Cloning and Expression of Vaccine Peptide Containing NS3, E2, NS5A Genes of Hepatitis C Virus in Pichia Pastoris
Received: 27-Dec-2021 / Manuscript No. jbtbm-22-50645 / Editor assigned: 29-Dec-2021 / PreQC No. jbtbm-22-50645 (PQ) / Reviewed: 12-Jan-2022 / QC No. jbtbm-22-50645 / Revised: 19-Jan-2022 / Manuscript No. jbtbm-22-50645(R) / Accepted Date: 19-Jan-2022 / Published Date: 24-Jan-2022 DOI: 10.4172/2155-952X.1000252
Abstract
Introduction: Hepatitis C is one of the most important causes of chronic hepatitis in developed countries. So far, no useful studies have been performed to design a cost-effective, high-immunity immunization vaccine for hepatitis C virus (HCV) infection. The aim of this study was to clone and express the vaccine peptide containing the NS3, E2, NS5A genes of hepatitis C virus in the yeast of Pichia pastoris.
Materials and methods: In this study, we used the methylotrophic yeast Pichia pastoris as our host to make the recombinant protein. In the next step, the dominant immuno-peptide genes NS3, NS5A and E2 were selected. The mouse IgG2a was selected as desired protein fraction. Linker peptide -GGGGS- was used to make the fusion peptide. The peptides were optimized using Geneious and Gen Script software before synthesis. Then, these genes were cloned into the expression vector pPICZαA. Pastoris strain GS115 strain was transferred to P. pastoris host cells.
Results: Spot blotting and SDS-PAGE techniques confirmed the expression of high levels of NS3-NS5A-E2 -Fc fusion peptide. The results of the other part of the study showed that the possibility of high protein expression is associated with the Codon Compatibility Index (CAI). The CAI of FC-NS3 -E2-NS5A components for Pichia pastoris expression host was 0.68. The other part of the study also showed that after optimizing the GC content became more uniform during the coding sequence. The results also showed that 39% of FC-NS3 -E2-NS5A codons were in the range of 91-100 codons (higher frequency codons) before optimization and this percentage reached 82% after gene optimization.
Conclusion: As a result, this study showed that fusion peptide is expressed in GS115 strains, as confirmed by dot blot technique.
Keywords
Hepatitis C virus; Development; Vaccine; Delivery system
Introduction
Hepatitis C Virus (HCV) is a small enveloped positive singlestranded RNA virus belonging to the Hepacivirus genus in the Flaviviridae family. Its diameter is nearly 60-50 nm, and the virion has nearly 10,000 nucleotides in length. The inner envelope capsule of HCV consists of a nucleocapsid, surrounded by a lipid bilayer where the two envelope glycoproteins, E1 and E2, are anchored [1]. Hepatitis C virus is heterogeneous and has six genotypes and more than 50 subtypes. Due to the genome nucleotide sequence variability, an approximate 31-34% genomic difference is observed among genotypes and genomic difference 23-20% is seen among subtypes. In 1989, this virus was discovered in the USA, and it infects 3-4 million people annually [2]. According to the World Health Organization (WHO) report, 3% of the world population has had an acute HCV infection. Eventually, HCV infection may lead to liver cirrhosis and hepatocellular carcinoma (HCC). Moreover, it induced a successful immune response in patients infected with HCV, produced a strong and persistent cytotoxic T lymphocyte (CTL) response, and maintained a boost immune response in CD8+ T cells [3]. Therefore, HCV clearance has been associated with direct stimulation of CD8+ by T helper 1 (Th1). The predominance immune Cytokine in CD4+ and CD8+ are IFN-γ and TNF-α. The previous study has shown which type of T cell responses controls the pro-viral load. Moreover, the effective cellular immune response against HCV