CDKN2B-AS1 ceRNA Network and KRAS-Dependent Tumorigenicity in Colorectal and Pancreatic Cancer
Received: 30-Oct-2024 / Manuscript No. DPO-24-151450 / Editor assigned: 04-Nov-2024 / PreQC No. DPO-24-151450 (PQ) / Reviewed: 18-Nov-2024 / QC No. DPO-24-151450 / Revised: 25-Nov-2024 / Manuscript No. DPO-24-151450 (R) / Published Date: 02-Dec-2024
Abstract
Abstract Objective: Kirsten Rat Sarcoma Viral Oncogene Homolog (KRAS) exhibits the highest mutation rate in Colorectal Cancer (CRC) and Pancreatic Cancer (PC), highlighting the need for a comprehensive understanding of KRAS-dependent pathogenesis. Given the regulatory role of long noncoding RNAs (lncRNAs) in gene expression, this study focused on constructing a competing endogenous RNA (ceRNA) network of a selected KRAS-related lncRNA. Methods: Differentially Expressed Genes (DEGs) were identified using sequencing data from the sequencing read archive database by analyzing the transcriptional profiles of CRC and PC cell lines with and without KRAS mutations. LncTarD, miRWalk and ToppCluster servers were used to construct ceRNA network of selected lncRNA to elucidate the interactions between corresponding miRNAs and target genes. Results: Notably, the analysis revealed 42 common upregulated DEGs (uDEGs), including differentially expressed lncRNAs and protein-coding genes, between KRAS-mutant and KRAS wild-type cells. Among them, CDKN2B-AS1 emerged as a key KRAS-related lncRNA for constructing the ceRNA network. The ceRNA network of CDKN2B-AS1 included 21 miRNAs and 34 genes selected from common uDEGs. Enrichment analysis of ceRNA target genes validated their involvement in critical cancer-related pathways and biological processes. Important, expression and survival analysis underscored the prognostic significance of some target genes within the CDKN2B-AS1 ceRNA network. Conclusions: Consistent with the key regulatory role of lncRNAs, the identification of CDKN2B-AS1 as a KRAS-related lncRNA and the construction of its ceRNA network improve our understanding of the potential contribution of lncRNAs to KRAS-associated pathogenesis and their application as potential diagnostic and prognostic biomarkers for KRAS-mutant cancers.
Keywords: KRAS mutation; CDKN2B-AS1; Colorectal cancer; Pancreatic cancer; ceRNA network; Long noncoding RNAs; Differential expression analysis
Abbreviations
KRAS: Kirsten Rat Sarcoma viral oncogene homolog; CRC: Colorectal Cancer; PC: Pancreatic Cancer; lncRNAs: long non-coding RNAs; ceRNA: competing endogenous RNA; SRA: Sequencing Read Archive; DEGs: Differentially Expressed Genes; uDEGs: Overlapping upregulated DEGs; DELs: Differentially Expressed lncRNAs (DELs); mutKRAS: KRAS mutant; wtKRAS: KRAS wildtype; GTPase: Small Guanosine Triphosphatase; GDP: Guanosine Diphosphate; GTP: Guanosine Triphosphate; GAPs: GTPase Activating Proteins; GEFs: Guanine Nucleotide Exchange Factors; MAPK: Mitogen-Activated Protein kinase; miRNAs: microRNAs; log2FC: log2 Fold Change; CDKN2A: Cyclin-Dependent Kinase Inhibitor 2A; HTRs: 5-Hydroxytryptamine Receptors; HMGA2: High Mobility Group Protein 2; CCND1: Cyclin D1.
Introduction
Mutations in KRAS have been identified as the most common oncogenic events in 25% of all endodermal carcinomas [1-3]. The KRAS protein is a small Guanosine Triphosphatase (GTPase) that serves as a molecular switch by cycling between inactive Guanosine Diphosphate (GDP)-bound and active Guanosine Triphosphate (GTP)-bound states in response to extracellular signals to induce intracellular responses [4]. These off/on molecular states based on GDP/GTP exchange are controlled by GTP hydrolysis reactions stimulated by GTPase- Activating Proteins (GAPs) and RAS-specific Guanine nucleotide Exchange Factors (GEFs) [5,6].
While GTP-bound KRAS transduces signals to its downstream effectors and activates multiple signaling pathways, somatic mutations favor a constant active state by impairing GTP hydrolysis and resistance to GAP function. High levels of the active form lead to hyperactivation of downstream oncogenic signaling pathways, including the Mitogen- Activated Protein Kinase (MAPK) pathway, which is involved in cell growth, proliferation, development, inflammation, differentiation, survival and apoptosis to initiate and promote malignant transformation [7].
Although recent advances in the understanding of the KRAS oncoprotein structure have led to the clinical development of novel selective anti-KRAS inhibitors, preclinical data and clinical translational series have recently revealed multiple mechanisms of resistance to these inhibitors [8-10]. Therefore, a deeper understanding of these factors, including histological characteristics, the immune microenvironment and the transcriptional landscape of tumor cells harboring KRAS mutations, is important. In this regard, additional studies are needed to elucidate the molecular and cellular mechanisms, including transcriptional alterations and pathway-related strategies responsible for modulating KRAS tumorigenesis.
Perturbations in lncRNAs, key regulators of gene expression, have been reported in the progression of many human cancers [11-13]. Identifying the relationships between KRAS mutations and abnormal expression of some lncRNAs is expected to significantly improve our knowledge of the mechanisms of tumorigenesis controlled by mutKRAS [14]. Abnormal levels of KRAS, a known mediator of many cellular signaling pathways, reciprocally cause various molecular alterations, such as dysregulation of lncRNA expression. Shi et al., showed that the levels of a KRAS-responsive lncRNA called KIMAT1 were positively correlated with KRAS levels in both cell lines and lung cancer specimens [15]. In addition, the role of KIMAT1 in maintaining a positive feedback loop to sustain KRAS signaling during lung cancer promotion has been reported as a strategy to ameliorate KRAS-induced tumorigenesis. Another study showed that Orilnc1 can be regulated by the RAS-RAF-MEK-ERK pathway, which is required for cell proliferation in RAS/BRAF-dependent human malignancies [16].
The association of lncRNAs with various regulatory apparatuses, including chromatin remodeling factors, transcription factors, splicing machinery and nuclear trafficking modulators, underscores the diversity and complexity of their associated regulatory mechanisms [17,18]. The function of lncRNAs as competing endogenous RNAs (ceRNAs) has been proposed as one of their main approaches to regulate gene expression [19-21]. Recent evidence has shown that many lncRNAs are upregulated in cancer tissues with oncogenic activity through the sponging of tumor suppressor microRNAs (miRNAs) [22,23]. The binding of lncRNAs (as ceRNAs) to miRNAs prevents the latter from recognizing their targets, resulting in mRNA upregulation. Thus, during malignant transformation, oncogenic lncRNAs enhance cancer promotion by downregulating miRNAs targeting various driver oncogenes [24,25].
