Research Article Open Access
Culture-Independent Analysis of Bacterial Diversity during Bioremediationof Soil Contaminated with a Diesel-Biodiesel Blend (B10)S
| Luisa WM1, Letícia T1, Francielle B1, Raquel D2, Patricia Dörr de Q1, Kateryna Z3, Jennifer D2, Robson A4, Eric WT2, Ana Paul GF5, Flavio AOC1 and Fatima MB1* | |
| 1Department of Microbiology, Immunology and Parasitology Federal University of Rio Grande do Sul, 500, Sarmento Leite Street, Porto Alegre, 90050-170, RS, Brazil | |
| 2Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, 1355 Museum Road, Gainesville, FL 32611-0700, USA | |
| 3Department of Ecology, Earth Sciences Division, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Mail Stop 70A3317 Berkeley, CA 94720, USA | |
| 4Center of Engineering, Federal University of Pelotas, 1734 Almirante Barroso, Pelotas, 96010-280, RS, Brazil. | |
| 5Department of Soil Science, Federal University of Rio Grande do Sul, Bento Gonçalves Avenue, 7712, Porto Alegre, 90040-000, Rio Grande do Sul, Brazil | |
| Corresponding Author : | Fatima MB Department of Microbiology, Immunology and Parasitology Federal University of Rio Grande do Sul 500, Sarmento Leite Street, Porto Alegre, 90050-170, RS, Brazil E-mail: fatima.bento@ufrgs.br |
| Received September 16, 2015; Accepted October 07, 2015; Published October 10, 2015 | |
| Citation: Luisa WM, Letícia T, Francielle B, Raquel D, Patricia Dörr de Q, et al. (2015) Culture-Independent Analysis of Bacterial Diversity during Bioremediation of Soil Contaminated with a Diesel-Biodiesel Blend (B10)S. J Bioremed Biodeg 6:318. doi:10.4172/2155-6199.1000318 | |
| Copyright: © 2015 Luisa WM. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
| Related article at Pubmed, Scholar Google | |
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
This study evaluated and compared the degradation of a B10 blend (90% diesel: 10% biodiesel) by native (autochthonous) soil bacteria and exogenous (allochthonous) bacteria. This experiment simulated a surface spill followed by different methods of bioremediation: natural attenuation, bioaugmentation with autochthonous or exogenous bacteria, and biostimulation. The bioremediation process in soil contaminated with B10 (at a rate of 36 g of total petroleum hydrocarbon (TPH) kg-1 of soil) was evaluated for 28 days and analyzed by chromatography (degradation). The heterotrophic and degrading population and fuel mineralization (respirometry) were estimated. The abundance, composition, and diversity of the microbial community resulting from each treatment method were assessed with an ultra-high-throughput sequencing system (Illumina HiSeq). Samples were analyzed at three time points: 1, 15 and 28 days after the contamination. The natural attenuation strategy reduced TPHs by 19%, which suggests a degradation capability of the autochthonous microbial population even when not previously exposed to the contaminant. This genetic feature of the autochthonous population may be due to TPH-degrading plasmids and operons. In bioaugmentation with autochthonous and exogenous bacteria strategy, TPH degradation was similar to that in the other treatments. Proteobacteria, Firmicutes, Acidobacteria, Actinobacteria, Bacteroidetes, and Armatimonadetes were the most abundant phyla post remediation. Natural attenuation presented the highest Fisher’s [alpha] diversity index (at the genus level) at the 28th day post-spill.
| Introduction |
| Diesel oil is one of the most commonly used fuels in Brazil and worldwide. Diesel oil leakages from underground storage tanks, distribution facilities, and various industrial operations represent an important source of soil and aquifer contamination [1-3]. In Brazil, diesel has been blended with biodiesel since 2008, and currently it is mandated that biodiesel comprise 7% of the blend (B7), with the biodiesel contribution expected to increase over time. Biodegradability studies on these blends are important for risk simulations of potential environmental spills. Diesel is a complex mixture consisting essentially of aliphatic and aromatic hydrocarbons obtained from fractional distillation of petroleum refining [4]. Biodiesel can be produced from a variety of sources such as vegetable oils and fats (tallow) and is most commonly obtained by transesterification. Due to its composition of mainly methyl or ethyl esters of fatty acids, biodiesel has benefits over petroleum-derived oil in that engines burning biodiesel emit considerably lower levels of particulates, CO2, and volatile compounds [5]. Because biodiesel consists of natural compounds, it is considered more biodegradable than petroleum hydrocarbons (diesel) and promotes microbial growth [2-9]. Environmental agencies across the world are promoting the disposal of hydrocarbon residues at suitable sites and acceptable contamination levels. According to the Environmental Agency of South of Brazil, (FEPAM-Portaria N° 016/2010), areas exposed to more than 5 gKg-1 of soil with Total Petroleum Hydrocarbons (TPH) should be subjected to a decontamination treatment, and bioremediation is one of the recommended ecofriendly treatments. Bioremediation uses microbial populations to degrade fuel through in 3 ways: natural attenuation (native microbiota, with no stimulants), biostimulation (with supplying of nutrients, biosurfactants, oxygen, etc) to the native microflora, and bioaugmentation, introducing specific pre-selected microbiota, with degradation abilities. Enhancing the natural degradation potential of contaminated soils through bioremediation strategies represents an important aspect of environmental research [10]. |