includes T cell response that enables them to identify and target multiple MHC class I- restricted epitopes. A previous study has shown that HCV contains two Tran’s membrane glycoproteins E1 and E2. E2 glycoprotein is known to induce a cellular response [4]. Moreover, E1 comprises 4 or 5 N- linked glycosylation sites, while E2 has 11 N-linked glycosylation sites. We focused on E2, which binds putative CD81, which critical for viral entry. In this study, E2 is used as a conserved region, making it a suitable target for designing peptide vaccines. Although we used NS3 and E2 peptides to design peptide vaccines, numerous studies have shown that NS5A is a known region for HCV replication complex, which has a critical role in the survival of the HCV virus. Moreover, it has been explained that NS5A interacts with cellular proteins such as Raf-1, p53, PI3K, and Grb2, which are involved in host cell signaling. Though, the function of HCV RNA is not yet completely understood. NS5A contains several immunogenic domains of viral protein. NS5A consists of 447 amino acid in three distinct complex structures, including domain I (aa1-213) is defined by crystallography as a dimer which is vital for RNA replication. Domain II (aa 250-342), and domain III (AA 356- 447) which are less conserve structures beside, natively unfolded [5]. Also, domain III is necessary for the proper assembly of the viral particle. One of the significant points mentioned in the recent articles is that NS5A stimulate both cellular and humoral immune response. According to recent findings, NS5A affects the IFN signaling pathway leading to the production of pro-inflammatory cytokines. Hence, NS5A is associated with adaptive immune response. Previous research has shown that NS5A is presented by MHC class I and plays an essential role in the production of IFN-γ. Also, recent studies showed that Fc receptors in mice play an essential role in regulating lymphocytes and inflammatory responses. Amongst mouse IgG subclasses, IgG2a is considered the most potent activator in the experiment of murine infection and tumor models. A recent study showed that IgG2a exerts a protective role in reducing tumor size in mice. Recent evidence confirmed the methylotrophic yeast, Pichia pastoris, as an alternative for the high-level recombinant protein production rather than E. coli. Moreover, Pichia pastoris is well documented as a post-translation modification of recombinant protein expression. The recombinant protein was produced in Pichia pastoris via methanol as the sole carbon source. The activity of Pichia pastoris relies on two genes: AOX1 and AOX2, which encoded two enzymes with AOX activity. Furthermore, AOX1 plays a vital role in the production of recombinant protein since 30% of the total soluble protein in the extracts of Pichia pastoris is grown solely on methanol. The aim of this study was to clone and express the peptide of a vaccine containing the NS3, E2, NS5A genes of hepatitis C virus in the yeast of Pichia pastoris.
Materials and Methods
Design and bioinformatics analysis of NS3, NS5A and E2 sequences
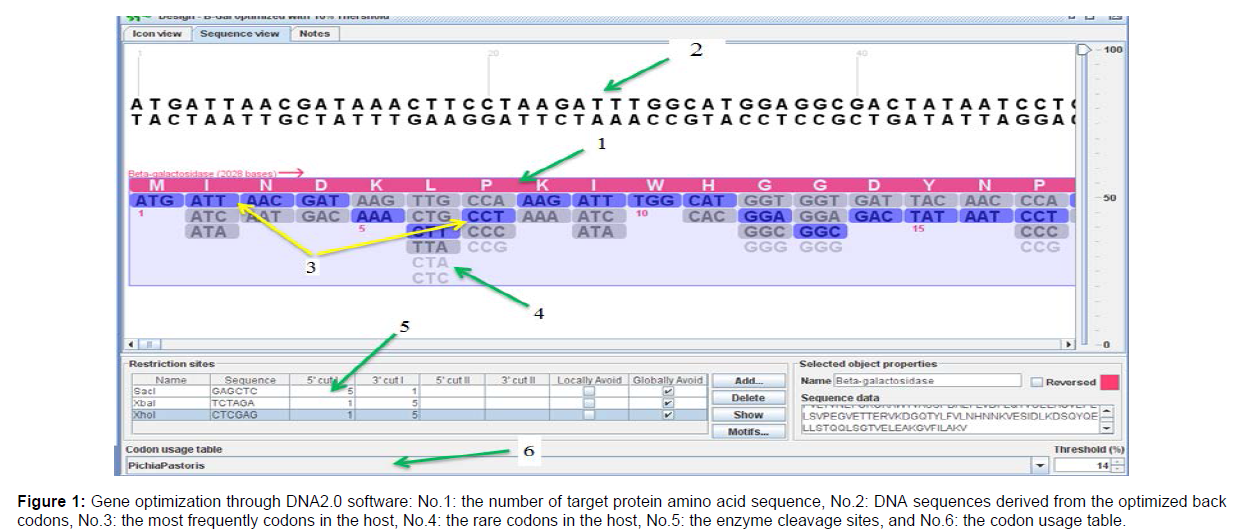
In the first step, sequences of the hepatitis C virus that effectively stimulate the immune system were examined. For this purpose, the published articles on the produced vaccines were reviewed and according to the multiplicity of genes studied against hepatitis C virus, the desired sequences were selected. The desired peptides were obtained through the Swiss-port / Uniprot KB sites and the National Center for Biotechnology Information (NCBI). NS5A protein contains sequence of amino acids 1992 and 2221-2231. FC peptide with a length of 718 bp was added to the HCV peptides. At the end of ´3 sequences, 6 histidine amino