In this study, we investigated abnormally overexpressed lncRNAs associated with KRAS mutations by analyzing the transcriptional profiles of CRC and PC cell lines with and without KRAS mutations. Overexpressed lncRNAs, known as oncogenic KRAS-related lncRNAs, were identified, and among them, CDKN2-AS1 was selected to construct the ceRNA network. The possible function of CDKN2B-AS1 through its associated ceRNA network target genes was determined by performing functional enrichment analysis. In addition, expression and survival analyses of the target genes in the CDKN2B-AS1 ceRNA network were performed to evaluate their prognostic performance as potential biomarkers in KRAS-mutant cancers. The role of ceRNAs and their associated networks in KRAS-dependent tumorigenesis is still unclear. Therefore, this study aimed to further explore the molecular and cellular mechanisms involved in the pathogenesis of KRAS-driven cancers by analyzing of the lncRNA-associated ceRNA network. Collectively, this study identified CDKN2B-AS1 as a promising KRASrelated lncRNA, which might be a potential diagnostic biomarker and therapeutic target which contributes to further understanding of the ceRNA pathogenesis in KRAS-driven cancers.
Materials and Methods
Samples and data collection
In this study, raw RNA sequencing data were extracted from the Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/ sra) [26]. The sequencing data of three human CRC cell lines, namely, HCT-116 (SRR1030462, SRR1030463, SRR1756569 and SRR8615282) and LoVo (SRR1756570, SRR8532655 and SRR8616185), which are the KRAS mutant (mutKRAS) samples and SW48 (ERR208907, SRR3228439 and SRR8615504), as the KRAS wild-type (wtKRAS) control sample, were downloaded. In addition, transcriptomic data of PC cell lines, including Capan-2 (SRR2313117, SRR2313118 and SRR2313119) as the mutKRAS sample and BXPC3 (SRR2313123, SRR2313124 and SRR2313125) as the wtKRAS control sample, were obtained.
Workflow of the study
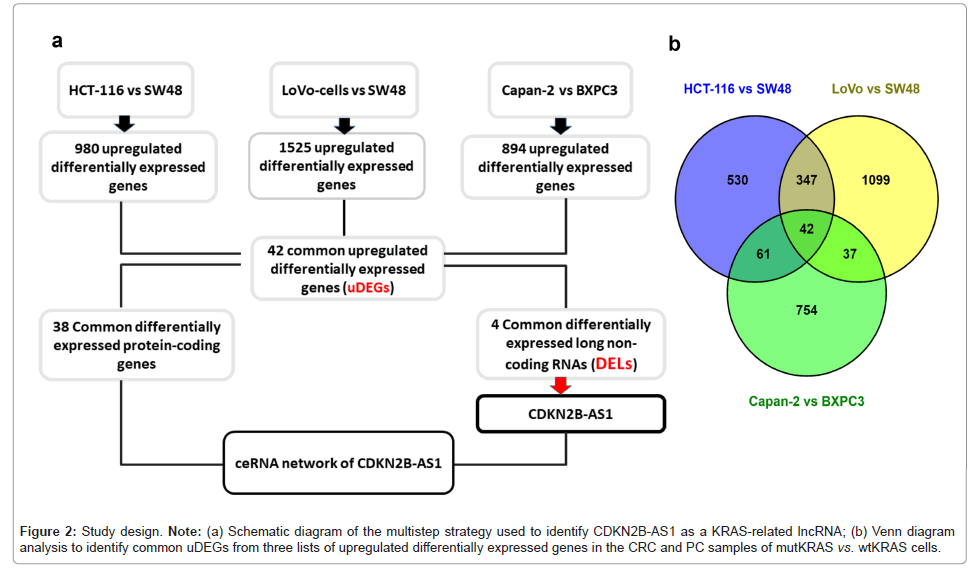
Due to the higher prevalence of KRAS mutations in pancreatic and colorectal cancer, CRC and PC cell lines were used in this study. Transcriptional profiling analysis of PC cell lines was performed to analyze the differential expression of genes between Capan-2 (mutKRAS) cells and BXPC3 cells, which were used as wtKRAS samples. In addition, differential expression analysis of CRC cells, including HCT-116 and LoVo (mutKRAS) vs. SW48 (wtKRAS) cells, was performed previously [27]. Transcriptional profile analysis of the samples revealed DEGs between the mutKRAS and wtKRAS cells. A venn diagram analysis (https://bioinfogp.cnb.csic.es/tools/venny/ index2.0.2.html) revealed 42 common upregulated DEGs (uDEGs), including common Differentially Expressed lncRNAs (DELs) and protein-coding genes [28]. According to the workflow of the study, detected DELs could be mapped to KRAS-related lncRNAs; among them, CDKN2B-AS1 was selected for further analysis.
Data preprocessing and differential expression analysis
RNA sequencing data were downloaded as SRA files and fastqdump from the SRA toolkit (v2.8.2) was used to convert the SRA to FASTQ format [26]. The sequencing quality of the FASTQ files was monitored using FastQC (v0.11.5) and modified using quality control software, including FLEXBAR (v3.0) and Trimmomatic (v0.39) [29- 31]. The human reference genome was downloaded from the Ensemble database (http://ftp.ensembl.org/pub/release95/fasta/homo_sapiens/ dna/Homo_sapiens.GRCh38.dna.toplevel.fa.gz) and indexed using Bowtie2 (v2.3.4.1) prior to mapping [32,33].
The filtered reads were aligned to the reference genome using the Bowtie2 software. The output files of the mapping in the SAM format were processed by the HTSeq-count program (v0.11.4) for simultaneous read counting and annotation using an annotated human reference genome downloaded from Ensemble [34]. Normalization and differential expression analysis were performed using the DESeq2 package (version 1.38.0) from Bioconductor in the R environment (version 3.6.1, https://www.rproject.org/) [35]. Significantly upregulated DEGs were identified using log2- Fold Change (log2FC) and adjusted p-value as screening criteria (|log2FC|>3, adjusted p-value<0.01). DEGs were annotated using Ensembl Biomart (https://asia.ensembl.org/biomart/martview) and the GRCh38.p13 reference genome for partitioning into proteincoding genes and DELs [36]. All the commands and scripts used for data processing and differential expression analysis were uploaded to the GitHub platform and are publicly available at https://github. com/mahsa1985/R-scripts.git and https://github.com/mahsa1985/ Linux-Commands.git.
Output visualization of differential expression analysis
Hierarchical clustering analysis was performed to visualize the results of the differential expression analysis related to KRAS mutations based on the normalized read counts of the mutKRAS and wtKRAS samples. Heatmap plots were generated using the gplots package in R and Variance Stabilizing Transformation (VST) was applied to the normalized count data before clustering. Linkage analysis and distance measurement were based on full linkage and Euclidean distance, respectively. According to the lowest adjusted p-value, the expression of 1000 genes was represented by heatmap plots based on expression data indicated as normalized values (Z-scores). MA plots were generated using the plotMA function of the DESeq2 package, with log2FC on the y-axis and the average of the normalized counts over all samples on the x-axis. Each gene is represented by a dot and the points in blue are genes with significant differential expression and adjusted p-values less than 0.01.