| Degradation of hydrocarbons is often the result of the interaction of different microbial communities, and the potential of bioremediation depends on the ability of these organisms to adapt to new environmental conditions [11]. It has been shown that microorganisms with hydrocarbon- degrading genes are commonly present even in uncontaminated environments, but under normal circumstances, the relative abundance of these microorganisms may be marginal [9,10,12]. Studies on the effect of biodiesel on the microbial community have mainly focused on biodegradation of diesel/ biodiesel blends with particular emphasis on bioremediation [1-9,13]. Understanding how bioremediation influences the diversity of the soil microbial community is important to gain insights into the behaviour and functions of these populations [14]. |
| Among the culture-independent methods used to monitor changes in the microbial population during bioremediation, analysis of denaturing gradient gel electrophoresis (DGGE) of 16S rRNA has proven successful [2,3,6,14-17]. The analysis of amplified and sequenced 16S rRNA genes has become the most important approach to rapid identification and classification of microorganisms from DNA samples. Amplicons from high-throughput sequencing of DNA can generate many thousands of 16S rRNA sequences per sample to identify organisms and describe population structures in many oilimpacted environmental samples [18-20]. |
| In this study, high-throughput Illumina sequencing was used to examine the microbial community diversity and composition under different bioremediation strategies of B10-contaminated soil for 28 days. The heterotrophic and degradation populations were estimated by the Most Probable Number (MPN) technique, and B10-degradation was analyzed by respirometry and chromatographic analyses (TPH). |
| Methods |
| Soil: Composite soil samples from the surface layer (0-20 cm) were collected in an agricultural area of the Faculty of Agronomy, UFRGS, Porto Alegre, Brazil (30°06'91"S 51°14'27"). The soil was sieved (<2 mm), and dried at room temperature. The physicochemical analysis was performed at the Soil Analysis Laboratory of the Federal University of Rio Grande do Sul. The experimental soil is acidic (pH 5.0) with sandy texture (68% sand, 13% silt and 21% clay). The nutrients were quantified as follows: 0.93% organic-carbon; 0.07% total nitrogen (N); 1.6% organic matter; 60 mg dm-3 phosphorus (P); 81 mg dm-3 potassium (K); and 7.3 mg dm-3 sulfur (S). |
| Fuels: The blend B10 was prepared in the laboratory by mixing 50 ppm of sulfur diesel oil (B0) and biodiesel (B100) (blend with 70% soybean oil and 30% tallow oil). Both fuels were provided by Ipiranga Products of Petroleum S.A. The fuels were sterilized by filtration through a 0.22 μm pore size filter. Thereafter, the fuels were stored at room temperature in sterile glass, and protected from the light to avoid photo-oxidation. |
| Bioprospection and isolation: Soil contaminated with 50 g of TPH kg-1 [21] was used to isolate active microbial strains according to the enrichment procedure described by [12]. First, 10 g soil samples were added to flasks containing minimal mineral medium (MM1) (0.7 g L-1 KCl, 2.0 g L-1 KH2PO4, 3.0 g L-1 Na2HPO4, 1.0 g L-1 NH4NO3, and 1 mL of micronutrients solution containing 4.0 g L-1 MgSO4, 0.2 g L-1 FeSO4, 0.2 g L-1 MnCl2, and 0.2 g L-1 CaCl2) [21] plus 1% B10, with a final volume of 100 mL. Every seven days, 10 mL of the cell culture was transferred to a new medium, and after 21 days (3 transferences), the culture grown was serially diluted and subjected to surface spreading and streak plating on nutrient agar. Purified isolates were pre-selected for their ability to metabolize B10 at different concentrations as carbon source. The consortium selected for soil bioaugmentation assays consisted of three bacterial strains. The bacterial strains were identified based on partial sequencing of the region between residues 331 and 797 on the Escherichia coli 16S rRNA gene, using the primers 515 F 5’TCCTACGGGAGGCAG-CAGT 3’ and 806 R 5’ GGACTACCAGGGTATCTAATCCTGTT 3’, described by Nadkarni et al. [22]. The polymerase chain reactions (PCR) were optimized in 25 μL deionized sterile water containing 1 μL genomic DNA, 0.5 μL Taq DNA polymerase (5 U/μl) (Invitrogen), 2.5 μL buffer (Invitrogen, SG, Milanese, Italy), 1.5 μL MgCl2 50 mM, 1 μL deoxynucleotides (25 mM) and 1 μL of each primer (10 μM). Amplifications were performed using an automated thermal cycler (Techne TC 312) as follows: 40 cycles (30s at 94°C; 30s at 37ºC and 1 min at 72ºC) and a final extension step at 72°C for 5 min. The PCR products were analyzed with 1% agarose gel electrophoresis. The amplicons obtained were purified (PureLink® Genomic DNA Mini Kit, Invitrogen) and sequenced by DNA Analyser ABI 3730xl (Applied Biosystems). Nucleotide sequence similarity searches were conducted using GenBank nucleotide collection BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST) and deposited in the GenBank® genetic sequence database. |
| Bioremediation microcosms: The bioremediation strategies used in this project are summarized in Table 1. Experiments were carried out in triplicate, using 1.0 L hermetically sealed glass flasks containing 200 g of soil (microcosms) adjusted to moisture content of 70% of the field capacity. The pH level was adjusted to 7.0 by applying 2.1 g of CaCO3 kg-1 to the soil, and a solution of NH4NO3 and KH2PO4 at a C:N:P ratio of 100:10:1. Thereafter, the flasks were incubated for two days at room temperature (23 ± 7°C) before adding the fuel. The soils were then contaminated with a B10 blend by adding 45 mL of fuel per kilogram of humid soil, simulating a surface spill, at a contamination rate corresponding to 36 g of Total Petroleum Hydrocarbons (TPH) kg-1 of soil. During the 28 days of experimental analysis, the systems were maintained at controlled temperature (28 ± 1°C) and lightprotected. |