acids (His-Tag) and c-myc epitops were placed, followed by the end codon (TAA). Finally, the enzyme cleavage sites for XhoI, XbaI and BamHI restriction enzymes were engineered between NS3, NS5A, E2 and Fc sequences. In this study, all peptide fragments were ligated using the general GGGGS linker. The HCV genotype and the synthesized gene sequences NS3, NS5A, E2 belong to genotype 1a. In this study, pPICZαA vector was used to express the selected sequences. DNA2.0 software was used to predict the expression of selected fragments of codon optimization for yeast hosts, to obtain nucleotide sequences from selected peptides, loci of restriction enzymes (XhoI, XbaI, SacI and BamHI) (Figure 1).
Cloning simulation in the plasmid pPICZαA
After optimizing the desired sequences in DNA2.0 and Genescipt software and in order to simulate the cloning process in the pPICZαA plasmid, Geneious software (Biomatters Ltd, Newealand) was used. For the process of cloning the selected fragments in the plasmid, 5 nucleotide sequences corresponding to the NS3-NS5A-E2 fragments of the XhoI enzyme cleavage site were added to the 5' end and 5 nucleotide was added also to the 3' end of BamHI enzyme cleavage site. A BamHI enzyme cleavage site was also added to the FC5 end and XbaI enzyme cleavage site was added to the 3' end. In this study, the expressed peptide secretion of alpha secretory factor (factor-α) was used. Therefore, the relevant sequence was added to the end of 5'. At the end of the simulation, the ORF framework, NS3-NS5A-E2, FC, c-myc epitope and 6xHis tag were examined to ensure correct translation of factor-xα. Sequence synthesis of the desired parts was performed by Nedaye Fan Company. NS3-NS5A-E2 and FC sequences were cloned into pGH plasmid and lyophilized. In this study, lyophilized DNA was solubilized using nuclease-free water. Then the desired solution was stored at -20°C until use. The final DNA concentration was 4 μg, and then add 8 μL of nuclease-free water to sample, for a total volume of 0.5 μL.
Obtain components containing NS3-NS5A-E2 and FC sequences
In this study, sequences of NS3-NS5A-E2 and Fc fragments were cloned into the pGH plasmid. At this stage, using the appropriate restriction enzyme, the desired fragments were removed from the plasmid. For this purpose, the following steps were performed:
Preparation of susceptible bacteria
In order to amplify the pGH plasmid containing the fragments, JM109 (E. coli strain) was used. At this stage, JM109 was cultured in ampicillin-free medium. After 24 hours in the incubator, a single colony was selected and inoculated into 5 ml of liquid LB medium without ampicillin and then overnight in the shaker incubator to allow bacteria to grow. 200 μl of the above medium was inoculated into 25 ml of fresh LB medium and placed in a shaker incubator for 2 to 3 hours. Suitable turbidity (absorption 0.5 to 0.8 at 600 nm) was considered. Distribute the cultured bacteria in 2 falcon tubes with 12.5 ml volume and place on ice for 10 minutes. The falcons were centrifuged at 8000 rpm for 5 minute at 4 ºC. Discard the supernatant and place the tubes on absorbent paper to completely remove the supernatant. The bacterial precipitate was suspended in 4 ml of 0.1 M sterile calcium chloride solution. The Falcons are centrifuged at 8000 rpm for 5 minutes at 4 ºC. The precipitate was suspended in 12.5 ml of 0.1 M sterile calcium chloride solution. The tube containing the cells was incubated on ice for 30 minutes. The centrifuge tubes were centrifuged at 4000 rpm for 5 minutes at 4 ºC. Discard the supernatant and dissolve the precipitate in 1 ml of sterile calcium chloride solution containing 20% glycerol. We divided these bacteria into volumes of 100 microliters and kept them at -80ºC.
