Conformational Dynamics in the Endonuclease Domain of Colicin E
Received: 11-Apr-2017 / Accepted Date: 19-Apr-2017 / Published Date: 26-Apr-2017
Abstract
The endonuclease domain of colicin E9 binds Zn2+ in a multi-step process with a substantial increase in stability that has been suggested to result partly from changes in the dynamic properties of the protein as detected by tryptophan fluorescence. We have used NMR and molecular dynamics to investigate the effect of zinc binding on the structure and dynamics of the backbone of the colicin E9 DNase through 15N chemical shift and relaxation parameter analyses. Significant differences in the experimental chemical shifts between the metal-free and Zn2+-containing forms were limited to the region of the metal binding site, with the largest difference of 12.7 ppm seen for Val 98. Analysis of the relaxation data was carried out using the Lipari-Szabo model-free formalism. The effective rotational correlation time of 10.8 ns for the Zn2+ bound E9 DNase is similar to that of the metal-free form, 11.0 ns. The local dynamics of both the metal-free and Zn2+ bound E9 DNase, were indicative of a rigid protein. Average order parameters for the metalfree and Zn2+ bound E9 DNase are uniformly high, averaging in excess of 0.9 for all the helices and for the E9 DNase as a whole. No significant differences were seen in S2 values for residues at the metal binding site. No significant differences could be seen in the Cα backbone rmsd fluctuations between the metal bound and metal free structures and no evidence for increased deviations between calculated and experimental order parameters. We conclude that the binding of zinc only leads to changes in the protein around the binding site and that the conformation and dynamics of the rest of the E9 DNase are little affected by the presence of zinc. The dramatic effect of zinc on the stability of the protein can be understood in terms of the enthalpy changes associated with metal binding.
Keywords: Colicin; E9 DNase; 15N relaxation; NMR; Protein dynamics; Molecular dynamics
Abbreviations
E9 DNase: Endonuclease Domain of Colicin E9; Im9: Immunity Protein for Colicin E9 DNase; NMR: Nuclear Magnetic Resonance
Introduction
Colicins are a heterogeneous group of anti-bacterial proteins that are produced by Escherichia coli under nutrient stress to kill competing bacteria [1,2]. Cell killing normally occurs either by pore-formation in the inner membrane or, as in the case of the nuclease E-type colicins, by enzymatic cleavage of nucleic acids in the cytoplasm. In common with most colicins, the E-type colicins consist of three functional domains: the killing activity is contained in the 134 residue C-terminal endonuclease domain (E9 DNase), while the central section contains the receptor-binding domain, and the N-terminal region is responsible for translocation of the cytotoxic domain into the target cell. Protection of the producing cell from the cytotoxic effects of the E9 DNase occurs through co-expression of an immunity protein, Im9. X-ray structures for the metal- free E7 [3] Zn2+-containing E9 DNase domains [4] and Im9-E9 DNase [5]. as well as Im7-E7 DNase complexes metal free [6] and metal containing [7] have been reported, in addition to NMR studies of the E9 DNase free [8] and bound to Im9 [9]. These show that Im9 binding does not cause a substantial conformational change to the E9 DNase.
The E9 DNase domain in solution is conformationally heterogeneous [8] with the rate of exchange between the two conformers present, a major and minor form, found to be 1.61 ± 0.5 s-1 at 288 K. Although described as open and closed conformations [10,11], such a description is misleading since there is no evidence for large scale structural differences between the two forms. One hypothesis for the two conformers found in solution by NMR is reorientation of tryptophan 22 about its Cα-Cβ and/or Cβ-Cγ bonds [9]. Notably the two conformers are retained for the E9 DNase domain when in a complex with Im9, with a similar rate of exchange ~1.2 s-1 at 298 K. The present paper deals principally with the major form, but where appropriate comparison will be made to the minor form.
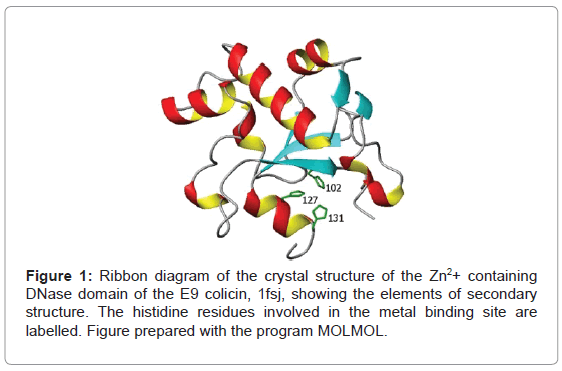
Another feature of the E9 DNase domain is the multi-step binding of transition metal ions, including zinc, nickel and cobalt [12], through the H-N-H motif leading to a ββα-metal topology (Figure 1). The function of the bound metal ion is not known. It may be catalytically active, by binding and stabilizing the DNA phosphate [13,14], or it may simply play a structural role. Equally, the question of whether the metal binding induces more wide ranging conformational changes facilitating the binding of the DNA or enhancing the action of the bound metal ion is unclear. On the one hand, ESI-MS results [15] suggest zinc binding leads to the presence of only a closed conformation implying a dramatically altered conformational heterogeneity while on the other hand NMR still shows two conformers remain in solution for the E9 DNase when metal bound. Taken together these imply the two conformers seen by NMR cannot be identified with the open and closed conformations proposed on the basis of ESI-MS data but they do leave open the possibility that one of the NMR conformers is actually the open and closed forms in fast exchange [10].
The role of the transition metal ion in the E9 DNase remains intriguing since Mg2+ is thought to be the essential cofactor for E9 DNase action and yet it does not bind to the H-N-H domain [16] while Zn2+ binds strongly and in so doing induces phosphate binding, yet is inactive [17]. An argument for the metal playing a structural role can be made on the basis that the melting temperature, Tm, is over 22 K greater when zinc is bound compared to the metal-free E9 DNase [16]. Yet, crystal structures of the E9 DNase:Im9 complex in the presence [5] and absence of Ni2+ [4], show only minor differences in structure in the region of the metal-binding site, a site of tetrahedral coordination based on three histidine ligands; 102, 127 and 131, and a phosphate molecule connected to histidine 103 [5,18,19]. Furthermore, metal binding has no effect on the conformational heterogeneity of the E9 DNase as seen by NMR since it is present in both the Zn2+ and Ni2+ bound forms. Evidence for a marked structural effect on metal binding is therefore absent and attention has turned to the dynamics of the E9 DNase domain in order to rationalize the change in properties of the E9 DNase accompanying zinc binding. Fluorescence measurements on the metal-free E9 DNase and Zn2+ containing E9 DNase as well as an E9 DNase-Im9 complex have shown significant changes in the dynamics of the fluorescent tryptophan probes (Trp22 and 58) on complexation [15]. On the basis of these measurements it was proposed that zinc binding immobilizes Trp22 and has a transmitted effect leading to more widespread conformational changes affecting Trp58 and the overall compactness of the protein. However, such measurements by their very nature, being dependent on just the two tryptophans present, offer only a limited view of the overall molecular dynamics. In addition questions remain unanswered about the extent to which replacing one of the tryptophans, by phenylalanine, to create the singleton Trp mutants modifies the dynamics of the protein. More complete dynamical information can be obtained over a wider range of timescales, including the nanosecond range encompassed by the fluorescence measurements, by analysis of the 15N NMR relaxation parameters, R1, R2 and the heteronuclear (1H)-15N NOE, of the backbone NH [20-22]. Furthermore molecular dynamics simulations are capable of exploring a wide range of protein dynamics [23,24], and through order parameters determined from the dynamics the flexibility of the protein backbone. Indeed simulations have been used to study protein-protein interactions in the colicin system looking at the changes seen when the immunity protein Im9 is bound to the endonuclease domain of E9 [25]. The purpose behind the current simulations is to assess the intrinsic flexibility of the backbone, especially around Trp22 and Trp58, by looking for any marked disagreement between an experimental order parameter found by NMR and the calculated one by molecular dynamics over a long 100 ns run. It has been established that force fields are poor at characterising inherently flexible or poorly defined parts of a backbone consequently these differences would be indicative that conformational changes are plausible in these regions [26]. Direct comparison with the fluorescence measurements is also possible through the indole Nε resonance. Indeed the role of tryptophan in inhibitor binding to a metallo-β-lactamase has been expressly studied by 15N relaxation measurements [27]. The aim of the present study was thus to determine the conformational dynamics of the metal-free E9 DNase and its complex with Zn2+ through molecular dynamics simulations, backbone 15N chemical shifts and relaxation parameters of the E9 DNase to find evidence or otherwise for more wide ranging conformational effects caused by complexation of the E9 DNase domain that might account for the increase in stability of the E9 DNase on zinc binding. Measurements on the Zn2+ containing form were specifically made at 319 K, a temperature at which the Zn2+ containing E9 DNase remains structured while the metal-free E9 DNase is unstructured.
Materials and Methods
Sample preparation
The E9 DNase protein was prepared using the methods previously described [28,29] while the methods described in Hannan et al. [18] were employed to ensure the removal of any Ni2+ which the protein could have been contaminated with from the Ni2+ affinity column. The E9 DNase samples were all 15N labelled. The Zn2+ E9 DNase sample was prepared by the addition of stoichiometric amounts of ZnCl2 to the metal-free E9 DNase.