Construction of the ceRNA network
The ceRNA network was constructed based on the ceRNA hypothesis that lncRNAs and mRNAs can coregulate each other by sharing MREs (miRNA response elements). The ceRNA network of CDKN2B-AS1 was constructed based on previous studies on the ceRNA function of CDKN2B-AS1 and the results of the present study. Table 1 shows the miRNA-CDKN2B-AS1-mRNA interactions, for which the LncTarD database (https://lnctard.bio-database.com/) was used to determine the miRNA-CDKN2B-AS1 and miRNA-mRNA interactions based on previous publications [37]. The list of the miRNAs in Table 1 was mapped in miRWalk (http://mirwalk.umm.uni-heidelberg.de/) and ToppCluster (https://toppcluster.cchmc.org/) to search for their mRNA targets [38,39]. According to the ceRNA hypothesis, the genes obtained from miRWalk and ToppCluster, which were also among the list of uDEGs, were considered as the target genes of CDKN2B-AS1 to construct the ceRNA network, while considering their related miRNAs, as shown in Table 1. Finally, the CDKN2B-AS1-miRNA-mRNA ceRNA network was constructed and visualized using the Cytoscape tool (version 3.9.1) (https://cytoscape.org/) [40].
Table 1: The list of the experimentally supported interactions between CDKN2B-AS1 and its miRNA targets and miRNA-mRNA interactions is based on the ceRNA hypothesis extracted from LncTarD [47-66].
Gene ontology and pathway analysis
To better understand the biological functions of the target genes in the CDKN2B-AS1 ceRNA network, Gene Ontology (GO) and pathway analyses were performed to underscore the potential tumorigenesis of CDKN2B-AS1 as a KRAS-related lncRNA. In this study, enrichment analysis was performed using the comprehensive gene set enrichment analysis web server EnrichR (https://maayanlab.cloud/Enrichr) [36]. GO analysis was based on enriched terms in the biological process and molecular function Pathway enrichment analysis was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database [41-43]. In addition, the Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics tool (https://david.ncifcrf.gov/) and the Gene Ontology Resource (www. geneontology.org) were used to validate enrichment analysis results [35,37]. GO terms and KEGG pathways with a p-value<0.05 were considered significantly enriched. The most significantly enriched GO terms and KEGG pathways were ranked based on the p-value. Finally, the results obtained from EnrichR were visualized using http://www. bioinformatics.com.cn/srplot, an online platform for data analysis and visualization.
Evaluation of the prognostic performance of ceRNA-related target genes
The prognostic power of the target genes in the CDKN2B-AS1 ceRNA network was evaluated by survival analysis using the interactive web-based tool GEPIA (Gene Expression Profiling Interactive Analysis), which is based on the gene expression RNA-seq datasets of The Cancer Genome Atlas (TCGA) (https://www.cancer.gov/tcga) and Genotype-Tissue Expression (GTEx) [44,45]. Using these databases for gene survival analysis, we set the group cutoff to the median and 95% Confidence Interval (CI) of the Yes. All analyses were considered statistically significant with a log-rank p-value<0.05. Correlation analysis between gene expression and sample type (tumor and normal samples) was performed using the UALCAN online dataset (https://ualcan.path.uab.edu/index.html) based on the TCGA database [46]. All parameters were set to default values to examine differential expression between tumor and normal samples, considering a p-value less than 0.05 to indicate statistical significance.
Results
Differential expression analysis and visualization
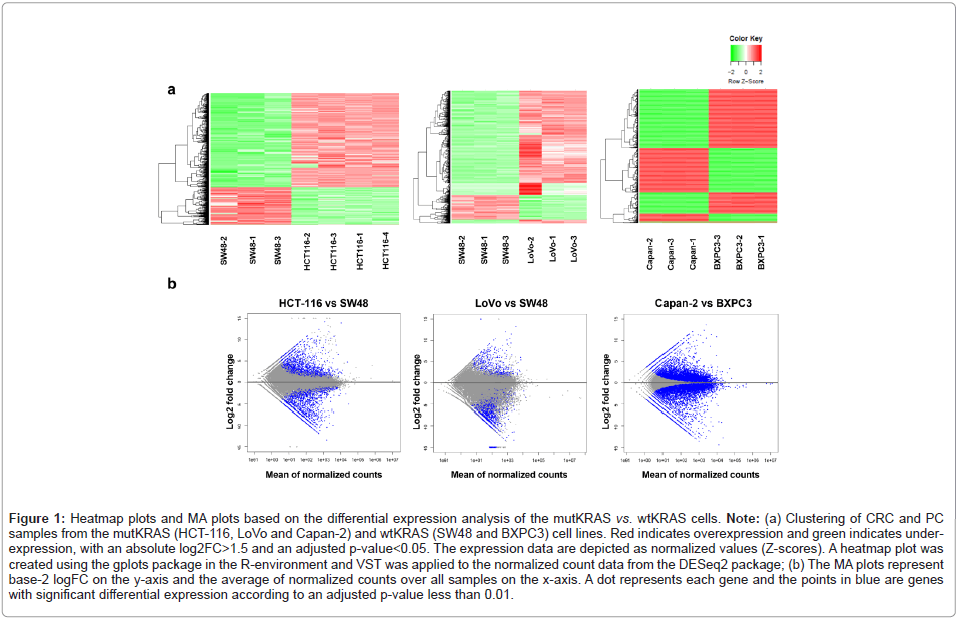
In this study, transcriptional profile analysis was performed on the mutKRAS and wtKRAS CRC and PC cell samples to identify DELs related to KRAS mutation. Hierarchical cluster analysis was used to visualize differential expression. Heatmap plots show DEGs in CRC and PC samples with and without KRAS mutations (Figure 1a). MA plots also show the log2FC of genes compared with their mean normalized counts (Figure 1b). The results of the RNA sequencing analysis revealed upregulated DEGs (|log2FC|>3, adjusted p-value<0.01). For the CRC cell lines, 980 and 1525 DEGs were upregulated in HCT-116 and LoVo (mutKRAS samples) vs. SW48 (wtKRAS control sample), respectively. In addition, transcriptional analysis of PC cell lines revealed a total of 894 upregulated DEGs in Capan-2 (mutKRAS) compared with BXPC3 as the wtKRAS control sample (Figure 2a).
Figure 1: Heatmap plots and MA plots based on the differential expression analysis of the mutKRAS vs. wtKRAS cells. Note: (a) Clustering of CRC and PC samples from the mutKRAS (HCT-116, LoVo and Capan-2) and wtKRAS (SW48 and BXPC3) cell lines. Red indicates overexpression and green indicates underexpression, with an absolute log2FC>1.5 and an adjusted p-value<0.05. The expression data are depicted as normalized values (Z‑scores). A heatmap plot was created using the gplots package in the R-environment and VST was applied to the normalized count data from the DESeq2 package; (b) The MA plots represent base-2 logFC on the y-axis and the average of normalized counts over all samples on the x-axis. A dot represents each gene and the points in blue are genes with significant differential expression according to an adjusted p-value less than 0.01.
Identification of CDKN2B-AS1
In the present study, a multistep strategy was applied to select CDKN2B-AS1 as a lncRNA with differential expression in the context of KRAS mutation (Figure 2a). Upregulated genes with significant differential expression were identified by comparing the transcriptomes of CRC mutKRAS samples (HCT-116 and LoVo) with wtKRAS CRC sample (SW48) and PC mutKRAS cell (Capan-2) with BXPC3 as the wtKRAS control PC sample. Venn diagram analysis revealed 42 uDEGs, including protein-coding genes and DELs, associated with the KRAS mutation (Figure 2b). In the next step, overlapping DELs were identified as shown in Figure 2a, which could be classified as KRASrelated lncRNAs. The upregulation of the overlapping DELs in the mutKRAS cell lines compared to the wtKRAS cell lines is consistent with the ceRNA hypothesis. Among the overlapping and upregulated DELs, some with less annotation, such as LINC00471, LINC01842 and DNAH17-AS1, were excluded and CDKN2B-AS1, as a KRAS-related lncRNA, was selected to construct its ceRNA network for further investigation.