Transfer of pGH plasmid to susceptible bacteria
We first melted 1 microliter of susceptible bacterial suspension on ice. We added 2 microliters of plasmid pGH to it. Incubate the mixture on ice for 30 minutes. The mixture was placed in a pan at 42 ºC for 70 seconds and immediately transferred to ice for 5 minutes. The mixture was then added to 200 μl of ampicillin-free LB medium and incubated at 37 ºC for 1.5 h. Pour 6 μl of the above culture on an LB ampicillin agar plate, then the plates were placed in a 37 ºC incubator for 18 to 20 hours. After the required time, the growth of colonies (containing ampicillin-resistant components) was examined.
PGH plasmid amplification
After colony formation, a single colony was isolated and transferred to 5 ml LB of ampicillin fluid. The above medium was placed in a hot shaker for 24 hours at 37 ºC and at a suitable speed.
Extraction of plasmid pGH
The extraction of plasmid is based on alkaline lysis and subsequent adsorption of DNA by silica and then separation of the plasmid from the column. For this purpose, Bionier company's plasmid extraction kit was used. The procedure was to add 500 μl of BL buffer to the CB3 column. Centrifuge the column at 12,000 rpm for 1 minute and discard the effluent. Centrifuge 1.5 ml of the culture medium containing the transformed bacteria incubated overnight at 37ºC for 1 minute at 12,000 rpm. We discarded the supernatant. Dissolved the bacterial cell plate in 250 μl of PR1 buffer. Add 250 μl of PL2 buffer and mix thoroughly by gently inverting the micro Centrifuge at 12,000 rpm for 10 minutes. We placed a CS column (filter) on a clean microtube. Transfer the supernatant to a CS column filter. We centrifuged at 12,000 rpm for 2 minutes. We took the cell lysis product on the balanced CB3 column. Centrifuge at 12,000 rpm for 1 minute and discard the liquid below. 500 μl of PD buffer was added to the CB3 column and centrifuged at 12000 rpm for 1 min. Discard the drained liquid. 700 μl of PW buffer was added to the CB3 column to wash and remove the salts and centrifuged at 12000 rpm for 1 minute. Discard the underlying liquid and centrifuge for another 2 minutes at 12,000 rpm to remove the remaining wash buffer. Hold the column with the door open for several minutes to allow the membrane surface to dry. To dissolve the DNA, place the column on a clean microtype. Pour 50 μl of PEB buffer (Tris.Cl 10 mM) or water into the center of the CB3 column, incubate for 2 minutes at room temperature, and centrifuge at 12000 rpm for 1 minute until Dissolved DNA is removed.
Enzymatic digestion of NS3-NS5A-E2 and FC components
After plasmid extraction, suitable restriction enzymes were used to isolate NS3-NS5A-E2 and FC fragments. In this study, the pGH plasmid containing NS3-NS5A-E2 fragments was digested by the restriction enzymes XhoI and BamHI, and the pGH plasmid containing the FC fragment was digested by the restriction enzymes XbaI and BamHI. Also, in order to clone the fragments in pPICZαA vector, this vector was digested by XhoI and XbaI enzymes after extraction. To separate the fragments resulting from enzymatic double digestion, agarose gel electrophoresis of the fragments was used. For this purpose, to prepare the X10TAE buffer first, 48.4 g of Tris, 20 ml of EDTA (0.5 M) with pH = 8, combined with 11.42 ml of glacial acetic acid and brought to a final volume of 1000 ml with sterile water. It should be noted that 1X concentration of this buffer was used for electrophoresis of parts.