The sample conditions for the NMR measurements were: 0.5 ml of 3 mM E9 DNase (either metal-free E9 DNase, or E9 DNase + Zn2+ (1:1)) in 50 mM KPO4 buffer with 90% H2O/10% D2O and ~0.01% sodium azide at pH 6.2 (Table 1).
| R1 | R2 | NOE | |
|---|---|---|---|
| Trp 22 NeH metal-free | 0.929 ± 0.028 | 12.871 ± 0.395 | 0.755 ± 0.023 |
| Zn2+ | 1.023 ± 0.030 | 12.782 ± 0.300 | 0.786 ± 0.040 |
Table 1: 15N relaxation parameter data, R1, R2 and hetero nuclear {1H-15N} NOE for the Trp 22 indole Nε in the metal-free and Zn2+ containing form of the E9 DNase domain at 288 K.
NMR spectroscopy
All spectra were acquired on a 600 MHz Varian Unity Inova spectrometer, with an operating frequency of 599.162 MHz for 1H and of 60.72 MHz for 15N. Assignments of the backbone amide NH have been made previously [30]. Backbone NH 15N spin-lattice and spinspin relaxation time measurements were carried out on the metalfree and Zn2+ containing E9 DNase at 288 K and at 319 K for the Zn2+ containing E9 DNase using modified HSQC pulse sequences [31-33]. All the experiments were carried out with spectral widths of 8000 Hz (1H) and 2500 Hz (15N). In the 1H dimension 1024 complex data points were measured, with 64 increments in the T1 dimension and 32 scans per t1 increment, except for the Zn2+ E9 DNase T2 measurements at 288 K in which case there were 128 increments, and 32 scans for each t1 increment. Eleven relaxation delays were used ranging from 10 ms to 4000 ms for R1 and 10 ms to 250 ms for R2 including three duplicate delays for estimation of the uncertainties in the peak intensities. A recycle delay of 3.5 s-4 s was used. Data were acquired in each set of experiments using a random order of relaxation time delays to average out effects such as sample heating. Heteronuclear NOE values were measured using three pairs of ‘with NOE’/‘without NOE spectra’, the ‘with NOE’ experiments including a proton presaturation period of three seconds. The spectra were acquired with 1024 complex points and a spectral width of 8000 Hz in t2 and 128 complex points, 2500 Hz in t1, with 32 scans per t1 point. NOE experiments were performed on the metal-free and Zn2+ containing at 288 K and 319 K. Temperature calibration of the sample was carried out using 100% (v/v) methanol for all measurements. All spectra were processed using NMRpipe [34] and analysed with FELIX 95.0 (Biosym/MSI, California).
Chemical shift calculations
The 15N chemical shifts were predicted using the program SHIFTX2 [35] and the E9 DNase crystal structures, 1bxi [5], 1emv [4] and 1fsj [4] from the Protein Data Bank. 1bxi and 1emv correspond to the metal bound and metal free forms of the Im9:E9 DNase complex while 1fsj is a Zn2+ containing oligomeric form. No crystal structure has been reported for the metal-free E9 DNase free of the immunity protein. Refinement of the structure for Val 98 was carried out using SHIFTS 4.1 [36].
NMR relaxation time analysis
15N relaxation rates R1 and R2 were determined by fitting the measured peak intensities to two or three parameter single exponential decay curves using the program curvefit (A.G Palmer III Columbia University). Errors were estimated by Monte Carlo methods using the same program. The steady state {1H}-15N NOE values were calculated from the peak intensity ratios obtained from the spectra in the presence and absence of proton saturation, errors were estimated from the triplicate set of experiments.
The NH 15N relaxation data were analyzed by the Lipari-Szabo model free analysis [37,38] in which the dynamics is partitioned between a global rotational diffusion and local dynamics. Model free simply means no motional models are assumed for the local dynamics in deriving the spectral densities. In fact the method relies critically on a number of nested models corresponding to various combinations of model free parameters, as well as a preliminary determination of the anisotropic rotational diffusion. Model free analysis was performed using the in-house programs R2R1Win and LSWin using protocols adapted from Mandel [39]. R2R1Win was used to determine the anisotropic rotational diffusion and LSWin the model free parameters. To avoid spurious changes in the model assignments in the Model free analysis resulting from differences in the errors in the experimental data, all the data were analysed using the errors found for the Zn2+ containing form of the E9 DNase at 288 K. Errors in the fitted parameters were estimated using 500 Monte Carlo calculations.
Direct comparison of the 15N relaxation data is complicated by the dependence of these data on motion at a number of different frequencies, as shown by the presence of the spectral densities.
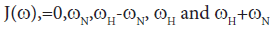 [40]
[40]
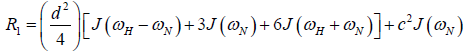
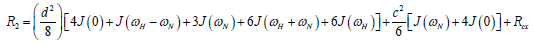
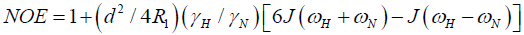
where 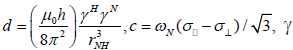 is the magnetogyric ratio, h is Planck’s constant μ0 the vacuum permeability, rNH the NH bond length taken as 1.02 Å and σ|| σ┴ are the parallel and perpendicular components respectively of the axially symmetric chemical shift tensor of the backbone amide 15N nuclei with (σ|| σ┴ ) taken as -160 ppm. In the case of the indole Nε the chemical shift anisotropy was taken as -89.6 ppm [27].
is the magnetogyric ratio, h is Planck’s constant μ0 the vacuum permeability, rNH the NH bond length taken as 1.02 Å and σ|| σ┴ are the parallel and perpendicular components respectively of the axially symmetric chemical shift tensor of the backbone amide 15N nuclei with (σ|| σ┴ ) taken as -160 ppm. In the case of the indole Nε the chemical shift anisotropy was taken as -89.6 ppm [27].
In the model-free theory of Lipari-Szabo [37,38] the spectral densities J(ω) for a protein are defined in terms of a global motion, the rotational diffusion of the protein, and a local motion characterized by the correlation times τm and τf respectively leading to the relationship:
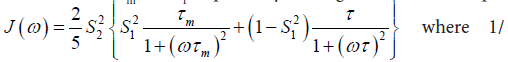
 The spatial restriction of the local motion is described by the order parameter
The spatial restriction of the local motion is described by the order parameter 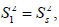 where S2 can vary from 0.0 meaning no restriction to 1.0 where the restriction is complete. Slower segmental motions on the milli- to micro-second timescale can be incorporated through an exchange term, Rex, to R2. In the extended model [41], an additional slower internal motion is introduced which is characterized by an order parameter
where S2 can vary from 0.0 meaning no restriction to 1.0 where the restriction is complete. Slower segmental motions on the milli- to micro-second timescale can be incorporated through an exchange term, Rex, to R2. In the extended model [41], an additional slower internal motion is introduced which is characterized by an order parameter 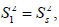 and a correlation time τs, while the faster internal motion is assumed to have a correlation time of 1 ps and an order parameter
and a correlation time τs, while the faster internal motion is assumed to have a correlation time of 1 ps and an order parameter 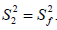 A modified methodology was adopted for the analysis of the relaxation data in order to obtain acceptable χ2 in the determination of the global rotational diffusion tensor. Residues having rigid NH were initially identified using the criteria of Tjandra and Bax [42], (An NOE>0.650 and the absence of exchange contributions to R2). Using these residues and their R2/R1 ratio the global rotational diffusion was determined with the program R2R1Win and the E9 DNase crystal structure co-ordinates. Lipari-Szabo model free analysis was then carried out using the program LSWin together with the rotation diffusion tensor components deduced from the previous step, with model selection based on the Mandel approach. Model free in this instance indicating no specific choice of the dynamics describing the NH bond vector. In practice a model, corresponding to the actual parameters used in the model-free expressions, was chosen on the basis of statistical evidence. The five models are: (1) S2 (2) S2, τf
A modified methodology was adopted for the analysis of the relaxation data in order to obtain acceptable χ2 in the determination of the global rotational diffusion tensor. Residues having rigid NH were initially identified using the criteria of Tjandra and Bax [42], (An NOE>0.650 and the absence of exchange contributions to R2). Using these residues and their R2/R1 ratio the global rotational diffusion was determined with the program R2R1Win and the E9 DNase crystal structure co-ordinates. Lipari-Szabo model free analysis was then carried out using the program LSWin together with the rotation diffusion tensor components deduced from the previous step, with model selection based on the Mandel approach. Model free in this instance indicating no specific choice of the dynamics describing the NH bond vector. In practice a model, corresponding to the actual parameters used in the model-free expressions, was chosen on the basis of statistical evidence. The five models are: (1) S2 (2) S2, τf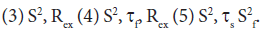 Model selection was carried out using statistical analysis using a combination of χ2 and F-test, a hierarchical selection tree and the principle of parsimony. Further refinement of the rigid NH set was achieved by using the Lipari-Szabo analysis as a filter, taking only those NH which fitted to models 1 and 2 and recalculating the anisotropic rotational diffusion [43].
