Research Article Open Access
Effect of Antibiotics Combination and Comparison of Methods for Detection of Synergism in Multiresistant Gram-Negative Bacteria
| Gleice Cristina Leite1,2, Lauro Vieira Perdigão Neto1,2, Juliana Januário Gaudereto1,2, Cláudia Maria Dantas de Maio Carrilho3, Flavia Rossi5, Anna Sara Levin1,4 and Silvia Costa1*,2,6 | ||
| 1Department of Infectious Diseases, University of São Paulo, Brazil | ||
| 2Laboratory of Medical Investigation 54 (LIM-54), Hospital Das Clínicas of FMUSP, São Paulo, Brazil | ||
| 3Hospital Universitário, Londrina State University, Londrina, Brazil | ||
| 4Department of Infection Control, Hospital das Clínicas, University of São Paulo, Brazil | ||
| 5Laboratory of Microbiology, Hospital Das Clínicas, University of São Paulo, Brazil | ||
| 6Institute of Tropical Medicine, University of São Paulo, Brazil | ||
| Corresponding Author : | Silvia Figueiredo Costa Laboratory of Medical Investigation 54 (LIM-54) Hospital Das Clínicas of FMUSP, São Paulo, Brazil Tel: 55-11-30617030 Fax: 55-11-30617043 E-mail: costasilviaf@ig.com.br |
|
| Received December 17, 2014; Accepted March 18, 2015; Published March 25, 2015 | ||
| Citation: Leite GC, Neto LVP, Gaudereto JJ, Carrilho CMDdM, Rossi F, et al. (2015) Effect of Antibiotics Combination and Comparison of Methods for Detection of Synergism in Multiresistant Gram-Negative Bacteria. J Infect Dis Ther 3:207. doi:10.4172/2332-0877.1000207 | ||
| Copyright: © 2015 Leite GC, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | ||
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
||
Visit for more related articles at Journal of Infectious Diseases & Therapy
Abstract
Background: Because of the rapid spread of antimicrobial resistance and the slow development of novel antimicrobials, Gram-negative infections treatments are challenging clinicians and the Public Health. We aimed to evaluate the activities of antimicrobial combination against MDR Gram-negative bacteria.
Methods: Twenty-eight P. aeruginosa, 20 A. baumannii and 17 K. pneumoniae carbapenem-resistant clinical isolates had MIC determined by broth microdilution. Synergistic effect was investigated using checkerboard method and time-kill assay, the gold standard method. PCR for carbapenemase genes and PFGE clonality assessment were done.
Results: All P. aeruginosa isolates were resistant to meropenem, but susceptible to colistin; the genes blaSPM and blaKPC were found in 82% and 25% of them, respectively; synergistic effect was seen only in combinations of colistin with meropenem (43%), meropenem with amikacin (36%) and colistin with amikacin (7%), by time-kill. Twenty-five P. aeruginosa isolates belonged to the same clonal profile. All 20 isolates of A. baumannii were resistant to meropenem, rifampicin, fosfomycin and harbored blaOXA-51-like; the genes blaOXA-23-like, blaOXA-143-like and blaIMP were found in 50%, 35% and 15% of them, respectively. Eleven in thirteen colistin-susceptible A. baumannii isolates showed distinct profiles and six in seven colistin-resistant isolates belonged to the same clone; synergistic effect was observed in almost all combinations by time-kill. Resistance to polymyxin B and resistance to imipenem were found in 100% and 91% of K. pneumoniae isolates. The blaKPC gene was found in 82% of them; five clusters were identified (each one with two isolates) and other seven different isolates; synergistic effect occurred in most of threedrug combinations.