Purification of NS3-NS5A-E2 and FC fragments and plasmid pPICZαA from agarose gel
The procedure was as follows: first, using a sterile scalpel, we separated the area of the gel where the DNAs were located and placed each in a sterile 1.5 micro tube. We weighed each of the gel-containing micro tube and added the same amount of weight to the GB buffer. Incubate at 50 degrees until the gel is completely melted. The sample was transferred to a transfer column on a clean micro tube and centrifuged at 100g for 1 minute. Discard the effluent. Repeat this step to remove the remaining sample from step 2. 750 μl of washing solution was added to the column, and then centrifuged at 10000g for 1 minute, the output solution is harvested. The column was centrifuged at 10000g for 1 minute for removing the remaining ethanol. For washing the DNA, the column was placed on a clean micro tube. Pour 30 to 50 microliters of solvent buffer, TE buffer or sterile water on the column filter and leave for 2 minutes. The 10000g centrifugation was performed for 1 minute until dissolved DNA. We kept the DNA at -20 degrees.
Clones of NS3-NS5A-E2 and FC fragments in the plasmid pPICZαA
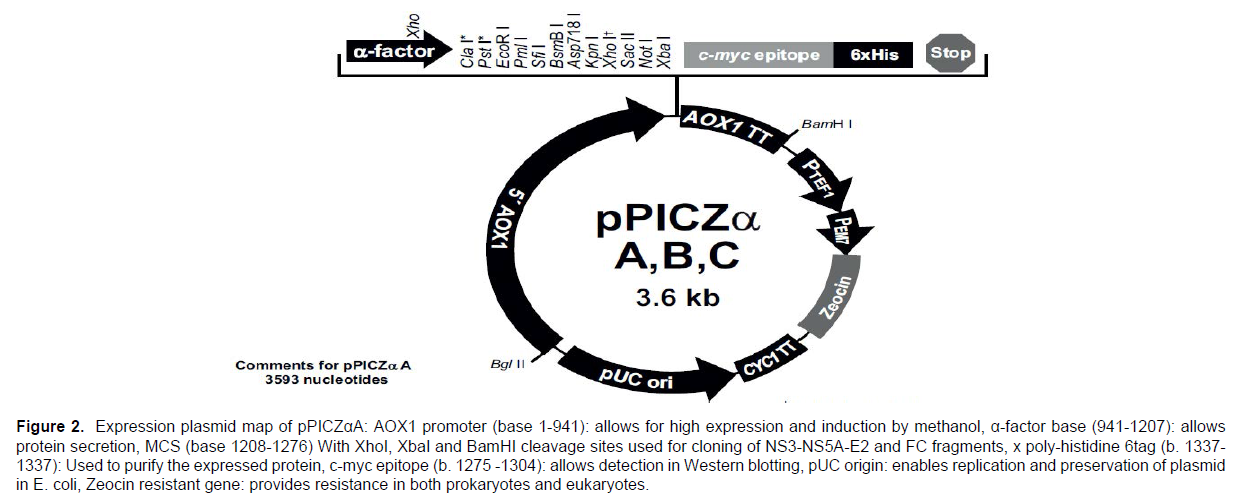
In this study, the expression plasmid (pPICZαA) of I nitrogen Company was used. This plasmid has different loci for different types of restriction enzymes that can be used to insert the desired gene into the plasmid. It also contains the zeocin antibiotic resistance gene, which allows transformed bacteria to survive in the environment containing zeocin. (Figure 2) shows the gene map of the plasmid pPICZαA.
After obtaining pure NS3-NS5A-E2 and FC fragments, we ligated the fragments using ligase enzyme to clone them in plasmid pPICZαA.