Figure 2: Study design. Note: (a) Schematic diagram of the multistep strategy used to identify CDKN2B-AS1 as a KRAS-related lncRNA; (b) Venn diagram analysis to identify common uDEGs from three lists of upregulat ed differentially expressed genes in the CRC and PC samples of m utKRAS vs. wtKRAS cells.
ceRNA network of CDKN2B-AS1
Guided by the ceRNA hypothesis, CDKN2B-AS1-miRNA and miRNA-mRNA interactions confirmed by previous studies are shown in Table 1. The data presented in Table 1 were extracted from LncTarD as a comprehensive resource of lncRNA-target interactions to report experimentally supported findings. A total of 21 miRNAs were identified from Table 1 and used as candidate miRNAs to construct the ceRNA network of CDKN2B-AS1. The results obtained from miRWalk and Toppcluster indicated that the candidate miRNAs were able to target many of the 42 uDEGs in the mutKRAS vs. wtKRAS cells according to the differential expression analysis. Therefore, based on the ceRNA hypothesis, the constructed network with 21 miRNAs and 34 mRNAs predicted that our upregulated DEGs could be positively correlated with upregulated CDKN2B-AS1 and negatively correlated with the miRNA expression levels involved in the ceRNA network (Table 1 and Figure 3).
GO and signaling pathway enrichment analysis
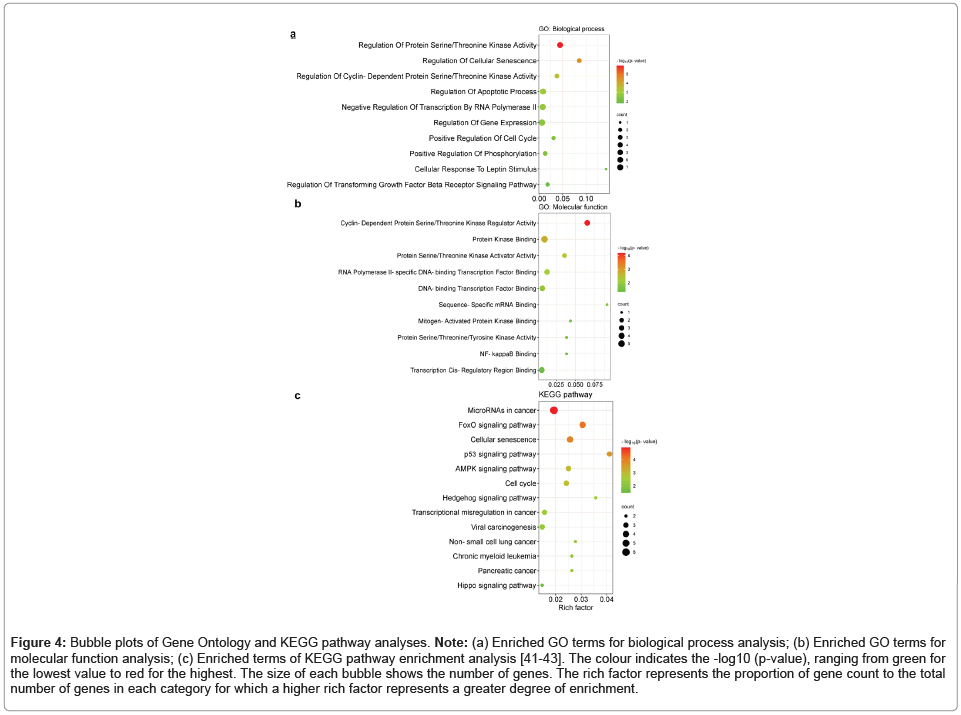
GO analysis and pathway enrichment analysis were performed for the genes in the ceRNA network as the target genes of CDKN2B-AS1. All the genes were computationally uploaded to the DAVID, EnrichR and Gene Ontology resources to better reveal the carcinogenicity of CDKN2-AS1 as a KRAS-related lncRNA. The results of enrichment analysis showed the involvement of the genes in the most significant and relevant enriched GO terms and KEGG pathways ranked by p-value in each category (Figure 4). In the biological process group, genes were mainly enriched in terms related to the regulation of protein serine/ threonine kinase activity, regulation of cellular senescence, regulation of the apoptotic process, and positive regulation of the cell cycle (Figure 4a). In the molecular function category of GO, genes were mainly enriched in the terms cyclin-dependent protein serine/threonine kinase regulator activity, protein kinase binding, NF-kappaB binding and protein serine/threonine kinase activity terms (Figure 4b). The results of pathway enrichment analysis indicated that the genes were mainly enriched in pathways such as miRNAs in cancer, pancreatic cancer, colorectal cancer, and the p53 signaling pathway (Figure 4c).
Figure 4: Bubble plots of Gene Ontology and KEGG pathway analyses. Note: (a) Enriched GO terms for biological process analysis; (b) Enriched GO terms for molecular function analysis; (c) Enriched terms of KEGG pathway enrichment analysis [41-43]. The colour indicates the -log10 (p-value), ranging from green for the lowest value to red for the highest. The size of each bubble shows the number of genes. The rich factor represents the proportion of gene count to the total number of genes in each category for which a higher rich factor represents a greater degree of enrichment.
Evaluation of the prognostic performance of ceRNA-related target genes
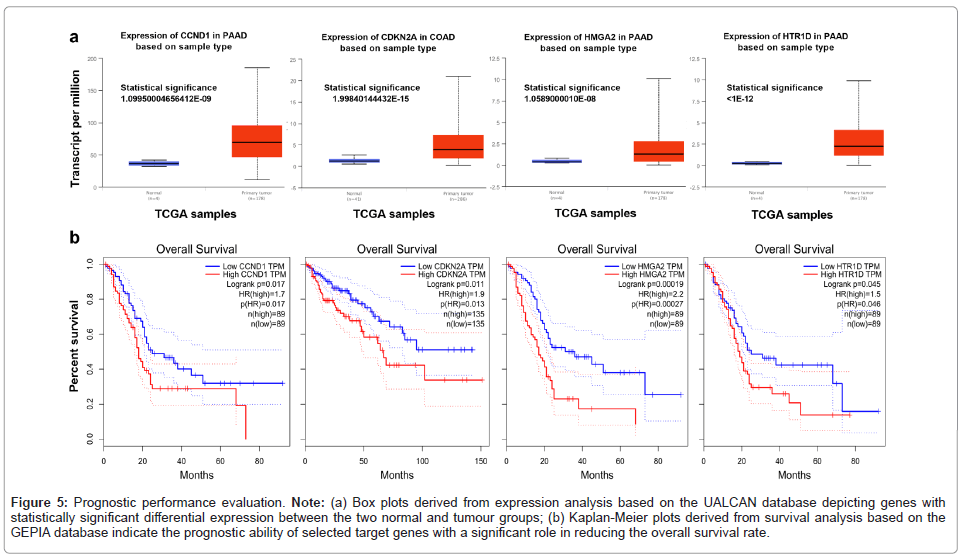
The prognostic power of the target genes in the CDKN2B-AS1 ceRNA network was evaluated based on survival and gene expression analysis across tumor and normal samples. The significant differences between the gene expression levels of the normal and tumor groups were evaluated for the target genes of the ceRNA network. The higher expression of the CDKN2A, CCND1, HTR1D and HMGA2 genes in the tumor samples, as determined by UALCAN, is due to the ceRNA hypothesis (Figure 5a). The GEPIA database was used for the survival analysis of target genes using RNA sequencing expression data of tumor and normal samples from the TCGA and GTEx datasets (Tang et al., 2017). Consistent with the results of expression analysis, the results of survival analysis showed that the CDKN2A gene in CRC patients and CCND1, HTR1D and HMGA2 in PC patients were significantly associated with unfavorable overall survival based on Kaplan-Meier plots (significance level at log-rank p-value<0.05) (Figure 5b).