Conclusion: We demonstrated that time kill assay must be considered the gold standard method to detect synergism in vitro, as it allows greater dynamic assessment and higher sensitivity, when compared to the other methods. We detected that colistin combinations are frequently synergic against A. baumannii and K. pneumoniae. For P. aeruginosa, the results were not that optimistic.
| Keywords | |
| Multidrug resistant microorganisms; Checkerboard; Time-kill; Synergism; Inhibitory concentration; Fractional inhibitory concentration index | |
| Introduction | |
| Infections caused by multidrug-resistant microorganisms have emerged as serious global health problem, and therapeutic options for treatment of infections caused by these pathogens are limited [1,2]. In the last decade, polymyxins B and E (colistin) have been used to treat infections due to multidrug resistant Gram-negative bacteria [2,3], although colistin-resistant clinical isolates have been reported more recently [3-5]. Old antibiotic agents, such as fosfomycin, are now being considered potential treatment alternatives due to the lack of new antibiotics against these agents [6]. Studies have demonstrated that fosfomycin is a promising agent, particularly in combination with other agents, for the treatment of infections due to MDR Gramnegative, specially Enterobacteriaceae; however, there is concern about its use against A. baumannii [6,7]. There are experimental evidences that even drugs active only against Gram-positive microorganisms, such as vancomycin, may have activity against Gram-negative bacteria when combined with other antibiotics [4]. In this scenario, treatment with combination therapy, using two or more antibacterial drugs, has increased in the last years [8-10]. Some authors have considered a greater benefit the combination of three drugs for treating K. pneumoniae infections [11-13]. | |
| The most widely used in vitro methods to assess drug-drug interactions are the checkerboard technique, using fractional inhibitory concentration index (FICI) and two-well interpretation criteria, and time-kill kinetics [14]. The checkerboard method is prone to error and, by necessity, results from the checkerboard are often confirmed with the more dynamic interaction provided by the timekill kinetic study format. However, there is no gold standard method, and few studies have evaluated pan-resistant microorganims. There is also doubt if the synergistic effect of antibiotics is related to the mechanism of resistance or to the clonality of isolates or both [14,15]. | |
| These data highlight the importance of evaluating antibiotic combinations that are effective in the treatment of infections caused by these bacteria. Thus, we evaluated the activities of some antimicrobial drugs in combination, against MDR gram-negative bacteria, including pan-resistant isolates, with different mechanisms of resistance and different clonal origins. | |
| Methods | |
| Bacterial isolates | |
| The twenty-eight isolates of Pseudomonas aeruginosa were obtained from bone marrow transplantation unit of Hospital das Clínicas of University of São Paulo from 2011 to 2013. The twenty Acinetobacter baumannii isolates belong to the bacterial collection of the Laboratory of Bacteriology (LIM-54) of the Department of Infectious Diseases of Faculty of Medicine of University of São Paulo: thirteen colistin-susceptible isolates from 2002 to 2004 and seven colistin-resistant isolates from 2011 to 2012. In addition, the seventeen isolates of Klebsiella pneumoniae were obtained from the Hospital Universitário of Londrina State University from 2011 to 2012. | |
| All P. aeruginosa isolates were obtained from blood. The 13 isolates of colistin-susceptible A. baumannii were acquired from blood samples and the colistin-resistant isolates were obtained from blood (n=4), secretion (n=1), ascites (n=1) and rectal swab (n=1). K. pneumoniae isolates were obtained from endotracheal secretion (n=7), blood (n=6) and urine (n=4). | |
| Susceptibility testing | |
