al. (2011) Partial genome sequence of murine gammaherpesvirus 72 and its analysis. Acta Virol 55: 317-325.
- LapunÃková B, LopuÅ¡ná K, Benkóczka T, Golais F, Kúdelová M, et al. (2015) Epigenetic modification of Rta (ORF50) promoter is not responsible for distinct reactivation patterns of murine gammaherpesviruses. Acta Virol 59: 405-412
- Jana S, Deb JK (2005) Strategies for efficient production of heterologous proteins in Escherichia coli. Appl Microbiol Biotechnol 67: 289–298
- Peti W, Page R (2007) Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expr Purif 51: 1–10
- Bauerová-Hlinková V, Hostinová E, GaÅ¡perÃk J, Beck K, Borko Ľ, (2010) Bioinformatic mapping and production of recombinant N-terminal domains of human cardiac ryanodine receptor 2. Protein Expr Purif 7: 33-41.
- Borko Ľ, Bauerová-Hlinková V, Hostinová E, GaÅ¡perÃk J, Beck K, et al. (2014) Structural insights into the human RyR2 N-terminal region involved in cardiac arrhythmias. Acta Cryst D Biol Crystallogr 70: 2897-2912.
- Berrow NS, Büssow K, Coutard B, Diprose J, Ekberg M, et al. (2006) Recombinant protein expression and solubility screening in Escherichia coli: a comparative study. Acta Crystallogr D Biol Crystallogr 62: 1218–1226
- Hlinkova V, Xing G, Bauer J, Shin YJ, Dionne I, et al. (2008) Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring assembly and opening. Acta Cryst D 64: 941-949.
- PanÄÃk P, Bauerová-Hlinková V, Kúdelová M (2013) Purification of recombinant M3 proteins of murine gammaherpesvirus 68 and 72 expressed in Escherichia coli Acta Virol 57: 59-68.
- Matúšková R, PanÄÃk P, Å tibrániová I, BelvonÄÃková P, Režuchová I, et al. (2015) Soluble M3 proteins of murine gammaherpesvirus 68 and 72 expressed in Escherichia coli: analysis of chemokine-binding properties. Acta Virol 59: 360-368.
- RaÅ¡lová H, Berebbi M, RajÄáni J, Sarasin A, Matis J, et al. (2001) Susceptibility of mouse mammary glands to murine gammaherpesvirus 72 (MHV-72) infection: evidence of MHV-72 transmission via breast milk. Microb Pathog 31: 47-58
- Greenfield EA (2014) Generating Monoclonal Antibodies (2014) In Antibodies: A Laboratory Manual, Greenfield EA ed., Cold Spring Harbor Laboratory Press 201-221.
- NemÄoviÄová I, Benedict ChA, Zajonc DM (2013) Structure of human cytomegalovirus UL141 binding to TRAIL-R2 reveals novel, non-canonical death receptor interaction. PLoSPathog 9: e1003224
- Speck SH, Ganem D (2010) Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe 8: 100-115
- Bridgeman A, Stevenson PG, Simas JP, Efstathiou S (2001) A secreted chemokine binding protein encoded by murine gammaherpesvirus-68 is necessary for the establishment of a normal latent load. J Exp Med 194: 301-312.
- van Berkel V, Preiter K, Virgin IV HW, Speck SH (1999) Identification and initial characterization of the Murine gammaherpesvirus 68 gene M3, encoding an abundantly secreted protein. J Virol 73: 4524-4529.
- Aldinucci D, Colombatti A (2014) The inflammatory chemokine CCL5 and cancer progression. Mediators of Inflammation 1-12.
- Raman D, Baugher PJ, Thu YM, Richmond A (2007) Role of chemokines in tumor growth. Cancer Lett 256: 137-165
- Inoue K, Slaton JW, Eve BY, Kim SJ, Perrotte P, et al. (2000) Interleukin 8 expression regulates tumorigenicity and metastases in androgen-independent prostate cancer. Clin Cancer Res 6: 2104-2119.
Citation: Šebová R, Lenhartová S, Štibrániová I, Nemčovičová I, Belvončíková P, et al. (2017) A Deletion of Signal Peptide in Recombinant Murine Gamma-herpesvirus 68 M3 Protein Enhances its Binding Affinity to CCL5 Chemokine and Increases its Yield from Insect Cells. J Infect Dis Ther 5: 347. DOI: 10.4172/2332-0877.1000347
Copyright: ©2017 Šebová R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 5661
- [From(publication date): 0-2017 - Dec 21, 2025]
- Breakdown by view type
- HTML page views: 4687
- PDF downloads: 974