Model selection was carried out using statistical analysis using a combination of χ2 and F-test, a hierarchical selection tree and the principle of parsimony. Further refinement of the rigid NH set was achieved by using the Lipari-Szabo analysis as a filter, taking only those NH which fitted to models 1 and 2 and recalculating the anisotropic rotational diffusion [43].
Molecular dynamics simulations
Molecular dynamics (MD) simulations were carried out with the OPLS-AA/L all-atom force field using the GROMACS 5.1 package [44]. Three structures for the endonuclease region for E9 DNase were used in the simulations 1bxi, 1emv and 1fsj. All crystallographic waters were removed but the metal ion was retained though replacing the nickel or zinc by magnesium because of the lack of suitable force field data for these metals. Calculations were repeated removing the metal ion from the metal ion containing structures to see whether any differences were observed.
Solvation was carried out by filling a cubic box, approximately 6.9 nm in dimension, with water molecules centring the protein to ensure it was at least 1 nm from the edge of the box. A three point model, spc216.gro, was used to represent the water molecule and roughly 9700 water molecules were required to fill the box. Eight chloride ions, replacing water molecules, were then added to balance the charges. Periodic boundary conditions were applied throughout the following simulations. Energy minimisation, to a target final maximum force of 1000 kJ mol-1 nm-1, was carried out on the solvated protein with a steepest descent minimisation followed by a two-step equilibration using a leap-frog integrator. Particle mesh Ewald (PME) was used to evaluate long range electrostatic interactions. The first step, with a fixed volume and temperature (NVT ensemble) using a modified Berenson thermostat and the second step, at a fixed pressure and temperature (NPT ensemble) with additional isotropic Parrinello-Rahman pressure coupling. In both cases all the bonds were constrained using LINCS at a temperature of 300 K for 100 ps in 2 fs steps. After equilibration was complete 100 ns production simulations were run at a temperature of 300 K with an NPT ensemble using the same couplings and constraints as before. Flexibility in the carbon backbone was established by determining the NH order parameters, S2, from the full 100 ns production molecular dynamics runs using the GROMACS routine g_rotacf. Global tumbling was first removed by a progressive fit to the reference crystal structure. Determination of order parameters using a full long molecular dynamics simulations is known to accentuate weaknesses in the crystal structure description of flexible or poorly defined regions consequently artificially low values of S2 will be found [26]. This approach was chosen deliberately to accentuate any flexibility resulting from the protein backbone structure since it has been shown experimental S2 as found by NMR can be grossly underestimated if the protein is flexible regardless of the force field used. The corollary being that if the S2 order parameter is not underestimated then that part of the structure is as rigid as implied by the experimental NMR order parameter.
Results
15N Chemical shifts
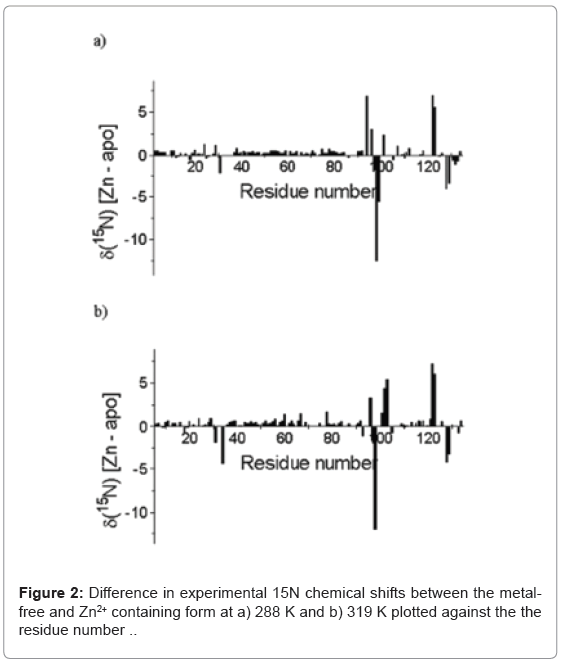
The 15N chemical shift between the metal -free and Zn2+-containing E9 Dnase at 288 K and 319 K are shown in (Figure 2). Calculations were carried out to predict the 15N chemical shifts using SHIFTX2 [35] and the E9 DNase crystal structures. These calculations gave a correlation coefficient of 0.880 between the predicted and observed 15N chemical shifts for the metal-free E9 DNase using the crystal structure (Protein Data Bank access code): 1emv. A similar comparison for the Zn2+-containing E9 DNase, gave correlation coefficients between the predicted and observed chemical shifts of 0.862 and 0.892 using the crystal structure 1bxi and 1fsj respectively. Mean absolute deviations between the predicted and observed chemical shifts were 1.76 ppm for metal-free (1 emv), 1.94 ppm Zn2+ containing (1bxi) and 1.79 ppm Zn2+ containing (1fsj) while the mean absolute deviation between the predicted 15N chemical shifts using 1emv and 1bxi was 0.76 ppm and 1.09 ppm for the two metal containing crystal structures 1bxi, 1fsj. A small number of residues gave deviations exceeding 5 ppm namely (Asp 36, -8.2 ppm; Val 98 -9.1 ppm Asp129, -6.4 ppm) for 1emv (Asn70, 8.55 ppm, Thr122, -6.66 ppm) for 1bxi and (Asp 24, -5.80 ppm, Thr122, 5.34 ppm, Thr123, 5.63 ppm) for 1fsj.
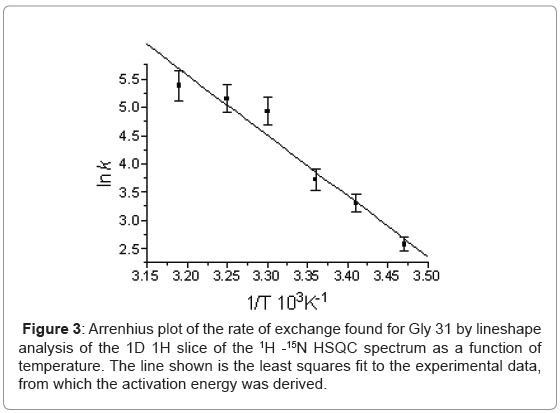
The temperature dependence of the 15N chemical shifts as determined from the 1H-15N HSQC NMR spectra obtained between 288 K and 319 K revealed two groups of resonances. In the first group, consisting of 80 residues, the average difference in chemical shift was only 0.125 ± 0.306 ppm implying an average temperature co-efficient of 4.2 ppb K-1. The largest difference in 15N chemical shift, of only 1.43 ppm, being seen for Leu 67. More complex behaviour was seen in the second group, which consisted of resonances from residues involved in the conformational heterogeneity. Here three types of behaviour were seen in the 15N NMR spectrum as a function of temperature depending on the chemical shift difference between the major and minor conformers at 288 K. When the chemical shift difference was small the resonances, 25 in all, merge together. When the chemical shift difference was large, two resonances remained corresponding to the major and minor forms, 15 in all; and finally a group in intermediate exchange which broaden and in some cases disappeared entirely, 9 in all. Line shape analysis on the 1D 1H slices of the 1H-15N HQSC for Gly 31, the resonances of which merged on increasing temperature, allowed the exchange rate for the conformational exchange to be determined as a function of temperature (Figure 3) and a ΔH‡ of 87.5 ± 9.7 kJ mol-1 to be derived assuming the exchange process is unimolecular. In addition to the resonances for which an exchange process was known to be present, another group of 16 resonances also disappear.
Relaxation time analyses
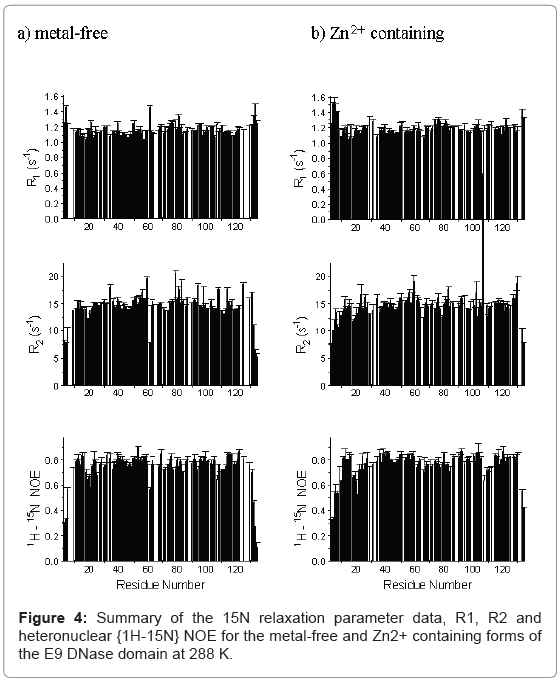
The backbone NH 15N spin-lattice and spin-spin relaxation rates, R1 and R2 and the {1H}-15N NOE for the metal-free and Zn2+ containing forms of the E9 DNase domain at 288 K are shown in Figure 4 and the relaxation parameters for the Trp22 indole Nε are listed in Table 1. The Trp58 indole Nε could not be identified in the Zn2+ containing form and is therefore omitted from the analysis. Only those residues for which a complete data set (R1, R2 and NOE) could be measured are shown, giving data for 102 and 105 backbone NH groups out of a possible 123 (134 residues less the N-terminus and 10 prolines) for the metal-free and Zn2+ containing forms, respectively, at 288 K. At 319 K only 68 residues of the Zn2+ containing E9 DNase could be assigned with a complete relaxation data set. The remaining residues of the metal-free and Zn2+-containing forms were excluded because peak overlap prevented the accurate measurement of one or more of their relaxation parameters. An added complication was the presence of two conformers in all samples of the E9 DNase associated with the major-minor conformational equilibrium which caused the NMR spectra to have many more peaks than would have been expected for a 134-residue protein. Data is only presented here for the major conformer as indicated by the intensity of the NMR resonance, but detailed comparison of resolved signals of the major and minor forms of the metal-free and Zn2+-containing E9 DNase revealed no consistent significant differences between the two conformers of either. For those residues where the two peaks were too close together to be measured separately, an average value for the two conformers was obtained using the overlapped peaks. All such peaks gave statistically acceptable fits to a single exponential in their relaxation decays. Since the exchange rate between the two conformers is in the order of 1-2 s-1 this will not affect the measured relaxation values significantly as only R2 processes might be affected by slow chemical exchange and the design of the pulse sequence excludes the effect of any exchange slower than the delay between pulses in the CPMG sequence [32]. The average errors in the individual measured values were: metal-free E9 DNase, 1.7% (R1), 1.8% (R2) and 3% (NOE); Zn2+-containing E9 DNase, 2% (R1), 4% (R2) and 3% (NOE) at 288 K and 1% (R1), 2% (R2) and 2% (NOE) at 319 K.