| Minimum inhibitory concentrations (MIC) of colistin, polymixin B (USP Reference Standard, Rockville, MD, USA), rifampicin, imipenem, gentamycin, amikacin, tigecycline, fosfomycin, vancomycin, teicoplanin (Sigma-Aldrich, St Louis, MO, USA), and meropenem (Astra Zeneca, Cotia, SP, Brazil) were determined using the broth microdilution method in duplicate on separate days, according to Clinical and Laboratory Standards Institute (CLSI) [16]. Susceptibility, MIC50 and MIC90 were determined and interpreted according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) for polymyxin and fosfomycin [17], Food and Drug Administration (FDA) for tigecycline[18], and CLSI criteria for the other drugs [16]. Pseudomonas aeruginosa ATCC27853, Escherichia coli ATCC25922 and Staphylococcus aureus ATCC29213 were used as controls. | |
| Polymerase chain reaction (PCR) | |
| For A. baumannii isolates, PCR techniques for carbapenemases genes bla OXA-23-like, bla OXA-51-like, bla OXA-58-like, bla OXA-24-like, bla IMP, bla SPM, bla VIM, bla SIM, blaNDM and bla OXA-143-like were performed [19,20]. | |
| For P. aeruginosa isolates, PCR to bla SPM, bla VIM, bla NDM and bla KPC were performed and K. pneumoniae isolates were submitted to PCR for blaKPC [21,22]. | |
| Pulsed-field gel electrophoresis (PFGE) | |
| Clonality of the isolates was evaluated by PFGE. The digestion of A. baumannii chromosomal DNA in Ultrapure Agarose (InvitrogenTM, Life technologies) was performed with ApaI endonuclase (Applied Biosystems, Foster City, California, USA), using method previously described by Durmaz et al. [22]. In turn, P. aeruginosa and K. pneumoniae chromosomal DNA digestion was done using SpeI endonuclease (Applied Biosystems, Foster City, California, USA) [23]. | |
| Restriction fragments were obtained by separation using a CHEF DR®III system (Bio-Rad, Hercules, Calif., USA) with 0.5x TBE buffer for 23 h at 14°C with pulse times of 5-30 s for A. baumannii , 5-90 s for P. aeruginosa and 5-60 s for K. pneumoniae [22,23] | |
| Patterns were interpreted using BioNumerics® version 7.1 (Applied Maths). | |
| Checkerboard microdilution | |
| Checkerboard microdilution synergism testing was performed in duplicate and evaluated after 20-24 h of incubation at 35°C. The isolates of A. baumannii and P. aeruginosa were submitted to combinations of two drugs. The FICI was then calculated, using fractional inhibitory concentration (FIC) for each drug, as FICI=FICA +FICB, where FICA=MIC of drug A in combination/MIC of drug A when alone, and FICB=MIC of drug B in combination/MIC of drug B when alone. Results were interpreted as follows: synergism if FICI ≤ 0.5, indifferent if FICI>0.5 and ≤ 4 and antagonism if FICI>4 [14,24]. | |
| The isolates of K. pneumoniae were submitted to combinations of three drugs. FICI was calculated as FICI=FICA+FICB+FICC, where FICC=MIC of drug C in combination/MIC of drug C when alone. Results were interpreted as follows: synergism if FICI<1.0, indifferent if FICI= ≥ 1.0 and ≤ 4 and antagonism if FICI>4.0 [25-27]. | |
| Time-kill assay | |
| Time-kill assays were performed in duplicate at concentrations based on the MIC determined from checkerboard testing of isolates: drugs alone and combined at 1x MIC and 0.5x MIC. Time-kill analysis was performed according to previously published techniques by Petersen et al. [15]. Flasks containing Müeller Hinton Broth and drug were inoculated with testing organism, at a density of ~106 cfu/mL and final volume of 10 mL, and incubated in a shaker at 35°C in ambient air. Aliquots were removed at time 0 and 2, 4, 6 and 24 h post-inoculation and serially diluted in 0.85% sodium chloride solution. Diluted samples of 0.01 mL were plated in duplicate on Müeller Hinton agar and the colonies were counted (log10 cfu/mL), after 20 h of incubation at 37°C. Synergism was interpreted as ≥ 2log10 decrease in colony count with the antimicrobial combination compared to the most active single agent; the drug combination was considered antagonistic for ≥ 2 log10 increase in cfu/mL and indifferent for <2 log10 increase or decrease in colony count with the combination compared with the most active drug alone. The results of the checkerboard method were compared to results of time-kill assay, considered as the gold standard for assessment of synergism [15]. | |
| Results | |