| Bacterial inoculum for Bioaugmentation: Bacteria isolated by [2] were used for bioaugmentation with exogenous bacteria. For bioaugmentation with authochthnous bacteria, bacteria were isolated as described in section 2.3. To prepare bacterial inoculum, we used 200 mL of sterile nutrient broth incubated for 24 h at 30°C and 190 rpm. The cells were centrifuged at 9,000 rpm for 10 min at 4°C. Subsequently, the cell extract was re-suspended in saline solution (0.85%) and incubated under shaking for 24 h at 100 rpm and 30°C to deplete energetic reserves (starvation). For standardization, the cell concentration of the bacterial isolates was determined with a spectrophotometer (Spectrumlab) at λ =600 nm. The microbial consortium (three strains) for bioaugmentation with autochthonous and exogenous bacteria (three strains) was obtained by mixing equal proportions of each bacterial strain (1.0×108 cells mL−1) [23]. |
| Indirect microbial growth: The total numbers of heterotrophic and degrading microorganisms were estimated using the MPN method in microtiter plates, according to the method described by [2]. The microbial population was determined using MPN tables [24]. The microbial population growth was monitored after 0, 8, 15, and 28 days. |
| Respiratory activity: To determine the metabolic activity in each microcosm, respirometric analysis was evaluated as cumulative CO2 release [2,3]. The carbon dioxide produced during microbial activity was captured with 0.5M NaOH solution (20 mL) in 50 mL flasks within the respirometric flasks. Periodically, the NaOH solution was replaced. To stop CO2 capture after the microcosms were opened, 2 mL of 30% BaCl2 solution was added. Four drops of 1% phenolphthalein indicator were added as color indicator for titration. The residual NaOH was titrated with 0.5 M HCl standardized solution. The carbonic gas produced was calculated using Eq.1: |
| Equation 1. C-CO2 generated (mg/Kg soil)=(VB – VA).(MC/2). MHCl. (FC/m) |
| Where: VB is the volume in mL of 0.5 M HCl used to titrate the control; VA the volume in mL of 1 M HCl used to titrate the treatment; MC the carbon molar mass in g/mol; MHCl the concentration in M of the standardized HCl solution; FC the correction factor for molarity (MHCl/ MNaOH) and m the dry soil mass in the flask in kg. |
| Fuel degradation |
| The degradation of B10 was evaluated by TPH (Total Hydrocarbons Petroleum) for each microcosm with soil. Two chromatographic analyses were carried out, after 0 and 28 days, of each bioremediation strategy, by analyzing hydrocarbons (C8 – C28). The procedures followed the terms established by the EPA method 8015 [12]. The results were compared to the initial fuel batches used in the biodegradation experiments. The degradation percentage was calculated according to Eq.2: |
| Equation 2. %TPH degradation=[(DTi−DTf)/ DTi]×100, |
| where DTi and DTf are the TPH values at the initial and the final time points, respectively. |
| TPH=Total Petroleum Hydrocarbons. |
| Microbial composition and diversity: Microbial diversity was analyzed by monitoring composition, abundance and diversity of the bacterial community during bioremediation. Samples were taken after 0, 15, and 28 days. |
| DNA extraction: Soil samples were collected at five equidistant points from inside each microcosm replicant; soil samples from three replicants were pooled, resulting in a total of 3 g. Genomic DNA was extracted from 0.5 g of each soil sample, using the PowerSoil DNA Isolation Kit (MoBio Inc.), following the manufacturer’s instructions. The purity of the extracted DNA was checked using a Nanodrop ND- 1000 spectrophotometer (Nanodrop Thermo Scientific, Wilmington, DE, USA) (260/280 nm ratio), and quantified by a Qubit® 2.0 fluorometer, using the dsDNA BR Assay kit (Invitrogen™), according to the manual. The DNA integrity was also confirmed by electrophoresis on 0.1% agarose gel with 1 X TAE buffer. |
| Illumina high-throughput sequencing: The bacterial and archaeal V4 regions of 16S rRNA genes were amplified using the universal prokaryotic primers 515F (5´-GTGCCAGCAGCCGCGGTAA-3´) and 806R (5´-GGACTACVSGGGTATCTAAT-3´) [25,26] with the addition of a barcoded sequence and the required Illumina adapters. The PCR was performed at an initial denaturation temperature of 94°C for 3 min, followed by 20 cycles at 94°C for 45 sec, at 53°C for 30 sec, and at 65°C for 90 sec. A final elongation step at 65°C was performed for 10 min. Sequencing was performed on a genome analyzer Illumina GAIIx (Illumina, Inc., CA, USA) with two paired-end read cycles of 101 bases each. The raw reads were separated according to their barcodes and the first seven bases corresponding to the barcode regions were trimmed using FASTX-Toolkit [27]. Paired reads were merged using a custom script (source available at https://github.com/Bioinfo- Tools/merge_ fastq_files.pl) . Reads were filtered for quality, based on a minimum percentage of bases with good quality of 70%, considering 20 as the minimum quality score in Phred + 33 encoding. The filtered reads were classified using the Basic Local Alignment Search Tool (BLAST) [28] checked against the ribosomal database SILVA (http://www.arb-silva. de) (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2175337/ ). For this step, reads were converted from FASTQ to FASTA format using FASTX-Toolkit. A parallel version of nucleotide BLAST, MPI- blastn, was used to minimize the execution time [29]. |
| Full taxonomic descriptions based on the SILVA database (http:// www.arb-silva.de ) were generated using a custom script (SILVAtaxcollector, source code available at: https://github.com/Bioinfo- Tools/SILVA-taxcollector). This tool consists of an adapted version of the algorithm introduced in NCBI-taxcollector [29], which includes an additional step before the taxonomic assignment. In this additional step, all sequence accession numbers from SILVA database were mapped with taxonomic identification numbers (TAXID) from NCBI taxonomy database [30].Sequence matches were classified at an 80% identity level for domain and phylum; 90% identity for class, order, and family; 95% identity for genus; and a 99% identity level for species. The total numbers of 16S rRNA classified sequences were converted into an OTU abundance matrix for each taxonomy level across the samples. Filtered abundances and OTU abundance matrix were generated using modified Megaclust and Megaclustable scripts (source code available at https://github.com/Bioinfo-Tools/PANGEA-plus ) [31]. |