The mean relaxation rates and their standard deviations are listed in Table 2. The values for the average R2/R1 exclude those at the termini (residues 3,4,5,130 and 134). To extract a fuller picture of the dynamical behaviour of the two forms of the E9 DNase from the relaxation parameters Model free analysis was used [37,38]. Initially the rotational diffusion was determined from the R2/R1 ratios for a full rigid set of residues chosen on the basis of the criteria proposed by Tjandra et al. [42]. These serve to remove residues undergoing slow internal motions or fast chemical or conformational exchange. The colicin E9 DNase coordinates from the crystal structure of the metal bound form (pdb 1fsj) were used for consistency for both the metal-free and Zn2+ containing E9 DNase given the absence of crystal data for the metal-free form. In order to get acceptable goodness-of-fits a further filter was used to prepare a reduced set of residues [43]. This filter consisted of selecting only those residues which gave model 1 and 2 fits in a subsequent Model free analysis with the initial best rotational diffusion tensor. With this reduced set of residues, and applying an F-test to successive diffusion models, the rotational diffusion tensor was found to be oblate axial for the metal-free and Zn2+ containing E9 DNase with goodness-of-fits better than 0.1 (Table 3).
| N | R1 | R2 | NOE | R2/R1 | |
|---|---|---|---|---|---|
| metal-free 288K | 102 | 1.163 ± 0.081 | 14.637 ± 2.318 | 0.746 ± 0.120 | 13.079 ± 1.49 |
| Zn288K | 105 | 1.198 ± 0.085 | 14.688 ± 2.898 | 0.765 ± 0.101 | 12.625 ± 2.37 |
| Zn319K | 68 | 1.962 ± 0.099 | 8.270 ± 1.781 | 0.783 ± 0.084 | 4.180 ± 0.859 |
Table 2: Average and error, expressed as the standard deviation of the 15N relaxation parameter data, for R1, R2 and heteronuclear {1H}-15N NOE as well as the ratio R2/R1 in the metal-free and Zn2+ containing form of the E9 DNase domain at 288 K and 319 K.
| Disoa | D||/D┴ | D|| | D┴ | φ | θ | χ2 | |
|---|---|---|---|---|---|---|---|
| (107 s-1) | (107 s-1) | (107 s-1) | |||||
| metal-freeb | 1.518 | 0.922 | 1.438 ± 0.015 | 1.558 ± 0.009 | 12.0 ± 3.9 | 76.2 ± 3.7 | 39.8 |
| Zn at 288 Kc | 1.545 | 0.897 | 1.437 ± 0.023 | 1.599 ± 0.015 | 12.9 ± 4.1 | 69.9 ± 4.1 | 58.1 |
| Zn at 319 Kd | 3.085 | 0.927 | 2.930 ± 0.058 | 3.162 ± 0.041 | 55.8 ± 5.0 | -70.7 ± 5.2 | 70.8 |
Table 3: Oblate axial rotational diffusion tensor components from the reduced set of aDiso=(2D┴ + D║ )/3. For comparison the isotropic model gives Diso=1.512 × 107 s-1; χ2=73.6 for the metal-free E9 DNase; and Diso=1.528 × 107 s-1 ; χ2=78.1 for the Zn2+ containing E9 DNase at 288 Kb 46 NH, c62 NH d57 NH.
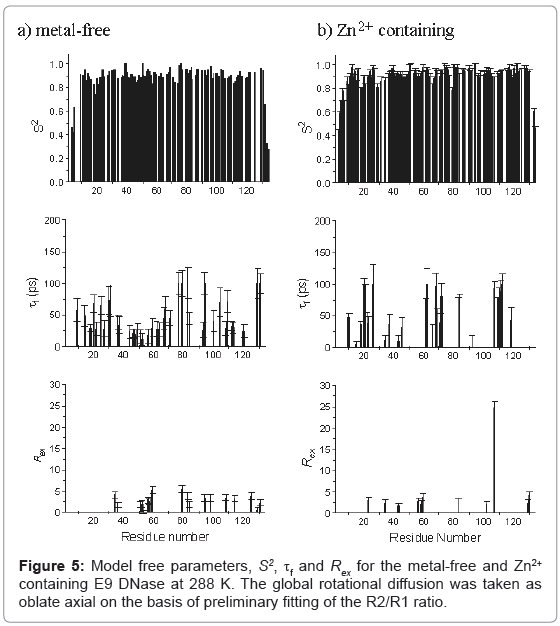
Model free parameters, (S2, S2 f, τf, Rex, and τs) were then found for metal-free and Zn2+ containing E9 DNase assuming oblate axial global rotational diffusion for the E9 DNase with the model selection approach modified to accept a lower critical value, α of 0.015 for the χ2- statistic than used in the original method. Monte Carlo simulations on noisy data sets constructed with relaxation parameters consistent with either model 4 (S2, τf,, and Rex) or model 5 (S2, S2 f and τs) demonstrated this led to fewer data sets not fitting to any model. At the same time the number of data sets under-fitting, that is fitting to models requiring fewer parameters than the three expected for models 4 and 5, was not affected. Although Akaike’s Information Criteria has been proposed as a better alternative to the χ2-statistic in model selection because it reduces the under-fitting of true complex synthesized relaxation data [45,46] in practice this is at the potential cost of over-fitting data represented by simpler models and therefore we did not use this approach. The model free parameters S2, τf, and Rex, for the metal free and Zn2+ containing E9 DNase at 288 K are shown in Figure 5. The numbers of residues fitting to each combination of model free parameters is shown in Table 4 with a comparison of the 65 residues of the Zn2+-containing E9 DNase at 288 and 319 K for which we have complete data in Table 5. The correlation coefficient for the model selection across the set of 91 residues of the metal-free and Zn2+-containing E9 DNase at 288 K for which we have complete data, and assuming the above models are grouped into fast picosecond motion (1), exchange term (3) and nanosecond (5) motions and no fit is assigned a value 0, is 0.443. With this classification 65 of the residues have the same type of motion. The correlation coefficient for the corresponding values of S2 was 0.811. Order parameters for the individual helices in the metal-free and Zn2+ containing E9 DNase are shown in Table 6. While the order parameters for the backbone NH and indole Nε of Trp22 are given in Table 7.
| S2 | S2, tf, | S2,Rex | S2, tf, ,Rex | S2,S2f , ts | No fit | |
|---|---|---|---|---|---|---|
| metal-free | 43 | 27 | 5 | 7 | 13 | 7 |
| Zn | 55 | 23 | 6 | 5 | 11 | 5 |
Table 4: Comparison of the numbers of residues fitting to particular models in the Lipari-Szabo analysis of the local dynamics for the metal-free and Zn2+ containing E9 DNase at 288 K.
| S2 | S2, tf, | S2,Rex | S2, tf, ,Rex | S2,S2f , ts | No fit | |
|---|---|---|---|---|---|---|
| Zn 288K | 39 | 11 | 4 | 2 | 7 | 3 |
| Zn 319K | 53 | 6 | 6 | 3 | - | - |
Table 5: Comparison of the numbers of residues fitting to particular models in the Lipari-Szabo analysis of the local dynamics for the Zn2+ containing E9 DNase at different temperatures.
| Helix | metal-free | Zn2+ |
|---|---|---|
| I 22-25 | 0.903 ± 0.030 | 0.899 ± 0.025 |
| II 36-42 | 0.912 ± 0.039 | 0.924 ± 0.020 |
| III 50-62 | 0.919 ± 0.040 | 0.947 ± 0.031 |
| IV 65-69 | 0.924 ± 0.032 | 0.920 ± 0.029 |
| V 73-80 | 0.951 ± 0.051 | 0.934 ± 0.072 |
| VI 124-129 | 0.949 ± 0.019 | 0.935 ± 0.015 |
Table 6: Comparison of the order parameter S2 for helices in the metal-free and Zn2+ containing E9 DNase.