| The MIC50 and MIC90 of the isolates were summarized in Table 1. All P. aeruginosa isolates were susceptible to colistin, resistant to meropenem and had MIC for teicoplanin>256 mg/L. The genes blaSPM and blaKPC were found in 82% and 25% of the isolates, respectively. The genes bla VIM and bla NDM were not identified. | |
| All 20 isolates of A. baumannii were resistant to meropenem, rifampicin, fosfomycin, had MIC for vancomycin>256 mg/L and harbored bla OXA-51-like. The genes blaOXA-23-like, bla OXA-143-like and bla IMP were found in 50%, 35% and 15% of the isolates, respectively. The genes bla SPM, bla VIM and bla SIM were not identified. | |
| Resistance to polymyxin B, using A. baumannii CLSI breakpoints, and resistance to imipenem were found in 100% and 91% of K. pneumoniae isolates. The bla KPC gene was found in 82% of them. | |
| Twenty-five P. aeruginosa isolates belonged to the same clonal profile. Eleven in thirteen colistin-susceptible A. baumannii isolates showed distinct profiles and six in seven colistin-resistant isolates belonged to the same clone. Five clusters were identified (each one with two isolates) and other seven different isolates. | |
| The FICI method detected indifferent effect for all P. aeruginosa isolates and the two-well method detected nine results of synergistic effect in meropenem with amikacin combination. The comparison between two-well and time-kill methods is shown in Table 2. | |
| The Table 3 shows the main synergy testing results according resistance mechanisms for 20 A. baumannii isolates with two-well method and time-kill. According interpretation by FICI, the synergism effect was detected in ten colistin-resistant isolates in the colistin with rifampicin combination and only one isolate in the colistin with vancomycin combination. Synergistic effect was observed more frequently for fosfomycin combined with amikacin (90% of the isolates) using two-well method, and for colistin with rifampicin using FICI and two-well methods (50% and 65% of the isolates, respectively), all confirmed by time-kill assay. Antagonism was not observed for any method. | |
| Six K. pneumoniae isolates were tested with two drug combinations (Table 4). The FICI method detected indifferent effect for all isolates and two-well method for five isolates; time-kill detected three isolates with synergistic effects (polymyxin B with tigecycline, polymyxin B with amikacin, and polymyxin B with imipenem). Eleven K. pneumoniae isolates were tested in three-drug checkerboard microdilution method, and synergism was detected for two isolates by FICI and time-kill assay (Table 5). Antagonism was observed in one isolate by FICI, however synergic result by time-kill. | |
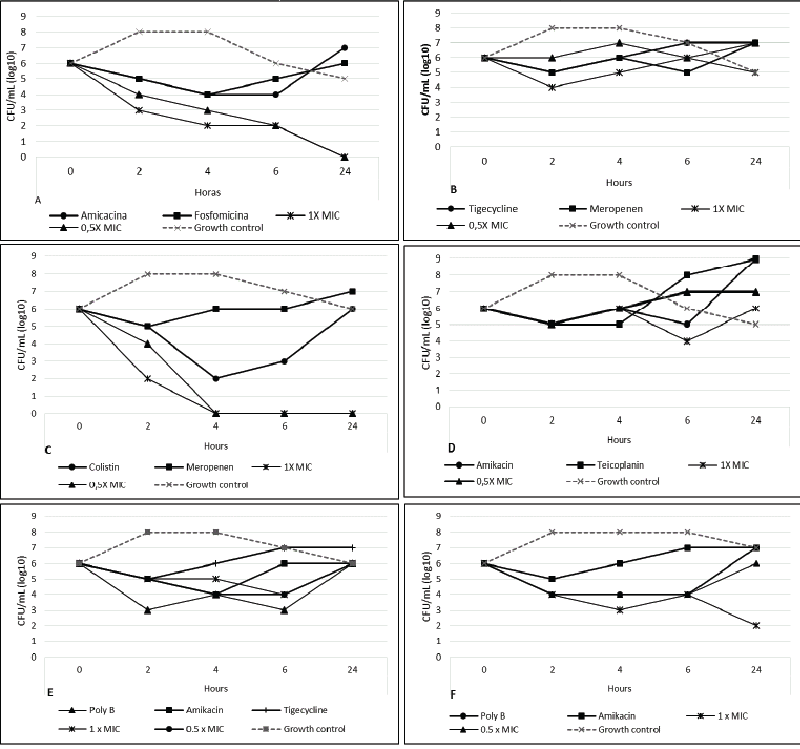
| Time-kill analysis was performed for all antimicrobial combinations. Colony counts of the isolates previously exposed for 0, 2, 4, 6 and 24 h to these combinations were then compared, after 20h of incubation, to the most active single agent (Figure 1). For P. aeruginosa isolates, the synergistic effect was observed in combinations between colistin with meropenem, colistin with amikacin and meropenem with amikacin (43%, 7% and 36% of the isolates, respectively). The use of two antibiotics in combination against A. baumannii showed synergistic effect for the most of the microorganisms, except for combinations with tigecycline, that showed regrowth before 24 hours. Combinations of fosfomycin with gentamicin, fosfomycin with meropenem, fosfomycin with vancomycin and imipenem with vancomycin showed regrowth in 20%, 50%, 45% and 45% of the isolates, respectively. At 1x MIC, two and three combined drugs showed synergism in 100% and 91% of the K. pneumoniae isolates, respectively. However, at 0.5x MIC, none of the isolates had synergistic effect in two-drug combinations and 64% of the isolates tested had synergistic effect when three drugs were combined. | |
| Discussion | |
| In this study, we evaluated the in vitro synergistic effects of different antimicrobial combinations against MDR P. aeruginosa , A. baumannii and K. pneumoniae using checkerboard and time-kill assay. | |