Figure 5: Prognostic performance evaluation. Note: (a) Box plots derived from expression analysis based on the UALCAN database depicting genes with statistically significant differential expression between the two normal and tumour groups; (b) Kaplan-Meier plots derived from survival analysis based on the GEPIA database indicate the prognostic ability of selected target ge nes with a significant role in reducing the overall survival rat e.
Discussion
Mutations in the KRAS oncogene with tumor-promoting activity have been identified in 25% of all cancers, with some cancers, such as pancreatic and colorectal cancer, having the highest mutation rates, at 90% and 45%, respectively. Despite developments in direct KRAS pharmacology, targeted therapies with direct inhibitors are followed by rapid reactivation of KRAS signaling, leading to resistance to longterm treatment [9]. Therefore, a comprehensive analysis of the various mechanisms and pathways associated with KRAS tumorigenic activity is critical to identify potential therapeutic strategies to inhibit its oncogenic behavior.
It has been reported that lncRNAs have an extensive ability to regulate gene expression, enabling intricate, multi-layered molecular interactions in numerous pathological conditions, including cancer [67]. The competitive endogenous activity of lncRNAs, as one of their posttranscriptional regulatory mechanisms, is conferred by their competitive binding to common miRNAs, freeing their targets from miRNA-induced degradation and thus significantly associated with gene upregulation [68].
During the process of malignant transformation, changes occur in the chromosome of cancer cells, including chromosomal rearrangements, truncated 3′UTRs and point mutations such as KRAS oncogenic mutations. Following these alterations, transcriptional changes and, as a consequence, dysregulation of lncRNAs and their related ceRNA network are closely linked to tumorigenesis [69]. Therefore, continuously updated studies on the role of lncRNAs and their ceRNA networks as multilayered intracellular communication have led to remarkable progress in this burgeoning hotspot to provide new insights into cancer pathogenesis.
This study investigated the KRAS-dependent dysregulated transcriptional profile in CRC and PC cells to identify upregulated DEGs and DELs to identify a ceRNA network associated with KRAS tumorigenesis. Comparison of the transcriptomes of the mutKRAS cell lines with those of their wtKRAS counterparts revealed 42 uDEGs, including protein-coding genes and DELs. We identified CDKN2BAS1 as a KRAS-related DEL. This lncRNA, also known as ANRIL, is located within the CDKN2B-CDKN2A gene cluster on chromosome 9p21, which is a significant genetically susceptible locus for several cancers. To identify the connection between CDKN2B-AS1 and uDEGs, a ceRNA network of CDKN2B-AS1 was constructed using uDEGs as target genes. The miRNA targets of CDKN2B-AS1 were determined according to previous publications on the ceRNA function of CDKN2B-AS1. The list of candidate miRNAs was submitted to the miRWalk and ToppCluster platforms to search for potential gene targets. Interestingly, 34 genes out of 42 uDEGs were found to be targets of candidate miRNAs. Finally, the ceRNA network of CDKN2B-AS1 was constructed from 21 miRNAs and 34 uDEGs.
To further understand the pathogenic mechanism of CDKN2B-AS1 as a KRAS-related lncRNA, the top enriched functional annotations of GO and KEGG pathway analyses were identified. The target genes were enriched in GO biological process categories, such as regulation of protein serine/threonine kinase activity, regulation of the apoptotic process and positive regulation of the cell cycle, which are closely related to tumorigenesis and cancer promotion. In addition, pathway enrichment analysis revealed several enriched pathways known as cancer-related pathways, including microRNAs involved in cancer, cell cycle regulation, pancreatic cancer and the p53 signaling pathway.
Furthermore, to determine the clinical value of CDKN2B-AS1, the prognostic power of the target genes of the ceRNA network was evaluated based on survival and expression analysis of tumor and normal patient samples. While the results showed a statistically significant association of CDKN2A in CRC patients and CCND1, HTR1D and HMGA2 in PC patients with survival, their higher expression in tumor samples was also confirmed. While Cyclin-Dependent Kinase Inhibitor 2A (CDKN2A) is well known as a susceptibility gene for melanoma and pancreatic cancer, its germline variants have also been associated with a wider range of malignancies, including tumor of neural system, breast cancer, head and neck squamous cell carcinoma and sarcomas [70,71]. While the expression level of Cyclin D1 (CCND1) is tightly regulated in normal cells, its increased activity has been observed in several types of neoplasms [72].
A positive correlation between CCND1 copy number in breast cancer and lymph node metastasis has been observed [73]. According to recent studies, 5-Hydroxytryptamine Receptors (HTRs), including HTR1D, are associated with several malignant tumors, such as melanoma, breast cancer, lung cancer and colon cancer [74-76]. The involvement of the HOXA10-AS/miR-340-3p/HTR1D axis in the progression of pancreatic cancer has been demonstrated [77]. Furthermore, the expression level of HTR1D in clinical samples of CRC adenocarcinoma suggested its role in the prognosis of patients [78]. The oncogenic role of High Mobility Group Protein 2 (HMGA2) in different types of cancer through diverse carcinogenic strategies have revealed that HMGA2 is a candidate for cancer diagnostic, prognostic and therapeutic purposes.
To date, an increasing number of dysregulated lncRNAs, key regulators of gene expression with critical roles in human neoplasms, such as CRC and PC, have been identified [79]. Based on the ceRNA phenomenon, the sequestration of tumor suppressor miRNAs from their mRNA target is one of the oncogenic mechanisms for gene expression regulation by lncRNAs [15,80,81]. In the present study, dysregulated lncRNAs were identified between the mutKRAS and wtKRAS samples of CRC and PC cell lines, using RNA-seq datasets from the SRA. Among the DELs, which were considered as the KRASrelated lncRNAs, some with less annotation were excluded and finally, CDKN2B-AS1 was selected for further analysis. In addition, we selected this lncRNA because of its indispensable role in several diseases, especially cancer [82,83].
Therefore, the ceRNA network of CDKN2B-AS1 was constructed from upregulated differentially expressed DEGs. The results of previous studies on the sponging effect of CDKN2B-AS1 were used to identify miRNAs that mediate the ceRNA function to identify all the elements necessary for the construction of the ceRNA network. The results of the GO and pathway analyses of the target genes included in the ceRNA network of CDKN2B-AS1 indicated their roles in cancerrelated pathways and biological processes. Furthermore, survival and expression analysis of the corresponding ceRNA target genes revealed the prognostic power of CDKN2A, CCND1, HTR1D and HMGA2.