Investigation of HCV and FC fragments in pPICZαA plasmid by colony PCR
We used polymerase chain reaction (PCR) to confirm the ligation action and insertion of two gene fragments into the pPICZαA vector. For this purpose, colonies grown in LB medium were dissolved in 10 μl of nucleoside-free distilled water and used 2 μl of it as a DNA template for PCR. For this purpose, PCR reaction of AOX1 and α-Factor primers in (Table 1) was performed. If the ligation and insertion of the fragments into the plasmid is performed correctly, the length of the fragment obtained by colony PCR will be 1434 bp.
| Product length | Sequence | Primer name |
|---|---|---|
| 1434 bp | 5´-GCAAATGGCATTCTGACATCC-3´ | 3´AOX1 sequencing primer(3´Pichia primer) |
| 5´-TACTATTGCCAGCATTGCTGC-3´ | α-Factor sequencing primer |
Table 1: Primers used in colony PCR.
Determination of cloned plasmid concentrations
The extracted plasmids were selected after confirmation of cloning by nano prop device and the highest concentration was selected from the samples for electroporation reaction.
Linearization of the recombinant plasmid pPICZαA
The pPICZαA plasmid is designed to enter the yeast genome through its AOX1 promoter and ensure permanent yeast expression. As a result, the recombinant plasmid pPICZαA was fermented using the SacI enzyme of the fermentase line to facilitate its integration into the Pichia pastoris genome. After completing the linearization process, the enzyme was inactivated for 20 minutes at 65ºC. For this purpose, according to (Table 2) the recombinant plasmid pPICZαA linearization reaction was performed. In this reaction, the restriction enzyme SacI of Fermentase Company was used.
| Materials required | Volume |
|---|---|
| Sterile water | 5.8 microliters |
| SacI enzyme | 1 microliter |
| Tango Buffer | 1 microliter |
| Recombinant plasmid pPICZαA | 4 microliter |
| the final Volume | 10 microliter |
Table 2: Line arization reaction of recombinant plasmid pPICZ αA.
Purification of linear plasmid by phenol-chloroform method
At this stage, the available solutes and additives were removed using phenol-chloroform method and pure plasmid was prepared for electroporation. At this stage, the linearly recombinant plasmid was transferred into the yeast using an electrical pulse. 3μg of linearly recombinant plasmid was mixed with 80 μl of competent cells of Pichia pastoris and then transferred to 0.2 cm electroporation cuvette. 150 μl of this falcon was cultured in a number of YPDs plates (2% agar, 2% peptone, 1% yeast extract, 2% dextrose and 1 sorbitol M1) containing 100 μg / ml zeosin. The plate was incubated for 2 to 3 days at 30ºC until colonies formed.
Screening of clones containing recombinant plasmids
After growing the colonies in the YPDs-Zeocine plate, several colonies were selected to find the colonies expressing the desired protein. These colonies were in 10 ml of BMGY medium (2% peptone 100 mM, 1% yeast extract (Yeast Nitrogen Base) with Ammonium), 1.34% YNB, 1% potassium glycerol phosphate, 4x10 biotin (sulfated and without amino acids) were cultured for 20 hours at 30°C in a shaker incubator to obtain OD600 equal to 1.5). Then 5 ml of these cultures were used to prepare glycerol storage and another 5 ml was used to optimize expression. 4 ml of BMMY medium was used to stimulate the expression of NS3-NS5A-E2-FC, which is similar in composition to BMGY medium and only methanol 5% instead of 1% glycerol. For 8 consecutive days, 1 ml of culture medium was removed each time and 1 ml of fresh BMMY medium containing 2% methanol was added to it. The samples were centrifuged at 1500 g for 5 minutes and their supernatants were kept at -80 for subsequent analyzes including dot blot and SDS-PAGE.
Evaluation of recombinant plasmid expression in yeast Pischia pasteuris
Perform the dot blot technique
At this stage, the method was used to detect, identify and analyze the protein produced in yeast. The procedure was as follows: 2μL of the samples were placed on nitrocellulose paper 4 mm apart and allowed to dry. The paper was incubated for one hour in the blocker solution at ambient temperature. Discard the blocker and wash the paper three times with TBS-T. The primary antibody (Anti-mouse IgG-HRP) was added (in TBS-T with 1 /. % BSA) and incubated for 3 minutes at room temperature. Discard the solution and wash the paper three times with TBS-T. The paper was incubated with HRP-conjugated secondary antibody (Anti-Rabbit IgG-HRP) for 30 minutes at room temperature. Discard the solution and wash the paper three times with TBS-T. The paper was incubated with ECL solution for one minute. We kept the paper in the dark and examined it with a chemo luminescence device. In the next step, SDS-PAGE analysis was performed to further confirm the peptide production in Pichia pastoris yeast. Proteins in Pichia pastoris yeast culture medium were isolated using SDS-PAGE and protein bands were stained using CBB-R250 dye.