| Using any interpretation criteria, synergistic effect was found infrequently in P. aeruginosa isolates in the present study. Meropenem with amikacin combinations had synergistic effect in only 36% of the isolates. Previous studies using multi-susceptible isolates reported frequent synergistic effect of meropenem with amikacin, reaching close to 60% of strains tested, in contrast to few studies that showed that this combination was less active against P. aeruginosa [28,29].More recently, despite reports that suggest an advantage of combination therapy over monotherapy, clinical data are scarce, and well-designed randomized trials with synergistic effect are needed to better elucidate the efficacy of the various combination regimens [30]. For colistin with meropenem combination, synergistic effect was observed in 43% of the isolates. One study showed that combined therapy of colistin with carbapenem in P. aeruginosa is more active when compared to the monotherapy in susceptible and resistant isolates to the drugs used. Another study conducted by the same author showed that the best activity of combination therapies was achieved with colistin against P. aeruginosa isolates [31,32]. | |
| To our knowledge, this the first study that evaluated the synergistic effect of fosfomycin with others antibiotics using time-kill assay, against well characterized A. baumannii isolates. Only few previous studies evaluated synergistic effect of fosfomycin against MDR A. baumannii , and reported low synergistic effect results [33,34]. In our study, all clinical isolates of A. baumannii harbored blaOXA-51-like, a gene that naturally exists in A. baumannii . The carbapenemase gene blaOXA-23-like was found in 50% of the isolates; this gene has been reported as an important carbapenem resistance mechanism in many countries, including Brazil [35]. It is important to notice that seven of ten isolates that harbored bla OXA-23-like were colistin-resistant. All isolates that harbored bla OXA-143-like showed a synergistic effect with fosfomycin and amikacin and 86% presented synergism with tigecycline and amikacin. Among the isolates harboring blaOXA-23-like, 100%, 80% and 80% presented a synergistic effect when exposed to the combination of colistin with vancomycin, colistin with imipenem and fosfomycin with amikacin. Discrepancies in resistance mechanisms between isolates from different centers may explain, at least in part, why combination regimens reported as synergic were not successful among our assays. | |
| Colistin is frequently used to treat carbapenem-resistant infections and considered the last treatment option, because it is often the only agent with in vitro activity. Few studies, however, have evaluated the synergism of colistin with other drugs as a potential option to treat infections due to colistin-resistant A. baumannii [32-34]. In our study, the combination of colistin with rifampicin showed the highest synergistic effect against colistin-resistant A. baumannii , followed by colistin with vancomycin. Colistin and rifampicin combination has already been suggested for treatment of MDR A. baumannii , by both in vitro and in vivo studies, mainly series of cases, and the combination of colistin with vancomycin has been reported in some studies against a few isolates of A. baumannii [33,34]. According to these authors, the colistin would disrupt the outer membrane and could facilitate the glycopeptide penetration across the outer membrane and expose the target site of the cell wall. A synergistic effect of colistin and vancomycin is an interesting result since, in the intensive care setting, the empiric combination for septic patient is a beta-lactam plus vancomycin and, in hospitals with high MDR rates, polymyxin is added [34-36]. | |