This study has several limitations that should be considered for a more precise interpretation of the results. The results are based on the analysis of transcriptional profiles of cancer cell lines, which should be validated by data from patient samples. Although there is agreement that lncRNAs are worthy of investigation and that there is still much to be done in this class of biomolecules, the mechanism of action of lncRNAs is often very complex and there is always uncertainty about their biological impact. Therefore, our findings are more predictions than certainties and more computational methods and molecular biology experiments should be used to increase the credibility of our results.
Conclusion
In conclusion, we analyzed the KRAS-dependent dysregulated transcriptional profiles of CRC and PC cells to identify DELs. As a result, CDKN2B-AS1 was identified as a KRAS-related lncRNA, and its ceRNA activity was further investigated as one of the major gene expression regulatory mechanisms of lncRNAs. The ultimate goal of this study was to highlight the significance of the ceRNA network of CDKN2B-AS1 in KRAS mutation-mediated tumorigenesis.
Funding Information
This study was supported and funded by the German Research Foundation (DFG; grant number: AH 92/8-1).
Availability of Data
The datasets generated and/or analyzed during the current study are available in the Sequence Read Archive (SRA) repository (SRR1030462, SRR1030463, SRR1756569, SRR8615282, SRR1756570, SRR8532655, SRR8616185, ERR208907, SRR3228439, SRR8615504, SRR2313117, SRR2313118, SRR2313119, SRR2313123, SRR2313124, SRR2313125).
Acknowledgements
The authors are thankful to Faezeh ghasemi and Niusha Gharehdaghi for their insightful comments.
Author Contributions
Mahsa Saliani conceptualized and designed the study, did the analysis and prepared the final draft; Ali Javadmanesh substantively revised the manuscript; Parisa Gonbadi interpreted data and improved introduction; Somayeh Rahimi conducted gene ontology and pathway analysis; Faezeh Dastgir and Hadise Mirahmadi Daryasary, prepared the figures and table and did literature search; Mohammad Reza Ahmadian. proofread and substantively revised the final draft. All authors reviewed and approved the manuscript.
Conflicts of Interests
The authors declare that they have no competing interests.
References
- Simanshu DK, Nissley DV, McCormick F (2017) RAS proteins and their regulators in human disease. Cell 170:17-33.
[Crossref] [Google Scholar] [PubMed]
- del Re M, Rofi E, Restante G, Crucitta S, Arrigoni E, et al. (2018) Implications of KRAS mutations in acquired resistance to treatment in NSCLC. Oncotarget 9:6630.
[Crossref] [Google Scholar] [PubMed]
- Saito Y, Koya J, Araki M, Kogure Y, Shingaki S, et al. (2020) Landscape and function of multiple mutations within individual oncogenes. Nature 582:95-99.
[Crossref] [Google Scholar] [PubMed]
- Mullard A (2019) Cracking KRAS. Nat Rev Drug Discov 18:887-892.
[Crossref] [Google Scholar] [PubMed]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmüller L, Lautwein A, et al. (1997) The RAS-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic RAS mutants. Science 277:333-339.
[Crossref] [Google Scholar] [PubMed]
- Scheffzek K, Ahmadian MR, Wittinghofer A (1998) GTPase-activating proteins: Helping hands to complement an active site. Trends Biochem Sci 23:257-262.
[Crossref] [Google Scholar] [PubMed]
- Hymowitz SG, Malek S (2018) Targeting the MAPK pathway in RAS mutant cancers. Cold Spring Harb Perspect Med 8:a031492.
[Crossref] [Google Scholar] [PubMed]
- Zeissig MN, Ashwood LM, Kondrashova O, Sutherland KD (2023) Next batter up! Targeting cancers with KRAS-G12D mutations. Trends Cancer 11:955-967.
[Crossref] [Google Scholar] [PubMed]
- Nussinov R, Jang H (2024) Direct K-Ras inhibitors to treat cancers: Progress, new insights, and approaches to treat resistance. Annu Rev Pharmacol Toxicol 64:231-253.
[Crossref] [Google Scholar] [PubMed]
- Bungaro M, Novello S, Passiglia F (2022) Targeting KRASp. G12C mutation in advanced non-small cell lung cancer: A new era has begun. Curr Treat Options Oncol 23:1699-1720.
[Crossref] [Google Scholar] [PubMed]
- Aprile M, Costa V, Cimmino A, Calin GA (2023) Emerging role of oncogenic long noncoding RNA as cancer biomarkers. Int J Cancer 152:822-834.
[Crossref] [Google Scholar] [PubMed]
- Zhan T, Cheng X, Zhu Q, Han Z, Zhu K, et al. (2023) LncRNA LOC105369504 inhibits tumor proliferation and metastasis in colorectal cancer by regulating PSPC1. Cell Death Discov 9: 89.
[Crossref] [Google Scholar] [PubMed]
- Zhang S, Shen C, Gu J, Li J, Jiang X, et al. (2023) Succinylation-associated lncRNA signature to predict the prognosis of colon cancer based on integrative bioinformatics analysis. Sci Rep 13:7366.
[Crossref] [Google Scholar] [PubMed]
- Saliani M, Mirzaiebadizi A, Javadmanesh A, Siavoshi A, Ahmadian MR (2022) KRAS-related long noncoding RNAs in human cancers. Cancer Gene Ther 29:418-427.
[Crossref] [Google Scholar] [PubMed]
- Shi L, Magee P, Fassan M, Sahoo S, Leong HS, et al. (2021) A KRAS-responsive long non-coding RNA controls microRNA processing. Nat Commun 12:1-19.
[Crossref] [Google Scholar] [PubMed]
- Zhang D, Zhang G, Hu X, Wu L, Feng Y, et al. (2017) Oncogenic RAS regulates long noncoding RNA Orilnc1 in human cancer. Cancer Res 77:3745-3757.
[Crossref] [Google Scholar] [PubMed]
- He RZ, Luo DX, Mo YY (2019) Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis 6:6-15.
[Crossref] [Google Scholar] [PubMed]
- Li Z, Zhao W, Wang M, Zhou X (2019) The role of long noncoding RNAs in gene expression regulation. Gene Expr Profil Cancer:1-17.
- Chu J, Jia J, Yang L, Qu Y, Yin H, et al. (2020) LncRNA MIR31HG functions as a ceRNA to regulate c-Met function by sponging miR-34a in esophageal squamous cell carcinoma. Biomed Pharmacother 128:110313.
[Crossref] [Google Scholar] [PubMed]
- Yang N, Liu K, Yang M, Gao X (2021) ceRNAs in cancer: Mechanism and functions in a comprehensive regulatory network. J Oncol 2021:4279039.
[Crossref] [Google Scholar] [PubMed]
- Yu B, Zhao Z, Chen Z, Xiang C, Wang P, et al. (2023) CD24-associated ceRNA network reveals prognostic biomarkers in breast carcinoma. Sci Rep 13:3826.
[Crossref] [Google Scholar] [PubMed]
- Qu C, Dai C, Guo Y, Qin R, Liu J (2020) Long non-coding RNA PVT1-mediated miR-543/SERPINI1 axis plays a key role in the regulatory mechanism of ovarian cancer. Biosci Rep 40.
[Crossref] [Google Scholar] [PubMed]
- Sun B, Liu C, Li H, Zhang L, Luo G, et al. (2020) Research progress on the interactions between long non-coding RNAs and microRNAs in human cancer. Oncol Lett 19:595-605.