Results
Bioinformatics analysis
Bioinformatics analysis
A codon-optimized α-MF along with the pPICZαA based vector was employed to obtain secretion of NS3-E2-NS5A-Fc fusion peptide at high levels in the culture medium. For pPICZαA-HCV synthesis and expression, the AOXI promoter sequence was used through homologous recombination and a codon-optimized α-factor from S. cerevisiae to drive the fusion peptide secretion. Pichia pastoris strain GS115 was used as the host and the pPICZαA was used as an expression vector. NS3-E2-NS5A-Fc segments were cloned in XhoI/XbaI site of MCS from pPICZαA. Amino acid sequences of the final recombinant protein are shown in (Figure 1 and Figure 2).
Figure 1: Gene optimization through DNA2.0 software: No.1: the number of target protein amino acid sequence, No.2: DNA sequences derived from the optimized back
codons, No.3: the most frequently codons in the host, No.4: the rare codons in the host, No.5: the enzyme cleavage sites, and No.6: the codon usage table.
Figure 2. Expression plasmid map of pPICZαA: AOX1 promoter (base 1-941): allows for high expression and induction by methanol, α-factor base (941-1207): allows
protein secretion, MCS (base 1208-1276) With XhoI, XbaI and BamHI cleavage sites used for cloning of NS3-NS5A-E2 and FC fragments, x poly-histidine 6tag (b. 1337-
1337): Used to purify the expressed protein, c-myc epitope (b. 1275 -1304): allows detection in Western blotting, pUC origin: enables replication and preservation of plasmid
in E. coli, Zeocin resistant gene: provides resistance in both prokaryotes and eukaryotes.
Construction and Confirmation of NS3-E2-NS5A-Fc
PCR and digestion beside linearized plasmid demonstrated that NS3-E2-NS5AFc segments were ligated with pPICZαA expression vector. The PCR and restriction digestion of the recombinant plasmid pPICZαA-NS3-E2-NS5A-Fc produced a fragment with the expected length of nearly 1444 bp based on the results of a typical PCR analysis by universal primers for the inserted segment. DNA fragments with the expected length of the target gene (1434 bp) were produced in the transformed yeast clones.
Expression of pPICZαA-HCV in Pichia pastoris
Digestion with Sa cI generates a fragment that may integrate into the AOXI region by homologous recombination. The recombinant pPICZαA-HCV vectors were linearized with Sa cI enzyme and transformed in GS115 cells of Pichia pastoris. After pPICZαAHCV expression in media, the media's supernatant was studied by dot blot and 10% SDS-PAGE. In dot blot, an antibody against c-Myc epitopes was used in the cassette gene's terminal segment. During the cultivation course, a fusion of the current peptide was shown using anti-cmy antibody in the supernatant, which was not found in the control extracts. The coomassie blue staining showed that 6 of 10 fusion peptide cloned produced in SDS-PAGE were present in the control strain with an expected molecular weight of (~60 kDa) for NS3-E2-NS5A-Fc.
Discussion
Recent studies have shown that an effective HCV vaccine should reduce the viral load of HCV. Moreover, dendritic cells play an important role in the antigen presentation to T cells and release IFN-γ along with activated T cells. Since IFN-γ triggers IgG2a expression and inhibits IgG3 and IgG1 production, finally, IgG2a expression may become recurrent a Th1- type immune response. Researchers believe that a successful immune response against HCV requires CD8+ T cells' activation since they can detect multiple sites of the viral genome indeed; one of the potentially significant obstacles in developing the HCV vaccine is that the virus (HCV) can escape the immune system due to the variability of HCV genotypes and subtypes. The investigation of HCV induction on chimpanzee showed that animals exposed to the same infection (homologous) HCV were protected, while chimpanzee was susceptible to heterologous strain. Usually, the virus that caused an infection may become advanced in the long run.