| Time-kill kinetics confirmed the synergistic effect demonstrated by the checkerboard method in our study and this was also shown by other studies, moreover, the high synergistic effect showed by our study was restricted for some combinations; these results were observed in seven in 25 antimicrobial combinations tested. Tan et al. evaluated the synergistic effect with polymyxin B and tigecycline, polymyxin B and rifampicin and tigecycline and rifampicin combinations; synergism was present in 40% of combinations by the method of time-kill, 17% by checkerboard and 2% by Etest, there was no agreement between time-kill and Etest methods for synergism testing [37]. In our study, although the high rate of susceptibility to tigecycline (95% of the isolates), few antibiotic combinations using tigecycline showed synergistic effect. The best synergistic tygecicline combinations results were obtained by time-kill interpretation criteria; in contrast, a previous study reported by Petersen et al. [15] showed as frequent as 56% of synergistic effect in tigecycline with amikacin combination by checkerboard, but not by time-kill. | |
| It is important to emphasize that K. pneumoniae isolates used in this study were resistant to polymyxin B. It is increasingly frequent the dissemination of this microorganism, with such sensitivity profile, in hospitals in developing countries [11,12]. This phenomenon hinders the possibility of further treatment of infections caused by this pathogen, which usually harbors carbapenemases encoding genes in its genome, which confers resistance to carbapenems, commonly used drugs for the treatment of nosocomial infections. The use of two or three-drug combination therapy in carbapenemase-producing K. pneumoniae has been reinforced by some authors. In addition to the scarce clinical research, several in vitro studies have shown the benefit of the drug combination of use of strains of K. pneumoniae [11,38]. | |
| In this study, we noticed frequent synergism in regimens containing polymyxin B (50% of the isolates in two-drug combinations and 91% in three-drug combinations). These data are similar to the results found in a recent study, when synergistic or bactericidal activity was demonstrated for double and triple-antibiotic combinations with colistin, using time-kill experiment in two VIM- and two NDMproducing K. pneumoniae strains; in this study, however, all strains tested were susceptible to colistin, unlike the strains tested in our study [38]. In other studies with colistin-resistant K. pneumoniae strains, antibiotic combinations with colistin showed potential therapeutical options against infections caused by multidrug-resistant carbapenemase-producing K. pneumoniae isolates [39,40]. | |
| A variety of methods for interpreting the results can be used and these methods may, lead to different results and conclusion, even the checkerboard technique that is well standardized. A study conducted by Bonapace et al. [15] that used different methods to interpret the results by checkerboard showed that the poorest agreement was found with FICI and time-kill assay. Standardization of interpretation would be desirable to decrease discordant results. It is a big challenge to perform synergism testing routinely in the clinical microbiology laboratory, especially due to the lack of accepted standards. The methods available are laborious, time-consuming, and require expertise in the specific procedures. | |
| In conclusion, our study demonstrates that time kill assay must be considered the gold standard method to detect synergism in vitro , as it allows greater dynamic assessment and higher sensitivity, when compared to the other methods. We detected that colistin combinations are frequently synergic against A. baumannii and K. pneumoniae , and probably not related to the clonality of isolates, and that fosfomycin combined with amikacin or with gentamicin were surprisingly active against A. baumannii . For P. aeruginosa , the results were not that optimistic. However, in the present scenario of multiresistance in which the choices of active drugs are scarce, combination therapy is an option to treat these infections, and in vitro experience is needed for the development of future clinical studies. | |
References
- Souli M, Galani I, Giamarellou H (2008) Emergence of extensively drug-resistant and pandrug-resistant Gram-negative bacilli in Europe. Euro Surveill 13.
- Falagas ME, Lourida P, Poulikakos P, Rafailidis PI, Tansarli GS (2014) Antibiotic treatment of infections due to carbapenem-resistant Enterobacteriaceae: systematic evaluation of the available evidence. Antimicrob Agents Chemother 58: 654-663.