Molecular dynamics simulations
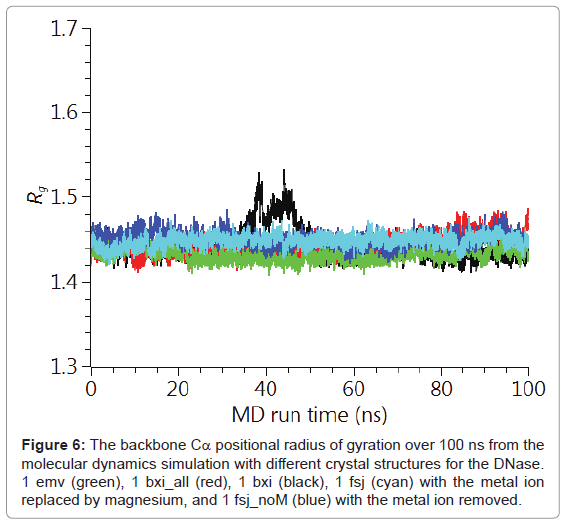
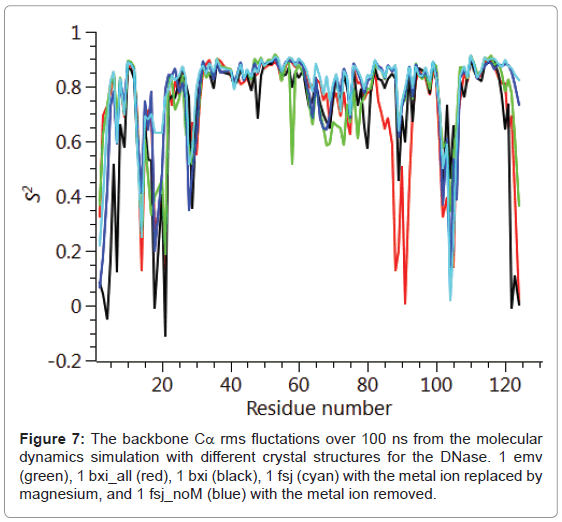
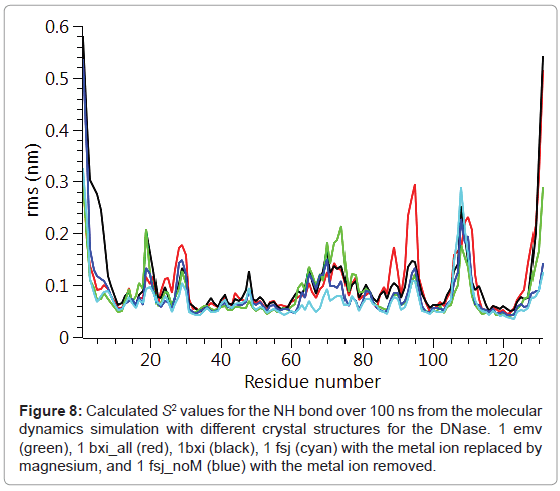
The backbone Cα positional radius of gyration over the 100 ns duration of the molecular dynamics simulation is shown in Figure 6. A stable fluctuation was seen over the entire period for all the crystal structures, 1emv, 1bxi and 1fsj with a mean value of ca. 0.44 nm in good agreement with previous simulations [25]. An indication of the overall backbone flexibility is revealed by the data in Figure 7 showing the Cα atom position root-mean-square fluctuations along the amino acid sequence. Calculated values for the generalised order parameters (S2) along the amino acid sequence are shown in Figure 8.
Figure 6: The backbone Cα positional radius of gyration over 100 ns from the molecular dynamics simulation with different crystal structures for the DNase. 1 emv (green), 1 bxi_all (red), 1 bxi (black), 1 fsj (cyan) with the metal ion replaced by magnesium, and 1 fsj_noM (blue) with the metal ion removed.
Discussion
Chemical shift analyses
15N chemical shifts are a sensitive guide to the local environment of the amide nitrogen reflecting the local structure, the torsion angles of adjacent residues and the presence of hydrogen bonding [47-49]. Although a precise interpretation of the actual differences in chemical shifts is not always possible, the absence of a significant change in a chemical shift on perturbing a protein, for example by metal binding, does mean that the local chemical environments are similar. Equally, when significant changes are seen, as for example in the case of a 4 ppm 15N deshielding on Ca2+ binding to an EF hand, distinct conformational changes, in this case from a compact to an open state, can be identified [50]. Thus in the case of zinc binding to the metal-free E9 DNase, when the vast majority of residues show a change in chemical shift of less than 1 ppm (Figure 2), the general backbone fold and relationship between the secondary structural elements must be preserved, which is consistent with the crystal structure [5]. Large differences in chemical shifts can be identified in three regions: residues 30, 94-104 and 120- 130. The latter two regions correspond to those containing the histidine ligands to the bound Zn2+ (His 102, His 127 and His 131) while the former corresponds to one strand of a β-sheet that forms part of the zinc binding site. Overall the largest difference in 15N chemical shift, 12.7 ppm, is seen for Val 98 arising from the unusually shielded value of 108.6 ppm seen in the Zn2+ form. An indication of the unusual nature of this 15N chemical shift in the Zn2+ form is the fact that only 3% of valine amide 15N nuclei have chemical shifts more shielded than 110 ppm in the RefDB database [51].
The observed correlation coefficients of 0.880 (1 emv: metal free) and 0.862, 0.892 (1bxi. 1fsj: Zn2+) between the predicted and experimental 15N chemical shifts were poorer than has been reported for other proteins [35] suggesting that there are small changes between the solution and crystal structures. However, only small differences in 15N chemical shifts are predicted between the metal-free and Zn2+ containing forms using SHIFTX2, with a mean absolute deviation of 0.76 ppm, and indeed none are observed experimentally outside of the zinc binding site, (Figure 2). Clearly the NMR data reflect a somewhat different solution conformation from the metal-free Im9- bound E9 DNase crystal structure 1emv, and the Zn2+ bound form, 1bxi, 1fsj, with the solution conformers being more similar to each other than the corresponding crystal structures. Importantly though, the experimental differences in chemical shift seen around Trp22 and Trp58, which fluorescence measurements indicate were affected by zinc binding [15] are of the same magnitude as seen elsewhere, suggesting little in the way of conformational change on metal binding. Also large chemical shift differences are not seen in the regions associated with the slow conformational exchange giving rise to the major and minor conformations, namely residues 18-25 and 66-71, consistent with the evidence from the NMR exchange rate data that the major-minor conformational equilibrium is unrelated to the metal binding [8]. Since the only significant changes in 15N chemical shifts are seen around the zinc binding site the open and closed conformations seen by ESI-MS must correspond to differences in the conformation around this part of the E9 DNase.
Only one residue, Val 98, stands out in the comparison between experimental and predicted 15N chemical shifts. Experimentally a difference in chemical shift of 12.7 ppm is seen and yet the program SHIFTX2 predicts essentially no difference. Consequently, some change in conformation in solution must take place around Val 98. Refinement of the side chain orientation using SHIFTS 4.1 demonstrates that the absence of shielding in the metal-free form can be understood simply in terms of changes in the χ1 angle of the Val 98 residue. Similarly the shielding in the Zn2+ form is consistent with the side-chain torsion angle χ1 of -62.4° [4]. A specific shielding effect caused by the presence of the nearby Zn2+ ion, for example an amide polarization effect, can be discounted in the Zn2+ bound form because this is likely to result in a deshielding rather than a shielding [50].
A notable feature of the temperature dependence of the 15N chemical shifts is that a substantial core of residues, over half in total, have a remarkably small variation in 15N chemical shifts. Indeed, the average temperature variation of 4.2 ppb K-1 is not out of line with that seen for proteins in mesophilic organisms which show a greater thermal stability [52]. Moreover, there is no evidence for a smaller temperature dependence for residues close to the metal binding side, despite the constraint caused by the co-ordination of the histidines to the zinc. Overall the data show that this core of residues in the E9 DNase maintains its structure up to 319 K and highlights the absence of specific structural features to account for the increase in melting temperature on binding zinc. Note that this core of residues is common to both the major and minor conformers.
Alongside this stable core of residues, more marked spectral changes arising from chemical exchange are seen for those resonances known to be associated with the major-minor conformational equilibrium. The value for ΔH‡, 87.5 ± 9.7 kJ mol-1, noted earlier in the Results, for this conformational equilibrium is comparable to those of other slow conformational changes in proteins, namely cistrans proline isomerisation, ~80 kJmol-1 [53] and slow flipping of aromatic rings in cytochrome c and 68 kJmol-1 [54]. It is worth noting that the disappearance of a further group of resonances on raising the temperature which do not show conformational heterogeneity at 288 K indicates that more wide ranging conformational exchange processes are present.
Relaxation time analyses
Qualitatively the 15N relaxation parameters profiles for the metalfree, and Zn2+ -containing E9 DNase as shown in Figure 4 are similar, as are the mean relaxation parameters, R1, R2, and heteronuclear NOE. Also of note is that the indole Nε of Trp22 has similar relaxation rates R1 and R2 in both forms, with the smaller values compared with the backbone NH reflecting the smaller chemical shift anisotropy of the indole Nε [27].