| Nucleotide sequence accession numbers: The sequences obtained were deposited in the GenBank database with the following NCBI's SRA Study accession: SRP059871 |
| Statistical analysis: The results from respirometry were analyzed by ANOVA at a confidence level of 95%, and, when significant, the Tukey test was applied using program Statistica, version 7.0. Principal component analysis was used to determine the relationship between bioremediation strategies and OTU abundances. Spearman correlations for non-normally distributed data were used to independently evaluate the correlation of each bioremediation strategy with the relative abundance of phylum OTU. All statistical analyses were performed using XLSTAT-Pro 2014. The diversity of microbial community was estimated by the Fisher’s [alpha] diversity index (FDI) using the R package (R version 3.1.0, 2014). |
| Statistical analysis: The results from respirometry were analyzed by ANOVA at a confidence level of 95%, and, when significant, the Tukey test was applied using program Statistica, version 7.0. Principal component analysis was used to determine the relationship between bioremediation strategies and OTU abundances. Spearman correlations for non-normally distributed data were used to independently evaluate the correlation of each bioremediation strategy with the relative abundance of phylum OTU. All statistical analyses were performed using XLSTAT-Pro 2014. The diversity of microbial community was estimated by the Fisher’s [alpha] diversity index (FDI) using the R package (R version 3.1.0, 2014). |
| Results and Discussion |
| Bacterial identification for bioaugmentation: The bacterial strains from the consortium used in this study on bioaugmentation with autochthonous microorganisms were identified by analysis of the 16S rRNA sequences (Table 2). |
|
The identified species are gram-negative rod-shaped bacteria, commonly associated with the nitrogen cycle. They may be involved in the fixation of atmospheric N [32,33] or denitrification [34,35] and are often isolated from the soil rhizosphere [36]. Soil bacteria often have an inherent capacity for degradation of recalcitrant organic compounds [9,10,37]. The ability to tolerate high concentrations of the blend B10 in a liquid medium was observed in these microorganisms as well as the degradation ability in the soil at the intervention concentration of 36 g.TPHKg-1of soil) of B10. |
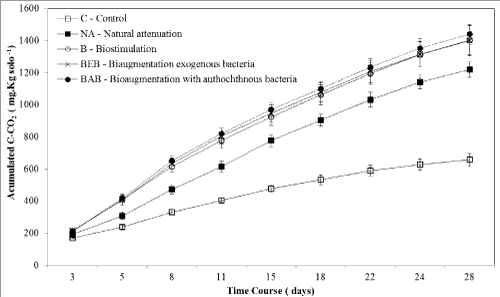
| Bioremediation strategies for blend B10 degradation: The bioremediation experiment was evaluated for 28 days by quantifying the heterotrophic populations and degraders (MPN estimate), CO2 release by respirometry, and degradation of blend B10 by gas chromatography microorganisms. The mineralization of blend B10 was assessed by quantification of C-CO2 released and cumulative over 28 days (Figure 1). |
| The values of cumulative C-CO2 production observed for all microcosms with B10 application were higher than in the control (soil without B10) over 28 days (p<0.01) (Figure 1). The values of C-CO2 in the microcosms of bioaugmentation and biostimulation were greater than the natural attenuation treatment (NA) and higher than in the control (p<0.01) after the 5th day. Among the strategies of bioaugmentation and biostimulation, no significant difference was observed in C- CO2 during the experiment indicating that nutrient addition was effective to stimulate native microorganisms. The production of C-CO2 microcosms compared with and without blend B10 suggests that the microorganisms were able to use the hydrocarbons and esters present in the mixture for growth. It is important to note that the bioremediation test was conducted at room temperature, which was not constant during incubation. On the 5th day, the temperature reached 40°C, and decreased to 30°C until the 15th day, remaining at 25°C until the 28th day. The temperature variation may have limited a more marked development of microbiota in all treatments. However, the release of C-CO2 from bioremediation treatments was greater than the negative control. This result suggests that the soil community was able to maintain and metabolize B10 despite being less active than in the preliminary bioremediation test. Alternatively, in a study on bioremediation of B20-contaminated soil, the authors found C-CO2 release rates of approximately 30 mg kg–1 of soil with maximum production on the 25th day [5]. Probably not only the type of contaminant, but other factors such as microbial metabolism and physicochemical soil profile may also have determined the behaviour observed in our study (Figure 1). |
| B10 degradation: The biodegradation of blend B10 was quantified by TPH at the end of the experiment. The results of degradation of different portions of diesel fuel showed that in all treatments, the most degraded fractions were in the range of C20-C40 (Table 3). For the BAB treatment (bioaugmentation with autochthonous bacteria), the reductions of TPH were highest than in all other treatments, for all TPH fractions analyzed (Table 3). However, no differences (p>0.05) were observed between TPH values of the bioremediation strategies. The results showed that all strategies stimulated metabolic activity suggesting that under appropriate abiotic conditions, the microbial metabolism of native (autochthonous) and exogenous (allochthonous) bacteria was stimulated to achieve similar levels of TPH degradation. For all bioremediation strategies, the cumulative C-CO2 was similar, indicating that the autochthonous bacterial species can potentially degrade B10. It was observed that autochthonous or exogenous bacteria plus nutrients supported and promoted hydrocarbon degradation indicating that in terms of pH, C: N ratio, and humidity, the environment had reached degradation-favorable conditions. The bioaugmentation strategy consisted of a single consortium application at the beginning of the treatment with autochthonous or exogenous microorganisms. The degradation in the microcosm with exogenous bacteria was lower than in the NA microcosm. In a study comparing degradation of autochthonous to degradation with exogenous bacteria, greatest degradation was observed when the consortium was inoculated into the soil of origin [12], similar to the results attained in this study. Some authors suggested that the introduction of inoculum immediately after contamination can have a negative impact on degradation rates [2,38,39]. Thus, the introduction of microbial cells in a community could also cause an imbalance in the resident microbiota and reduce the degradation of the contaminant by restoring the original population. |