[Google Scholar] [PubMed]
- Chen Y, Sheng HG, Deng FM, Cai LL (2021) Downregulation of the long noncoding RNA SNHG1 inhibits tumor cell migration and invasion by sponging miR-195 through targeting Cdc42 in oesophageal cancer. Kaohsiung J Med Sci 37:181-191.
[Crossref] [Google Scholar] [PubMed]
- Saliani M, Mirzaiebadizi A, Mosaddeghzadeh N, Ahmadian MR (2021) RHO GTPase-related long noncoding RNAs in human cancers. Cancers 13:5386.
[Crossref] [Google Scholar] [PubMed]
- Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database C (2011) The sequence read archive. Nucleic Acids Res 39:D19-D21.
[Crossref] [Google Scholar] [PubMed]
- Saliani M, Jalal R, Javadmanesh A (2022) Differential expression analysis of genes and long non-coding RNAs associated with KRAS mutation in colorectal cancer cells. Sci Rep 12:7965.
[Crossref] [Google Scholar] [PubMed]
- Oliveros JC (2007) VENNY. An interactive tool for comparing lists with Venn Diagrams. CiNii.
- Andrews S (2010) FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom.
- Dodt M, Roehr JT, Ahmed R, Dieterich C (2012) FLEXBAR-Flexible barcode and adapter processing for next-generation sequencing platforms. Biology 1:895-905.
[Crossref] [Google Scholar] [PubMed]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30:2114-2120.
[Crossref] [Google Scholar] [PubMed]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357-359.
[Crossref] [Google Scholar] [PubMed]
- Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, et al. (2018) Ensembl 2019. Nucleic Acids Res 47:D745-D751.
[Crossref] [Google Scholar] [PubMed]
- Anders S, Pyl PT, Huber W (2015) HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166-169.
[Crossref] [Google Scholar] [PubMed]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106.
[Crossref] [Google Scholar] [PubMed]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, de Moor B, et al. (2005) BioMart and bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 21:3439-3440.
[Crossref] [Google Scholar] [PubMed]
- Zhao H, Shi J, Zhang Y, Xie A, Yu L, et al. (2020) LncTarD: A manually-curated database of experimentally-supported functional lncRNA-target regulations in human diseases. Nucleic Acids Res 48:D118-D126.
[Crossref] [Google Scholar] [PubMed]
- Kaimal V, Bardes EE, Tabar SC, Jegga AG, Aronow BJ (2010) ToppCluster: A multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res 38:W96-W102.
[Crossref] [Google Scholar] [PubMed]
- Sticht C, de La Torre C, Parveen A, Gretz N (2018) miRWalk: An online resource for prediction of microRNA binding sites. PLoS One 13:e0206239.
[Crossref] [Google Scholar] [PubMed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498-2504.
[Crossref] [Google Scholar] [PubMed]
- Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27-30.
[Crossref] [Google Scholar] [PubMed]
- Kanehisa M (2019) Toward understanding the origin and evolution of cellular organisms. Protein Sci 28:1947-1951.
[Crossref] [Google Scholar] [PubMed]
- Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M (2023) KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res 51:D587-D592.
[Crossref] [Google Scholar] [PubMed]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, et al. (2013) The Genotype-Tissue Expression (GTEx) project. Nat Genet 45:580-585.
[Crossref] [Google Scholar] [PubMed]
- Tang Z, Li C, Kang B, Gao G, Li C, et al. (2017) GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45:W98-W102.
[Crossref] [Google Scholar] [PubMed]
- Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, et al. (2022) UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 25:18-27.
[Crossref] [Google Scholar] [PubMed]
- Zhang YW, Chen Q, Li B, Li HY, Zhao XK, et al. (2021) NAP1L1 functions as a tumor promoter via recruiting hepatoma-derived growth factor/c-Jun signal in hepatocellular carcinoma. Front Cell Dev Biol 9:659680.
[Crossref] [Google Scholar] [PubMed]
- Ma Y, Zhang H, Li G, Hu J, Liu X, et al. (2019) LncRNA ANRIL promotes cell growth, migration and invasion of hepatocellular carcinoma cells via sponging miR-144. Anti-Cancer Drugs 30:1013-1021.
[Crossref] [Google Scholar] [PubMed]
- Chen J, Huang X, Wang W, Xie H, Li J, et al. (2018) LncRNA CDKN2BAS predicts poor prognosis in patients with hepatocellular carcinoma and promotes metastasis via the miR-153-5p/ARHGAP18 signaling axis. Aging 10:3371-3381.
[Crossref] [Google Scholar] [PubMed]
- Liu P, Zhang M, Niu Q, Zhang F, Yang Y, et al. (2018) Knockdown of long non-coding RNA ANRIL inhibits tumorigenesis in human gastric cancer cells via microRNA-99a-mediated down-regulation of BMI1. Braz J Med Biol Res 51:e6839
[Crossref] [Google Scholar] [PubMed]
- Zhu L, Zhang Q, Li S, Jiang S, Cui J, et al. (2019) Interference of the long noncoding RNA CDKN2B‐AS1 upregulates miR‐181a‐5p/TGFβI axis to restrain the metastasis and promote apoptosis and senescence of cervical cancer cells. Cancer Med 8:1721-1730.
[Crossref] [Google Scholar] [PubMed]
- Ma J, Li T, Han X, Yuan H (2018) Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J Cancer Res Clin Oncol 144:205-214.
[Crossref] [Google Scholar] [PubMed]
- Dong X, Jin Z, Chen Y, Xu H, Ma C, et al. (2018) Knockdown of long non-coding RNA ANRIL inhibits proliferation, migration, and invasion but promotes apoptosis of human glioma cells by upregulation of miR-34a. J Cell Biochem 119:2708-2718.
[Crossref] [Google Scholar] [PubMed]
- Li K, Zhao B, Wei D, Cui Y, Qian L, et al. (2020) Long non‐coding RNA ANRIL enhances mitochondrial function of hepatocellular carcinoma by regulating the MiR‐199a‐5p/ARL2 axis. Environ Toxicol 35:313-321.
[Crossref] [Google Scholar] [PubMed]
- Wang J, Zhao SM (2021) LncRNA-antisense non-coding RNA in the INK4 locus promotes pyroptosis via miR-497/thioredoxin-interacting protein axis in diabetic nephropathy. Life Sci 264:118728.
[Crossref] [Google Scholar] [PubMed]
- Liu F, Xiao Y, Ma L, Wang J (2020) Regulating of cell cycle progression by the lncRNA CDKN2B-AS1/miR-324-5p/ROCK1 axis in laryngeal squamous cell cancer. Int J Biol Markers 35:47-56.
[Crossref] [Google Scholar] [PubMed]
- Zhang H, Wang X, Chen X (2017) Potential role of long non‐coding RNA ANRIL in pediatric medulloblastoma through promotion on proliferation and migration by targeting miR‐323. J Cell Biochem.
[Crossref] [Google Scholar] [PubMed]
- Gui D, Cao H (2020) Long non-coding RNA CDKN2B-AS1 promotes osteosarcoma by increasing the expression of MAP3K3 via sponging miR-4458. In Vitro Cell Dev Biol-Animal 56:24-33.
[Crossref] [Google Scholar] [PubMed]
- Xu C, Zhai J, Fu Y (2020) LncRNA CDKN2B-AS1 promotes the progression of ovarian cancer by miR-143-3p/SMAD3 axis and predicts a poor prognosis. Neoplasma 67:201-208.