Analysis of the full and reduced sets of R2/R1 ratios indicated a rotational diffusion with a small anisotropy in the form of an oblate axial tensor for both forms, with only minor differences between the metalfree and Zn2+ containing E9 DNase. The isotropic correlation time for the metal-free E9 DNase implied by the rotational diffusion tensor is 11.0 ns, rather similar to the value of 11.6 ns seen for the Trp22Phe and Trp58Phe mutants by fluorescence anisotropy decay [15,55].
On binding zinc the isotropic rotational diffusion coefficient seen for the metal-free E9 DNase by NMR shows a small increase of 4%, corresponding to a decrease in the correlation time to 10.8 ns. The effect seen by fluorescence on the single Trp mutants is more marked with a decrease in the correlation time to 9.4 ns Trp22Phe and 9.7 ns Trp58Phe. Overall though, the picture derived from the NMR data is of similar global rotational diffusion of the metal-free and Zn2+ containing E9 DNase with the presence of the metal ion having only a minor effect. Anisotropy of the global rotational diffusion is consistent with the ratio of the moments of inertia (1.0000: 0.85412: 0.69145) as well as with hydrodynamic calculations using HYDRONMR [56] based on the crystal structure of the E9 DNase component of the E9 DNase-Im 9 complex, 1bxi, which suggested fully anisotropic diffusion with an effective isotropic diffusion coefficient of 1.832 × 107 s-1 and a predicted diffusion anisotropy D||/D┴ of 0.847.
The general pattern of models selected for each form of the E9 DNase from the Model free analysis is unremarkable, with the large number of model 1 and 2 fits consistent with a relatively rigid protein backbone, and the numbers of model 3 and 4 fits indicative of slower dynamics, in line with the broad range of proteins. Model 5 fits are mainly seen at the termini, for which greater local dynamics is to be expected, in agreement with the qualitative interpretation of the R2/R1 ratios. Striking differences were seen, however, in the models selected for individual residues in the metal-free and Zn2+ containing E9 DNase, both in terms of the low correlation coefficient for the model selection, of less than 0.5, and the actual numbers having the same model with only 60 out of 91 residues common to the metal-free and Zn2+ containing E9 DNase having common models. This is all the more emphatic given the classification into broad categories of fast motion, exchange and slow motion. Within the exchange category there is consensus for only 5 residues; 34, 56, 59, 83 and 129. Note that the major-minor conformational exchange will not give rise to resonances falling within the exchange category because the rate of exchange for this process is too slow at 288 K. Both the metal-free and Zn2+ containing E9 DNase show a cluster of residues undergoing exchange around 56 to 59, while the metal-free form has another cluster around residues 79 to 84. In general the Rex terms are small, less than 3 s-1, and do not show significant difference between the forms. Caution must be attached to the significance of these smaller Rex terms since these may well be spurious, arising because Rex contributions only affect R2 in the fitting process. The one exception to this is Ile 107 with a value of 24.8 s-1 for the Zn2+-containing E9 DNase. Examination of the NMR spectra confirms this is not an artefact as the resonance appears broad and the {1H}-15N NOE is low at 0.63. Note Ile 107 lies close to the metal binding site and therefore does not give evidence for more wide ranging conformational effects on metal binding. A similar picture of low agreement is seen for the slow motions because, although in both the metal-free and Zn2+ containing E9 DNase there is a strong consensus at the termini, outside of these regions there is little commonality. The Zn2+ containing form shows a small cluster at residues 76-79 with the other residues isolated, while the metal-free form shows a small cluster at residues 72 and 74. Consequently the effect of zinc binding on the dynamics within the E9 DNase is complex and cannot be reduced to a simple increase in rigidity.
Confirmation of this is provided by the order parameters, S2. For both the metal free and Zn2+ containing E9 DNase the values for S2 are consistently high, in the range 0.8-1.0, indicative of a rigid backbone. Excluding the termini the average S2 is 0.903 ± 0.070 for metal-free E9 DNase, and 0.924 ± 0.044 for Zn2+ containing E9 DNase. One region, characterized by a series of residues having an S2 more than one standard deviation less than the mean, is seen for both metal-free and Zn2+-containing E9 DNase, namely residues 18-21. At the termini, the order parameter does drop significantly below 0.8, and this is associated with the flexibility of the ends of the protein chain. In contrast to the model selection, very good correlation is seen for S2 between the metalfree and Zn2+ containing E9 DNase with good agreement in the actual values, which have a mean difference of only -0.027 or 5.2%. Any possible mis-assignment of the motional model into an exchange category will not affect this conclusion because on average S2 is decreased by only 0.03 when an exchange model is used. Furthermore an examination of the residues involved shows that overall the difference in S2 between the metal free and Zn2+ containing forms becomes smaller when the residues with a small Rex term are fitted to models 1 or 2. The absence of significant differences in the model free parameters is not unusual; for example, a comparison of a thermophilic and mesophilic ribonuclease HI enzyme showed a similar poor agreement in the model selection, 55 out of 110 the same, and yet the correlation co-efficients between the derived parameters were high with a correlation coefficient of 0.92 [57]. No significant differences are seen in the order parameters for the different helices in the metal-free and Zn2+ containing E9 DNase, consistent with the chemical shift analysis indicating metal binding does not affect the conformation of the protein core. Taken together the mean and mean differences in S2 values for the NH vectors show some weak evidence for a minor general constriction in the dynamics of the E9 DNase on binding to zinc. In the variation of τf,s with residue, the most significant differences are apparent in the region of residues 76-79 and 107-111, where slower local dynamics are seen in the case of the Zn2+-containing E9 DNase and metal-free protein, respectively.
A direct comparison of the dynamics seen by the time-resolved fluorescence anisotropy decay and NMR measurements is possible through the NMR relaxation behaviour of the indole Nε which, in contrast to the backbone NH, reports directly on the side-chain indole ring dynamics like the fluorescence measurements. In case of the metalfree E9 DNase Trp22 the spatial extent of the local dynamics seen by the two techniques is rather similar, an angular displacement of 16° by fluorescence implying an order parameter of 0.887 and by NMR an order parameter of 0.929. Although the correlation times derived for the local component are quite different, 1 ps or less by NMR and 1.39 ns by fluorescence, nevertheless, the important consideration is that the motion of the indole side chain is heavily constrained. Furthermore, similar agreement between the two techniques is seen for the dynamics of Trp22 in the Zn2+ containing E9 DNase. Both reports a further reduction in the mobility of the sidechain, indeed no local motion is seen by fluorescence while by NMR the order parameter is 0.983. Consequently the NMR results support the conclusion reached using fluorescence measurements, namely that zinc binding constrains the dynamics of Trp22 itself. Despite this agreement the presence of more widespread ligand-induced conformational changes is not supported because only small changes are seen in the peptide backbone order parameters.
A doubling of the rotational diffusion rate is seen with temperature, consistent with the factor of 2.2 expected on the basis of the change in solution viscosity and temperature. Although, the anisotropy of the rotational diffusion tensor is largely maintained, the orientation of the tensor is different implying some change in the protein-solvent interactions and hence the manner in which the protein tumbles in solution. A similar pattern of models is seen for the local dynamics in the Zn2+ containing E9 DNase at the two temperatures, the majority being model 1 or 2, corresponding to picosecond motions. Somewhat more model 3 fits are seen at higher temperature, but for the most part these involve a Rex of less than 1 s-1. Indeed apart from the fast exchanging Ile 107 at low temperature which is too broad at 319 K to be seen, and Val 98 there is no evidence for an increasing exchange rate at higher temperature for those residues identified as having an exchange contribution in the lower temperature data. Thus it is tempting to see the small exchange rate contributions as an artefact arising from a default fit to higher R2 values. Those residues for which the majorminor conformational equilibrium might have given an Rex term because of the increased rate of exchange at higher temperature are not seen to do so because, as noted earlier, they are broadened extensively sometimes to the extent of disappearing altogether. A paired Students t-test of the average differences in S2 at the two temperatures reveals that the mean difference (S2288K-S2319K) of 0.011 does not differ from zero at the 95% confidence level (t=0.972, n=65) [58]. Thus although the dynamics are less constrained at higher temperatures, the effect is very small. Consequently, the effects of binding zinc on the dynamics of the E9 DNase are too small to be revealed fully by these experiments.
Molecular dynamics
The results seen for the molecular dynamics simulations in terms of the radius of gyration and sequence dependent Cα rms fluctuations are consistent with previous simulations [25]. In particular the radius of gyration was stable over the 100 ns run and common regions showing increased rms fluctuations for the backbone Cα, namely 20-30, 65- 75, 90-95 and 105-115. These same regions of increased flexibility are reflected in the NH order parameters S2 providing a consistent picture of the E9 DNase. Therefore it is likely that the intrinsic flexibility of the E9 DNase and its response to binding the metal ion is being properly represented by the simulations. Turning to the dynamics around the two Trp noted in the fluorescence measurements, Trp22 and Trp58. First, there is no evidence from the radius of gyration that the metal bound forms 1bxi and 1sfj show any changes in the compactness of the protein as proposed on the basis of the fluorescence measurements. Second, there is little evidence for a conformational change propagated from Trp22 towards Trp58 as a consequence of metal binding. For Trp22 looking at the NH S2 parameter, there is no evidence that the metal containing structures 1bxi and 1fsj have more restricted backbone dynamics than the metal free structure. While for Trp58 there is some evidence to suggest increased flexibility in the metal free form, 1 emv. Consequently as with the NMR results whatever changes are being seen in the side chain dynamics by the fluorescence measurements are not reflected in the backbone dynamics.