| The results of hydrocarbon biodegradation showed that the fraction with higher degradation was in the range C20-C40 (Table 3). Some authors suggested that the C8-C11 fraction is more easily degraded [12,40-42]. Our evaluation also showed that the degradation percentage of the C8-C11 fraction was around 30% in all strategies. The microcosm with greatest reduction of the heavy fraction was BAB (47.8%) (p<0.05); however, no significant differences to the treatment with exogenous bacteria (BEB) (p>0.05) were detected (Table 3). |
| Bento FM et al. [12] evaluated bioremediation of dieselcontaminated soil, and observed degradation of approximately 70% of the C20-C40 fraction after 110 days of bioaugmentation with a consortium of autochthonous microorganisms. In this experiment, the highest percentage of TPH degradation occurred in the heaviest fraction of petroleum hydrocarbons (C20-C40). These results corroborate results of other authors [2,3,12], demonstrating that both autochthonous and exogenous soil microbiota can potentially use this fraction. In the treatment bioaugmentation with autochthonous bacteria, the percentage of TPH degradation was 24.6%. This degradation percentage was similar to that reported by Colla TS et al. [2] in bioremediation assays with a lower B10-contamination level (30 g TPH kg-1 soil), and at controlled temperature (28ºC). Silva G et al. [6] analyzed the biodegradability of B5, B20 and B50 in a contaminated soil during 60 days of incubation, and observed TPH degradation rates of 51, 80 and 62%, respectively. Li H et al. [43] evaluated the biodegradation of diesel fuel under increasing fuel concentrations for 110 days and found that the two highest concentrations (30 and 50 g Kg-1 of soil) resulted in lower rates of TPH removal (49.5% and 36.8%, respectively) from the systems. |
| The complexity of the fuel compounds requires a microbial community with different biodegradation capabilities to act synergistically in degrading the polluting compounds in order to gradually reduce environmental contamination [44]. The combination of certain microorganisms with a diverse metabolic profile should be capable of degrading the various components of diesel fuel. It is essential that the microbiota be able to tolerate and metabolize recalcitrant compounds, e.g., long-chain hydrocarbons, to prevent their accumulation in the environment. The approach that uses autochthonous microorganisms from the corresponding environment can optimize the decontamination process. Since potentially degrading microbial communities are ubiquitously present, increasing the population by bioaugmentation may be a way of accelerating remediation [9,10,12]. Even if preliminary tests identify specific bacteria that efficiently degrade biodiesel at high concentrations in a lab setting, there is no guarantee that the microbes will degrade as efficiently in a soil environment [45]. |
| In this study, the levels of hydrocarbon reduction were relatively low when compared to other soil bioremediation studies [2,3,6]. However, the degradation percentages were consistent with studies that examined bioremediation of highly hydrocarbon-contaminated (≥ 30 gkg−1) soils analyzed for a long period (110 days) [43]. According to Li H et al. [43], hydrocarbon degradation proceeds faster when the microbes are exposed to lower contamination levels (e.g., 500; 1,000; or 5,000 mg kg−1 of soil). |
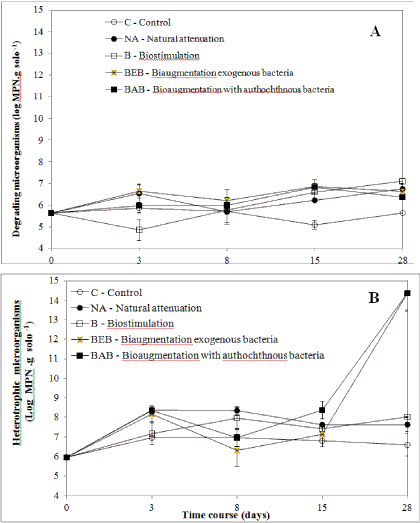
| Microbial estimate of heterotrophic and degradation microorganisms: Heterotrophic and degradation population’s growth were measured for all microcosms (Figure S1A and S1B), including the control, since the first 3 days post contamination. The increase of the microbial population in the microcosms with B10 added demonstrated that the initial TPH concentration (36 g kg−1 of soil) did not inhibit the population growth in all treatments. The BA, BEB, and NA treatments had increased in heterotrophic populations after 72 h, from 106 to 108 cells g-1 soil. This result was expected in BEB and BAB treatments, in which microorganisms with an initial concentration of 108 cells g-1 of soil were added. |
| After 72 hr, it was observed that the heterotrophic microbiota population increased about 100-fold, which suggests the use of fuel as carbon and energy source by the autochthonous community. |
| Except in the biostimulation (B) treatment, in which the population of degraders decreased in the first three days (Figure S1A,), an increase in population density was observed for most treatment groups. After this initial period, an increase in the microbial population of degraders in the B treatment group was also detected 28 days postcontamination. The population density in this treatment (B) was highest at the last sampling time (day 28th) (p<0.05). The population density in bioaugmentation treatments (BAB, BEB) increased initially and remained higher (p<0.05) than in the original soil. In the NA treatment, the population decreased on day 8, and thereafter, the concentration remained higher in the NA group than in the original soil as similarly observed in the bioaugmentation treatments on day 28. These results suggest that the autochthonous community, even though not previously exposed to contamination, showed adaptability and survival. Colla