[Crossref] [Google Scholar] [PubMed]
- Miao JT, Gao JH, Chen YQ, Chen H, Meng HY, et al. (2019) LncRNA ANRIL affects the sensitivity of ovarian cancer to cisplatin via regulation of let-7a/HMGA2 axis. Biosci Rep 39:BSR20182101.
[Crossref] [Google Scholar] [PubMed]
- Dasgupta P, Kulkarni P, Majid S, Hashimoto Y, Shiina M, et al. (2020) LncRNA CDKN2B-AS1/miR-141/cyclin D network regulates tumor progression and metastasis of renal cell carcinoma. Cell Death Dis 11:660.
[Crossref] [Google Scholar] [PubMed]
- Zhao X, Cheng S, Li S, Li J, Bai X, et al. (2020) CDKN2B-AS1 aggravates the pathogenesis of human thoracic aortic dissection by sponge to miR-320d. J Cardiovasc Pharmacol 76:592-601.
[Crossref] [Google Scholar] [PubMed]
- Wang G, Xu G, Wang W (2020) Long noncoding RNA CDKN2B-AS1 facilitates lung cancer development through regulating miR-378b/NR2C2. OncoTargets Ther 13:10641-10649.
[Crossref] [Google Scholar] [PubMed]
- Zheng Y, Zeng J, Xia H, Wang X, Chen H, et al. (2021) Upregulated lncRNA Cyclin-dependent kinase inhibitor 2B antisense RNA 1 induces the proliferation and migration of colorectal cancer by miR-378b/CAPRIN2 axis. Bioengineered 12:5476-5490.
[Crossref] [Google Scholar] [PubMed]
- Li Z, Cai X, Zou W, Zhang J (2021) CDKN2B-AS1 promotes the proliferation, clone formation, and invasion of nasopharyngeal carcinoma cells by regulating miR-98-5p/E2F2 axis. Am J Transl Res 13:13406-13422.
[Google Scholar] [PubMed]
- Zhang LM, Ju HY, Wu YT, Guo W, Mao L, et al. (2018) Long non-coding RNA ANRIL promotes tumorgenesis through regulation of FGFR1 expression by sponging miR-125a-3p in head and neck squamous cell carcinoma. Am J Cancer Res 8:2296-2310.
[Google Scholar] [PubMed]
- Goodall GJ, Wickramasinghe VO (2021) RNA in cancer. Nat Rev Cancer 21:22-36.
[Crossref] [Google Scholar] [PubMed]
- Yang N, Liu K, Yang M, Gao X (2021) ceRNAs in cancer: Mechanism and functions in a comprehensive regulatory network. J Oncol 2021:1-12.
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Hou J, He D, Sun M, Zhang P, et al. (2016) The emerging function and mechanism of ceRNAs in cancer. Trends Genet 32:211-224.
[Crossref] [Google Scholar] [PubMed]
- Chan SH, Chiang J, Ngeow J (2021) CDKN2A germline alterations and the relevance of genotype-phenotype associations in cancer predisposition. Hered Cancer Clin Pract 19:21.
[Crossref] [Google Scholar] [PubMed]
- Chen Z, Guo Y, Zhao D, Zou Q, Yu F, et al. (2021) Comprehensive analysis revealed that CDKN2A is a biomarker for immune infiltrates in multiple cancers. Front Cell Dev Biol 9:1-12.
[Crossref] [Google Scholar] [PubMed]
- Montalto FI, de Amicis F (2020) Cyclin D1 in cancer: A molecular connection for cell cycle control, adhesion, and invasion in tumor and stroma. Cells 9:1-15.
[Crossref] [Google Scholar] [PubMed]
- Valla M, Klæstad E, Ytterhus B, Bofin AM (2022) CCND1 amplification in breast cancer - associations with proliferation, histopathological grade, molecular subtype, and prognosis. J Mammary Gland Biol Neoplasia 27:67-77.
[Crossref] [Google Scholar] [PubMed]
- Liu Y, Zhang H, Wang Z, Wu P, Gong W (2019) 5-Hydroxytryptamine1a receptors on tumour cells induce immune evasion in lung adenocarcinoma patients with depression via autophagy/pSTAT3. Eur J Cancer 114:8-24.
[Crossref] [Google Scholar] [PubMed]
- Yasi EA, Allen AA, Sugianto W, Peralta-Yahya P (2019) Identification of three antimicrobials activating serotonin receptor 4 in colon cells. ACS Synth Biol 8:2710-2717.
[Crossref] [Google Scholar] [PubMed]
- Gwynne WD, Shakeel MS, Girgis-Gabardo A, Kim KH, Ford E, et al. (2020) Antagonists of the serotonin receptor 5A target human breast tumor initiating cells. BMC Cancer 20:724.
[Crossref] [Google Scholar] [PubMed]
- Wu W, Li Q, Zhu Z, Li C, Lu P, et al. (2022) HTR1D functions as a key target of HOXA10-AS/miR-340-3p axis to promote the malignant outcome of pancreatic cancer via PI3K-AKT signaling pathway. Int J Biol Sci 18:3777-3794.
[Crossref] [Google Scholar] [PubMed]
- Zeng C, Chen Y (2019) HTR1D, TIMP1, SERPINE1, MMP3 and CNR2 affect the survival of patients with colon adenocarcinoma. Oncol Lett 18:2448-2454.
[Google Scholar] [PubMed]
- Siddiqui H, Al-Ghafari A, Choudhry H, Al Doghaither H (2019) Roles of long non-coding RNAs in colorectal cancer tumorigenesis: A review. Mol Clin Oncol 11:167-172.
[Google Scholar] [PubMed]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 146:353-358.
[Crossref] [Google Scholar] [PubMed]
- López-Urrutia E, Montes LPB, de Guevara Cervantes DL, Pérez-Plasencia C, Campos-Parra AD (2019) Crosstalk between long non-coding RNAs, micro-RNAs and mRNAs: Deciphering molecular mechanisms of master regulators in cancer. Front Oncol 9:1-15.
[Crossref] [Google Scholar] [PubMed]
- Giaccherini M, Farinella R, Gentiluomo M, Mohelnikova-Duchonova B, Kauffmann EF, et al. (2023) Association between a polymorphic variant in the CDKN2B-AS1/ANRIL gene and pancreatic cancer risk. Int J Cancer 153:373-379.
[Crossref] [Google Scholar] [PubMed]
- Hjazi A, Ghaffar E, Asghar W, Alauldeen Khalaf H, Ikram Ullah M, et al. (2023) CDKN2B-AS1 as a novel therapeutic target in cancer: Mechanism and clinical perspective. Biochem Pharmacol 213:115627.
[Crossref] [Google Scholar] [PubMed]
Citation: Saliani M, Ahmadian MR, Gonbadi P, Dastgir F, Daryasary HM, et al. (2024) CDKN2B-AS1 ceRNA Network and KRAS-Dependent Tumorigenicity in Colorectal and Pancreatic Cancer. Diagn Pathol Open 9:242.
Copyright: © 2024 Saliani M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Open Access Journals
Article Usage
- Total views: 1223
- [From(publication date): 0-0 - Dec 09, 2025]
- Breakdown by view type
- HTML page views: 932
- PDF downloads: 291