Enhanced stability of Zn2+-containing E9 DNase
Attempts to seek a role for dynamics in the increased stability of the Zn2+- containing E9 DNase may well be misplaced because the extra thermal stability can be understood in terms of the enthalpy of zinc binding within the framework of the following energy cycle:
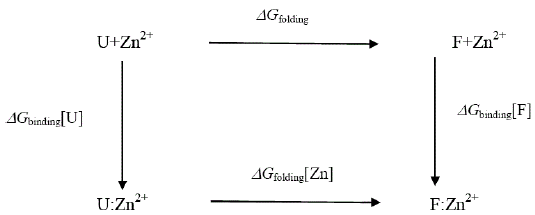
where U represents the unfolded E9 DNase and F the folded E9 DNase. Overall
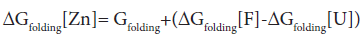
In the absence of complex formation between the unfolded E9 DNase and zinc this reduces to
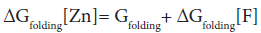
Isothermal titration calorimetry yields a value of -80.9 kJ mol-1 for ΔHbinding[F] at 298 K. This is consistent with the values seen for zinc binding in carbonic anhydrase II where the zinc similarly binds to three histidine ligands. Importantly the enthalpy changes in the case of carbonic anhydrase II are attributable to metal desolvation and the formation of metal-histidine bonds, not widespread conformational changes [59,60]. A good estimate for ΔGbinding[F] cannot be made because only an upper limit of 1 × 10-9 M can be placed on the equilibrium constant for the dissociation of the Zn2+ containing E9 DNase complex by calorimetry Pommer et al. [16]. Despite this uncertainty in ΔGbinding[F] the enthalpy of binding suggests ΔSbinding[F] must be negative because even in the event of a picomolar equilibrium constant, ΔSbinding[F] ≈ -50 J mol-1 K-1. A negative binding entropy is indicative of structural re-organization involving either solvent, the protein or both during zinc binding [57,58]. The observed changes in 15N chemical shift are consistent with a small re-organization in the protein limited to the Zn2+ binding site. Folding itself is entropically unfavourable, a rough estimate of ΔSbinding=-2.77 kJ mol-1 K-1 can be made on the basis of an average of -20.7 J mol-1 K-1 per residue [61,62]. This contribution will dominate the temperature dependence of ΔGfolding[Zn] through TΔS, with the binding entropy adding no more than 4% and in such a manner as to decrease Tm. Consequently ΔSbinding[F] can be neglected. To a first approximation then the increase in Tm must lie in the favourable ΔHbinding[F]. The stabilization by 80.9 kJmol-1 can be estimated as being equivalent to an increase in Tm of 29.2 K for the E9 DNase on binding zinc, assuming the enthalpy terms are temperature independent. Thus the increase in thermal stability of the Zn2+ containing DNase can be understood in terms of the enthalpy changes associated with metal desolvation and the formation of zinc- histidine bonds.
Conclusion
15N relaxation times and heteronuclear nOe have been used to study the dynamics of the E9-DNase domain and to examine changes in the dynamics on binding the metal ion Zn2+. The NMR measurements complement earlier fluorescence ones by providing a more comprehensive picture of the dynamics as well as looking specifically at the backbone of the wild type protein rather than side-chain dynamics of a mutant protein. A comparison of the raw data revealed only subtle differences between the metal-free and zinc containing forms. Chemical shifts indicated small differences between the solution structure for the E9 DNase and the crystal form of the Im9 bound E9 DNase. In particular, the binding of zinc caused dramatic changes in the 15N chemical shift of residues close to the metal binding side, notably Val 98. Yet despite this sensitivity, overall the zinc binding induced shifts were small indicating rather similar structures for the zinc-containing and metal-free forms in contrast to the fluorescence measurements. Model free analysis of the relaxation data gave an overall view of the dynamics in the metal-free E9 DNase as a slightly anisotropic rotational diffusion with a rigid backbone undergoing relatively complex motions. Globally the dynamics were in line with expectations based on the crystal structure and hydrodynamic calculations. Major differences were seen between the metal-free and Zn2+ containing form in terms of the local dynamics with a significant number of the NH involved in conformational exchange. Unlike the fluorescence measurements no evidence was found for local nanosecond motions for Trp22 in the metal-free E9 DNase. On metal binding a smaller decrease was seen in the correlation time for rotational diffusion than by fluorescence, however, the conclusion that the E9 DNase is more compact is not so clear cut since any surface perturbations caused by altering the solvent interactions need to be considered. Changes in the order parameters for the indole Nε support the idea that zinc binding immobilizes Trp22. The Zn2+ E9 DNase is characterized by the core of residues which were stable over a range of temperature as revealed by their chemical shifts and dynamics. Overall the conformation and dynamics of the E9 DNase are little affected in the folded state by zinc binding. The dramatic effect of Zn2+ on the stability of the protein can be understood in terms of the changes in the metal solvation and the formation of metal-histidine bonds.
Supporting Information Available
R1, R2, NOE of the metal-free, Zn2+- containing E9 DNase at 288 K and the Zn2+- containing E9 DNase at 319 K. Model free data for the Zn2+- containing E9 DNase at 319 K.
Acknowledgements
We thank the BBSRC for a research studentship (ESC) and the Wellcome Trust for its support of colicin NMR studies.
References
- James R, Penfold CN, Moore GR, Kleanthous C (2002) Killing of E coli cells by E group nuclease colicins.Biochimie84: 381-389.
- Papadakos G, Wojdyla JA, Kleanthous C (2012) Nuclease colicins and their immunity proteins. Quarterly Rev. Biophysics 45: 57-103.
- Cheng YS, Hsia KC, Doudeva LG, Chak KF, Yuan HS (2002) The crystal structure of the nuclease domain of colicin E7 suggests a mechanism for binding to double-stranded DNA by the H-N-H endonucleases.J Mol Biol 324: 227-236.
- Kühlmann UC, Pommer AJ, Moore GR, James R, Kleanthous C,et al.(2000) Specificity in Protein-Protein Interactions: The structural basis for dual recognition in endonuclease colicin-immunity protein complexes. J Mol Biol 301:1163-1178.
- Kleanthous C, Kühlmann U, Pommer AJ, Ferguson N, Radford SE, et al. (1999) Structural and mechanistic basis of immunity toward endonuclease colicins. Nature StructBiol 6: 243-252.
- Ko TP, Liao CC, Ku WY, Chak KF, Yuan HS, et al.(1999) The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure 7: 91-102.
- Sui MJ, Tsai LC,Hsia KC, Doudeva LG, Ku WY,et al. (2002) Metal ions and phosphate binding in the H-N-H motif: crystal structures of the nuclease domain of ColE7/Im7 in complex with a phosphate ion and different divalent metal ions. Protein Sci 11: 2947-2957.
- Whittaker SBM,Boetzel R, MacDonald C, Lian LY, Pommer AJ,et al.(1998)NMR detection of slow conformational dynamics in an endonuclease toxin. J Biomol NMR12: 145-159.
- Whittaker SBM, Czisch M, Wechselberger R, Kaptein R, Hemmings AM, et al. (2000)Slow conformational dynamics of an endonuclease persist in its complex with its natural protein inhibitor. Protein Sci9: 713-720.
- Van den Bremer ETJ, Jiskoot W, James R, Moore GR, Kleanthous C, et al. (2002)Probing metal ion binding and conformational properties of the colicin E9 endonuclease by electrospray ionization time-of-flight mass spectrometry. Protein Sci 11: 1738-1752.
- Van den Bremer ETJ, Keeble AH, Jiskoot W, Spelbrink RE, Maier CS,et al. (2004) Distinct conformational stability and functional activity of four highly homologous endonuclease colicins. Protein Sci13: 1391-1401.
- Keeble AH, Hemmings AM, James R, Moore GR, Kleanthous C, et al.(2002) Multistep binding of transition metals to the H-N-H endonuclease toxin colicin E9. Biochemistry41: 10234-10244.
- Galburt EA, Chevalier B, Jurica MS, Flick KE, Monnat RJ, et al. (1999) A novel endonuclease mechanism directly visualized for I-PpoI.Nat StructBiol 6: 1096-1099.
- Ku WY, Liu YW, Hsu YC, Liao CC, Liang PH, et al. (2002) The zinc ion in the HNH motif of the endonuclease domain of colicin E7 is not required for DNA binding but is essential for DNA hydrolysis. Nucleic Acids Res30: 1670-1678.
- Van den Bremer ETJ, Keeble AH, Visser AJWG, van Hoek A, Kleanthous C, et al. (2004)Ligand-induced changes in the conformational dynamics of a bacterial cytotoxic endonuclease.Biochemistry 43: 4347-4355.