TS et al. [2] observed that during bioremediation of blend B10, the MPN of the microorganisms was able to metabolize the fuel, increasing from 1.0 × 103 (initial levels) to 1.0 × 106 MPN g−1 of soil. This result indicates that this microbial profile would be interesting to apply in an oil contamination scenario in view of the survival ability and adaptive capacity of endogenous microorganisms to toxic conditions. In microcosms of bioaugmentation with exogenous bacteria (BEB), the population density was higher than in the other microcosms in most samplings. In the BEB treatment, the population increased in the first three days, with small reductions on the 8th and 15th day, and after that increasing population until the end of the experiment (Figure S1A). The maintenance of the exogenous bacteria population density may be due to the introduction of bacteria that can potentially degrade contaminants previously selected and tested by Colla TS et al. [2]. |
| For the bioaugmentation treatment with autochthonous bacteria (BAB), the degrader population size increased in the first 15 days, with a slight reduction in density after 28 days. Thus, probably after inoculation with autochthonous consortia, the population size increased approximately 100 times and continued growing at a higher rate than the levels observed at the baseline (p<0.05). The addition of nutrients to the soil in the treatments B, BEB and BAB had a positive effect on the growth of the heterotrophic population. |
| In biostimulation treatments, the effect of adding nutrients to indigenous microbiota to degrade the mixture was evaluated. On the 15th day, the estimates of degrader populations were higher in the treatments BEB, BAB and B. Only on the 28th day, biostimulation reached a higher population density than both bioaugmentation (p<0.01) treatments. Thus, the incorporation of nutrients combined with the introduction of microorganisms may be a promising alternative for the bioremediation process, since the exogenous as well as autochthonous microbiota are benefited by the regulation of some environmental factors [2,38]. |
| Microbial community composition by ILLUMINA HiSeq analysis of 16S rRNA gene |
| A total of 1,816,406 raw reads of V6 region of the 16S rRNA gene amplicons were obtained by the ILLUMINA HiSeq platform from the soil samples under different bioremediation strategies after 15 and 28 days of incubation (Table S1 and S2). The number of high quality sequences obtained after sequence processing in each bioremediation strategy is presented in Table S2, Supporting Information,. Within the classified reads, a total of 50 phyla; 60 Class; 114 Order; 217 Family; 1.204 Genus and 876 species were found within the soil samples. |
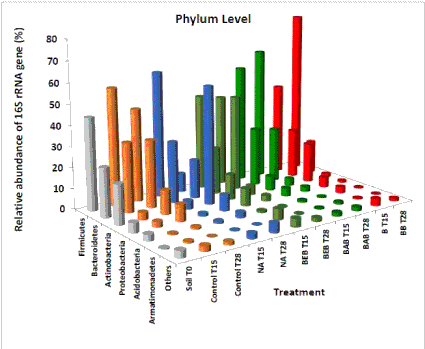
| The six most representative phyla (16S rRNA relative abundance >0.1% of total reads) Firmicutes, Bacteroidetes, Acidobacteria, Proteobacteria, Actinobacteria, and Armatimonadetes were determined (Figure 2). Among these, Firmicutes, Bacteroidetes and Actinobacteria were the most abundant, except in the NA treatment T28, in which Actinobacteria were most abundant, followed by Bacteroidetes and Firmicutes. In general, these three phyla might play an important role in hydrocarbon degradation. |
| The relative abundance of Archaea in these samples was very low during the bioremediation process. Less than 0.5% of the 1,044,698 high quality 16S rRNA gene sequences were classified as Archaea (Table S1). A study by Sutton NB et al. [19] examining dieselcontaminated soil after over 30 years in Poland showed a high relative abundance of Archaea, specifically of the phylum Euryarchaeota, in contaminated samples. According to the authors the high abundance of Euryarchaeota was much higher than what was found in clean soil samples. A study on archaeal diversity from 146 uncontaminated soil samples collected across six continents showed that the relative abundance of Archaea averaged 2% and was less than 16% in all sample [19]. The phylum Euryarchaeota have been observed to be the dominant archaeal phylum in heavily oil-contaminated environments [46]. Biases from DNA extraction, PCR amplification, primer choice, and sequencing, may confound the archaeal abundance found in these papers [46]. The primers used in this study are specific to the Archaea and have been used in the past so, it is unlikely that many archaeal taxa were missed [25,47]. Additionally, several bacterial and archaeal sequences remained taxonomically unresolved, indicating potentially novel microorganisms in this soil microcosms under bioremediation process. |
| We also reported a number of sequences that were related to uncultured microorganisms or that could not be classified, which might represent novel hydrocarbon degradation communities. Furthermore, shotgun metagenomic approaches do not suffer from most of the biases associated with culturing microorganisms and PCR-based methods since it involves direct sequencing of fragmented genomic DNA. |
| Control T28 had a much higher relative abudance of unclassified organisms (21.2%) compared to the Control T15 (3.2%) and Soil T0 (2.7%) samples. This increase of unclassified organisms was not observed in most of treatments except for the Natural attenuation T28, which had a 3-fold increase of uncultured organisms compared to Natural attenuation T15. The presence of unclassified organisms may represent unknown functions and interactions in the microbial community that may be missing in other treatments. Similar to the increase of unclassified organisms, natural attenuation was the only treatment with an increase in the relative abundance of Archaea (~19- fold, from 0.022 in Natural attenuation T15 to 0.411% in Natural attenuation T28). Archaea are known to be involved in the soil nitrification process through ammonia oxidation [48], which is an important component of the nitrogen cycle. |