Chimpanzees were the only model in the clinical hepatitis c infection, though several points make the investigation difficult such as low chronic infection rate and lack of liver fibrosis, besides costs and ethical concerns. As a result, a new mouse model has been identified in the evolution of the hepatitis c virus that is severed as an excellent model to study the virus. AFC8hu HSC/Hep mice supported HCV infection in the liver and generated human T cell response to HCV. Another mouse strain has been modulated, Fah–/–Rag2–/–Il2rg–/– mouse, whose liver can very efficiently repopulated by human hepatocytes .
Moreover, it has been demonstrated that the non-structural protein NS5A can improve the virus's survival through the interaction of innate immunity and acquired immunity. However, NS5A HCV protein is suggested as a target for antiviral treatment. Thus, a recent analysis showed that, at least in mice, the minimum region of NS5A requires mediating protection against HCV involving domain I and domain II, and the amino-terminal of domain II. The level of anti- NS5A immunoglobulin is more than 10^4 after immunization. The most important part of this response is IFN-γ production by T cells. A study showed that the CTL response could inhibit NS5A . Eventually, based on previous results, designing a peptide vaccine against HCV dramatically stimulates T cell and phagocytic cells, balances the immune response, and even decreases the HCV viral load
Conclusion
Variety of yeast expression systems are being used to produce recombinant proteins. Studies showed the great potential of the methylotrophic yeast Pichia pastoris for producing recombinant proteins due to lower costs and higher efficiency. The present study results indicated that the appropriate expression and peptide fusion is improved by Pichia pastoris. We hope that the findings of our study have a considerable effect on the development of HCV peptidevaccines.
Statements
Statement of Ethics
This study did not require ethics approval because it did not involve human subjects or animals.
Conflict of interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
This study was financially supported by a grant from the Islamic Azad University of Damghan, an institution involved in education and research. The funder had no role in study design, collection, analysis, and interpretation of data, and/or writing of the report.
Author Contributions Statement
R.K., A.E. and S.T. designed and performed the experiments. R.K., A.E. and S.T. analyzed the data. R.K. wrote the manuscript draft, and B.M. and S.A.J. critically revised the manuscript. S.A.J. supervised the study. All authors have read and approved the final version of the manuscript.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author [S.A.J.] upon reasonable request.
References
- Deleersnyder V, Pillez A, Wychowski C, Blight K, Xu J, et al. (1997) Formation of native hepatitis C virus glycoprotein complexes. Journal of virology 71:697-704.
- Helle F, Dubuisson J (2008) Hepatitis C virus entry into host cells. Cellular and molecular life sciences 65:100-112.
- Fytili P, Dalekos G, Schlaphoff V, Suneetha P, Sarrazin C, et al. (2008) Cross-genotype-reactivity of the immunodominant HCV CD8 T-cell epitope NS3-1073. Vaccine 26:3818-3826.
- Pasetto A, Frelin L, Brass A, Yasmeen A, Koh S, et al. (2012) Generation of T-cell receptors targeting a genetically stable and immune dominant cytotoxic T-lymphocyte epitope within hepatitis C virus non-structural protein . J Gen Viro 93:247.
- Rehermann B, Nascimbeni M (2005) Immunology of hepatitis B virus and hepatitis C virus infection. Nature Reviews Immunology 5:215-229.
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Citation: Jamehdar SA, Karimi R, Esmaili A, Tabaei S, Mashkani B (2022) Cloning and Expression of Vaccine Peptide Containing NS3, E2, NS5A Genes of Hepatitis C Virus in Pichia Pastoris. J Biotechnol Biomater, 12: 252. DOI: 10.4172/2155-952X.1000252
Copyright: © 2022 Jamehdar SA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 3912
- [From(publication date): 0-2022 - Nov 22, 2025]
- Breakdown by view type
- HTML page views: 3231
- PDF downloads: 681