- Pommer AJ, Kühlmann UC, Cooper A, Hemmings AM, Moore GR, et al. (1999) Homing in on the role of transition metals in the HNH motif of colicin endonucleases. J Biological Chem274: 27153-27160.
- Van den Bremer ETJ, Keeble AH, Kleanthous C, Heck AJR (2005) Metal induced selectivity in phosphate ion binding in E9 DNase. ChemCommun (Camb) 7: 1137-1139.
- Hannan JP, Whittaker SBM, Davy SL, Kühlmann UC, Pommer AJ, et al. (1999) NMR study of Ni2+ binding to the H-N-H endonuclease domain of colicin E9.Protein Sci 8: 1711-1713.
- Hannan JP, Whittaker SBM, Hemmings AM, James R, Kleanthous C, et al. (2000)NMR studies of metal ion binding to the zinc finger-like HNH motif of colicin E9.J Inorg Biochemistry79: 365-370.
- Henzl MT, Wycoff WG, Larson JD, Likos JJ (2002) 15N nuclear magnetic resonance relaxation studies on rat beta-parvalbumin and the pentacarboxylate variants, S55D and G98D. Protein Sci11: 158-173.
- Palmer AG (2004)NMR characterization of the dynamics of biomacromolecules. Chem Rev104: 3623-3640.
- Villanueva J, Hoshino M, Katou H, Kardos J, Hasegawa K, et al. (2004) Increase in the conformational flexibility of beta(2)-microglobulin upon copper binding: A possible role for copper in dialysis-related amyloidosis. Protein Sci13: 797-809.
- Dodson GG, Lane DP, Verma CS (2008) Molecular simulations of protein dynamics: New windows on mechanisms in biology. EMBO Reports 9: 144-150.
- Zwier MC, Chong LT (2010) Reaching biological timescales with all-atom molecular dynamics simulations. CurrOpinPharmacol10:745-752.
- Baron R, Sergio E, Wong SE, de Oliveira CAF, McCammon JA (2008) E9-Im9 colicinDNase -Immunity Protein Biomolecular Association in Water: A multiple-copy and accelerated molecular dynamics simulation study. J PhysChemB 112: 16802-16814.
- Koller AN, Schwalbe H, Gohlke H (2008)Starting structure dependence of NMR order parameters derived from MD simulations: implications for judging force-field quality. Biophys J 95: L04-L06.
- Huntley JJA, Scrofani SDB, Osborne MJ, Wright PE, Dyson HJ (2000)Dynamics of the metallo-b-lactamase fromBacteroidesfragilis in the presence and absence of a tight-binding inhibitor.Biochemistry 39: 13356-13364.
- Wallis R, Reilly A, Barnes K, Abell C, Campbell DG, et al. (1994) Tandem overproduction and characterisation of the nuclease domain of colicin E9 and its cognate inhibitor protein Im9.Eur J Biochem220: 447-454.
- Garinot-Schneider C, Pommer AJ, Moore GR, Kleanthous C, James R(1996)Identification of putative active-site residues in the DNase domain of colicin E9 by random mutagenesis. J Mol Biol 260: 731-742.
- Whittaker SBM,Boetzel R, MacDonald C, Lian LY, James R, et al. (1999)Assignment of 1H, 13C and 15N signals of the DNase domain of colicin E9. J Biomol NMR 14: 201-202.
- Kay LE, Keifer P, Saarinen T (1992)Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am ChemSoc 114: 10663-10665.
- Kay LE, Nicholson LK, Delaglio F, Bax A, Torchia DA (1992)Pulse sequences for removal of the effects of cross correlation between dipolar and chemical-shift anisotropy relaxation mechanisms on the measurement of heteronuclear T1 and T2 values in proteins.J MagnReson97: 359-375.
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM,et al. (1994) Backbone dynamics of a free and a phosphopeptide-complexedSrc homology 2 domain studied by 15N NMR relaxation. Biochemistry 33: 5984-6003.
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, et al. (1995) NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277-293.
- Han B, Liu Y, Ginzinger SW, WishartDS (2011) SHIFTX2: significantly improved protein chemical shift prediction. J Biomol NMR 50: 43-57.
- Xu XP, Case DA (2001) Automated prediction of 15N, 13Calpha, 13Cbeta and 13C' chemical shifts in proteins using a density functional database. J Biomol NMR 21: 321-333.
- Lipari G, Szabo A (1982) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules.1.Theory and range of validity.J Am ChemSoc104: 4546-4559.
- Lipari G, Szabo A (1982) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules.2.Analysis of experimental results. J Am ChemSoc104: 4559-4570.
- Mandel AM, Akke M, Palmer AG (1995)Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol246: 144-163.
- Abragam A (1961) The Principles of Nuclear Magnetism. Clarendon Press, Oxford, UK.
- Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, et al.(1990) Deviations from the simple two-parameter model-free approach to the interpretation of 15N nuclear magnetic relaxation of proteins.J Am ChemSoc112: 4989-4991.
- Tjandra N, Feller SE, Pastor RW, Bax A (1995)Rotational diffusion anisotropy of human ubiquitin from 15N NMR relaxation. J Am ChemSoc117: 12562-12566.
- Pawley NH, Wang CY, Koide S, Nicholson LK (2001) An improved method for distinguishing between anisotropic tumbling and chemical exchange in analysis of 15N relaxation parameters. J Biomol NMR 20: 149-165.
- Vander Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, et al. (2005) GROMACS: Fast, flexible, and free. J ComputChem 26: 1701-1718.
- d'Auvergne EJ, Gooley PR (2003) The use of model selection in the model-free analysis of protein dynamics. J Biomol NMR 25: 25-39.
- Chen J, Brooks CL III, Wright PE (2004)Model-free analysis of protein dynamics: Assessment of accuracy and model selection protocols based on molecular dynamics simulation. J Biomol NMR29: 243-257.
- de Dios AC, Pearson JG, Oldfield E (1993) Secondary and tertiary structural effects on protein NMR chemical shifts: an ab initio approach. Science260: 1491-1496.
- Le H, Oldfield E (1996) Ab Initio Studies of Amide-15N Chemical Shifts in Dipeptides: Applications to Protein NMR Spectroscopy. J PhysChem100: 16423-16428.
- Case DA (2013) Chemical shifts in biomolecules. CurrOpinStructBiol 23: 172-176.
- Biekofsky RR, Turjanski AG, Estrin DA, Feeney J, Pastore A (2004)Abinitio study of NMR 15N chemical shift differences induced by Ca2+binding to EF-hand proteins. Biochemistry43: 6554-6564.
- Zhang H, Neal S, Wishart DS (2003) RefDB: A database of uniformly referenced protein chemical shifts.J Biomol NMR 25: 173-195.
- Bougault CM, Eidsness MK, Prestegard JH (2003) Hydrogen Bonds in Rubredoxins from Mesophilic and Hyperthermophilic Organisms. Biochemistry 42: 4357-4372.
- Grathwohl C, Wüthrich K (1981)NMR studies of the rates of prolinecis-trans isomerization in oligopeptides.Biopolymers 20: 2623-2633.
- Campbell ID, Dobson CM, Moore GR, Perkins SJ, Williams RJP (1976)Temperature dependent molecular motion of a tyrosine residue of ferrocytochrome C. FEBS Lett70: 96-100.
- Damberg P,Jarvet J, Allard P, Mets Ü, Rigler R, et al. (2002) 13C-1H NMR relaxation and fluorescence anisotropy decay study of tyrosine dynamics in motilin.Biophys J 83: 2812-2825.
- Garcia de la Torre JG (2001)Hydration from hydrodynamics. General considerations and applications of bead modelling to globular proteins. BiophysChem 93: 159-170.
- Butterwick JA, Loria JP, AstrofNS, Kroenke CD, Cole R, et al. (2004)Multipletime scale backbone dynamics of homologous thermophilic and mesophilicribonucleaseHI enzymes. J Mol Biol 339: 855-871.
- Devore JL (1999) Probability and statistics for engineering and the sciences.(5th Edn), Duxbury, Pacific Grove, USA.
- DiTusa CA, Christensen T, McCall KA, Fierke CA (2001) Thermodynamics of metal ion binding. 1. metal ion binding by wild-type carbonic anhydrase. Biochemistry 40: 5338-5344.
- DiTusa CA, McCall KA, Christensen T,Mahapatro M, Fierke CA,et al. (2001) Thermodynamics of metal ion binding. 2. metal ion binding by carbonic anhydrase variants. Biochemistry 40: 5345-5351.
- Murphy KP, Xie D, Garcia KC, Amzel LM, Freire E (1993) Structural energetics of peptide recognition: Angiotensinll/antibody binding. Proteins StructFunc Genet 15: 113-120.
- Makhatadze GI, Privalov PL (1996) On the entropy of protein folding. Protein Sci5: 507-510.
Citation: Clayden NJ, Moore GR, Collins ES, Boetzel R (2017) Conformational Dynamics in the Endonuclease Domain of Colicin E. Biopolymers Res 1: 103.
Copyright: © 2017 Clayden NJ, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 5264
- [From(publication date): 0-2017 - Dec 19, 2025]
- Breakdown by view type
- HTML page views: 4176
- PDF downloads: 1088