| Firmicutes (approximately 60% of total sequences per sample), were the most dominant phyla in the bioremediation treatment samples. Proteobacteria, usually known as the most abundant phylum found in soils [49,50], seems to be less abundant in soil when contaminated with diesel and/or biodiesel. In a comparison over time (T0, T15-T28), the relative abundance of the phylum Firmicutes decreased with time in the treatments control and NA. On the other hand, the relative abundance of Firmicutes increased (from T15-T28) in the bio stimulation and bioaugmentation with autochthonous bacteria treatments. |
| Firmicutes, unlike the gram-negative Proteobacteria, consist of mainly gram-positive bacteria and have a low GC (guanine-cytosine) content. Many Firmicutes produce endospores that tend to resist adverse environmental settings (50). Thus, these microorganisms are often found to at least tolerate extreme conditions of temperature, radiation, desiccation or other abiotic factors. The Firmicutes are typically divided into the anaerobic Clostridia and the aerobic or facultative Bacilli. All Firmicutes detected in this study were associated with the family Clostridiaceae, which was the predominant family in all bioremediation treatments. The family Clostridiaceae includes recognized species of gram-positive, aerobic, spore-forming, rodshaped, non-motile bacteria, and both were isolated from soil [51,52]. |
| The phylum Bacteroidetes, the second most dominant in this study (approximately 30% of all sequences), only increased significantly in bioaugmentation with exogenous bacteria with increasing population size over time in relation to the initial population. Bacteroidetes were detected in clean (autochthonous community soil-control) and contaminated samples (other treatments). Roesch LF et al. [53] using pyrosequencing to study uncontaminated soils, identified Bacteriodetes as clearly predominant with a relative abundance from 15 to 25% in samples. A study by Sutton NB et al. [19] examining diesel-contaminated soil after over 30 years in Poland indicated that the phyla Firmicutes, Actinobacteria, and Acidobacteria were highly abundant while Bacteroidetes were not dominant in the study. Xu Z et al. [54] used 33 published available metagenomes from diverse soil sites (grassland, forest soil, desert, Arctic soil, and mangrove sediment) and showed that Firmicutes and Bacteroidetes, which are the two major most abundant phyla in the human microbiome, were not the most abundant in the evaluated soil microbial communities. Nacke H et al. [55] also used pyrosequencing to compare community structures of forests with grasslands, and placed Bacteriodetes in the rare-phylum group, with 1% overall relative abundance. |
| The relative abundance of Actinobacteria increased with time in the control treatment, but mostly under NA, from 4% to 55% of the total sequences. Actinobacteria was found to be the third most important bacterial phylum during B10-bioremediation. This group of microorganisms was easily detected in the sequences and associated with 12 different families of Actinobacteria. Actinobacteria are gram-positive aerobes with a high GC content (45%) in their DNA, a characteristic that distinguishes them from Firmicutes [49,56,57]. As a group, they tend to be widely distributed in nature and their representatives are known to survive for extended periods under adverse environmental conditions. Most of the other bacteria types detected were relatively less represented in the fuel microbial communities. |
| The phylum Proteobacteria was the fourth most abundant among the samples with Gamaproteobacteria and Alphaproteobacteria accounting, respectively, for 3.56 and 2.13% of the total reads. Betaproteobacteria and Desltaproteobacteria were less abundant with only 0.38 and 0.17% of total reads. Epsilonproteobacteria was least abundant in the class Proteobacteria with 0.04% of total reads. The abundance of Proteobacteria increased significantly with time in the treatments control and NA, and was dramatically reduced by bioaugmentation and biostimulation. The phylum Acidobacteria, although less abundant, had the same pattern as Proteobacteria, and with approximately 1% of abundance in all samples, increased to almost 6% of the sequences at the end of the treatment period with exogenous bacteria bioaugmentation. |
| The phyla Firmicutes, Actinobacteria, and Acidobacteria were studied previously in uncontaminated soil samples, and in contaminated soils with aliphatic or aromatic compounds, both were found in the polluted samples and the clean reference soil [19]. Previous amplicons analyses of the 16S rRNA gene showed that the bacterial phyla Acidobacteria, Actinobacteria, Bacteroidetes, and Proteobacteria are often abundant and ubiquitous in the soil [19]. |
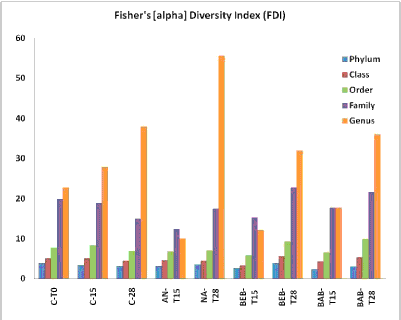
| The Fisher’s [alpha] diversity index (FDI) was calculated including the classified and non-classified operational taxonomic unit (OTU) to determine how microbial diversity is affected by different bioremediation strategies (Figure 3). To Phylum, Class, and Order level, it was not detected any significant difference. To all treatments, the Fisher’s [alpha] diversity index (FDI) increased with time (from T15 to T28) suggesting that the addition of blend B10 promoted the appearance of different groups, increasing the microbial diversity. The NA-T28 treatment presented the highest diversity index of all treatments at the 28th day. |
| During the bioremediation process by the studied strategies, the diversity of the microbial community seemed unaffected by the addition of 36 g TPHkg−1 of blend B10 for 28 days. In uncontaminated soil (control), the classification of taxonomic profiles was similar to the other bioremediation strategies tested with B10-contaminated soil. The effects of nutrient addition caused changes in the community structure after 15 and 28 days. Colla TS et al. and Quadros PD et al. [2,57] reported similar results. They stated that the population stabilized after 15 days in comparing profiles of microbial communities biostimulated with nutrient supplementation in the presence and absence of oil. They concluded that the changes resulted predominantly from the introduction of nutrients. The communities responded similarly to autochthonous and exogenous bacterial consortium addition. Community dynamics and structures were different from the initial conditions, but similar among the strategies. These results indicate that the additional inoculation caused no significant community change in relation to the control. |
| Many factors can affect microbial diversity in diesel-contaminated soils, e.g., carbon source availability, nutrients, and intrinsic ability of the microbial community to degrade petroleum hydrocarbons. In some cases, high TPH concentrations can be toxic and affect the C:N:P ratio. The increasing bacterial richness and diversity were probably due to the growth of diverse species on the degradable hydrocarbon fractions, or an increase in the number of ‘‘structures’’ (niches) in the system [58,59]. According to Li H et al. [43], oil (diesel oil) concentrations over 10 gkg−1 soil can significantly affect both soil microbial activity and community structure. Li H et al. [43] observed that the H index decreased slightly during the first 15 days of incubation and recovered to the control level on day 30. Aleer S et al. [60] investigated bioremediation effects on a microbial community from previously contaminated soil. Similarly to this study, in biodegradation studies of [2,3,14,43,60], they used soils without reports of pre-contamination by diesel oil. The presence of adapted microorganisms in naturally contaminated soil can result in the exclusion of an adaptation phase of remediation. Consequently, the biodiversity reduction in the initial period of the process would not have been detected if the Fisher alpha diversity index were not evaluated. |
| Responses of microbial community to bioremediation strategies |
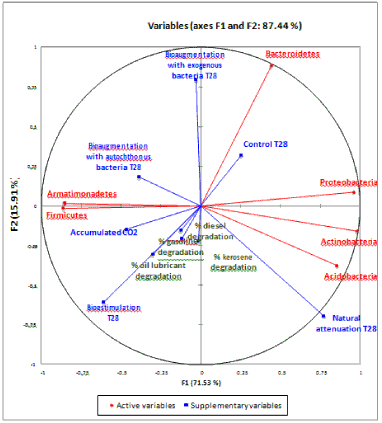
| The responses to the bioremediation strategies in the phyla of the microbial community were analyzed in this study by PCA (Figure 4). Natural attenuation was positively correlated with the abundance of Acidobacteria and Actinobacteria. It was observed that abundance of Firmicutes and Armatimonadetes had a positive correlation with cumulative C-CO2 released from the treatments bioestimulation and bioaumentation with autochthonous bacteria. After 28 days, there was no significant correlation between overall community compositions with the release of cumulative C-CO2 and different TPH fractions. Soil without contamination (control) and bioaugmentation with exogenous bacteria were positively correlated with Bacteroidetes phyla and negatively correlated with TPH contamination and bioestimulation (Figure 4). The PCA results showed differences between the tested strategies at the phylum level. By PCA, the strategies can be visualized in different quadrants, indicating a differentiated composition of the microbial communities. The occurrence of Firmicutes was highly correlated with abundance of Armatimonadetes, which may indicate some level of association between these phyla in hydrocarbon degradation. They were also correlated with CO2 production. The phylum Bacteroidetes correlated with bioaugmentation with exogenous bacteria treatment on the 28th day. The percentage of degradation of hydrocarbon compounds indicated some correlation with bioestimulation. Proteobacteria, Actinobacteria and Acidobacteria were significantly correlated, indicating some degree of association related to succession and co-metabolism. |
| In conclusion the total heterotrophic and degrading populations of microbial communities increased over time during bioremediation with different approaches of a B10 blend. The values of cumulative C-CO2 demonstrated no significant difference in soil microbial activity between biostimulation and bioaugmentation treatments. The TPH reduction (19%) by NA (with pH and moisture correction), suggests that the autochthonous microorganisms, even without previous exposition to the contaminant, are able to express degradation capability. Biostimulation and bioaugmentation with autochthonous and exogenous microorganisms had similar effects on biodegradation, reducing TPHs by 16; 24.5 and 14%, respectively. By Illumina sequencing, the soil community composition could be detected, showing different dynamics according to the treatments. Proteobacteria, Firmicutes, Acidobacteria, Actinobacteria, Bacteroidetes, Armatimonadetes were the most abundant phyla in all treatments. By all bioremediation strategies, the Fisher’s [alpha] diversity index (FDI) increased significantly after 28 days of the B10 addition. |
| *Supporting information: Tables listing percentage of total reads found in soil study (Table S1) and the number of raw reads and high-quality sequences for taxonomic groups (Table S2 and Figures S1A, S1B) Degrading (A) and Heterotrophic (B) microorganisms estimated by the Most Probable Number (MPN) in bioremediation of B10-contaminated soil. Fisher’s [alpha] diversity index to different taxonomic groups for each strategy of B10 remediation (Figure S2) and Principal component analysis (PCA) results between the tested strategies at the phylum level after 28 days of incubation (Figure S3). |
| Acknowledgements |
| The authors would like to thank Mr. Sérgio Luiz Viscardi and Dra. Roberta Miranda Teixeira from Ipiranga Products of Petroleum S.A, for providing the biodiesel and diesel samples. This project was supported by resources of the LABBIO- Laboratory of Biodeterioration of Fuels and Biofuels at Federal University of Rio Grande do Sul. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 | Table S1 | Table S2 |
Figures at a glance
 |
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 | Figure S1 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 15359
- [From(publication date):
November-2015 - Sep 03, 2025] - Breakdown by view type
- HTML page views : 10626
- PDF downloads : 4733