- Gales AC, Jones RN, Sader HS (2006) Global assessment of the antimicrobial activity of polymyxin B against 54 731 clinical isolates of Gram-negative bacilli: report from the SENTRY antimicrobial surveillance programme (2001-2004). ClinMicrobiol Infect 12:315-321.
- Percin D, Akyol S, Kalin G (2014) In vitro synergism of combinations of colistin with selected antibiotics against colistin-resistant Acinetobacterbaumannii. GMS Hyg Infect Control 9: Doc14.
- Kontopidou F, Giamarellou H, Katerelos P, Maragos A, Kioumis I, et al. (2014) Infections caused by carbapenem-resistant Klebsiellapneumoniae among patients in intensive care units in Greece: a multi-centre study on clinical outcome and therapeutic options. ClinMicrobiol Infect 20: O117-123.
- Falagas ME, Kastoris AC, Karageorgopoulos DE, Rafailidis PI (2009) Fosfomycin for the treatment of infections caused by multidrug-resistant non-fermenting Gram-negative bacilli: a systematic review of microbiological, animal and clinical studies. Int J Antimicrob Agents 34: 111-120.
- Lu CL, Liu CY, Huang YT, Liao CH, Teng LJ, et al. (2011) Antimicrobial susceptibilities of commonly encountered bacterial isolates to fosfomycin determined by agar dilution and disk diffusion methods. Antimicrob Agents Chemother 55: 4295-4301.
- Principe L, D'Arezzo S, Capone A, Petrosillo N, Visca P (2009) In vitro activity of tigecycline in combination with various antimicrobials against multidrug resistant Acinetobacterbaumannii. Ann ClinMicrobiolAntimicrob 8: 18.
- Klastersky J, Georgala A (2014) Strategies for the empirical management of infection in cancer patients with emphasis on the emergence of resistant gram-negative bacteria. Crit Rev OncolHematol 92: 268-278.
- Zavascki AP, Bulitta JB, Landersdorfer CB (2013) Combination therapy for carbapenem-resistant Gram-negative bacteria. Expert Rev Anti Infect Ther 11: 1333-1353.
- Tumbarello M, Viale P, Viscoli C, Trecarichi EM, Tumietto F, et al. (2012) Predictors of mortality in bloodstream infections caused by Klebsiellapneumoniaecarbapenemase-producing K. pneumoniae: importance of combination therapy. Clin Infect Dis 55: 943-950.
- Hirsch EB, Tam VH (2010) Detection and treatment options for Klebsiellapneumoniaecarbapenemases (KPCs): an emerging cause of multidrug-resistant infection. J AntimicrobChemother 65: 1119-1125.
- Zarkotou O, Pournaras S, Tselioti P, Dragoumanos V, Pitiriga V, et al. (2011) Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiellapneumoniae and impact of appropriate antimicrobial treatment. ClinMicrobiol Infect 17: 1798-1803.
- Bonapace CR, Bosso JA, Friedrich LV, White RL (2002) Comparison of methods of interpretation of checkerboard synergy testing. DiagnMicrobiol Infect Dis 44: 363-366.
- Petersen PJ, Labthavikul P, Jones CH, Bradford PA (2006) In vitro antibacterial activities of tigecycline in combination with other antimicrobial agents determined by chequerboard and time-kill kinetic analysis. J AntimicrobChemother 57: 573-576.
- Clinical and Laboratory Standard Institute (2013) Performance standards for antimicrobial susceptibility testing. Nineteenth informational supplement. CLSI document M100 S19. CLSI, Wayne, PA, USA.
- Hope WW, Cuenca-Estrella M, Lass-Flörl C, Arendrup MC; European Committee on Antimicrobial Susceptibility Testing-Subcommittee on Antifungal Susceptibility Testing (EUCAST-AFST) (2013) EUCAST technical note on voriconazole and Aspergillus spp. ClinMicrobiol Infect 19: E278-280.
- U.S. Food and Drug Administration Tygecycline Label. http://www.accessdata.fda.gov/drugsatfda_docs/label/2009/021821s016lbl.pdf.
- Mostachio AK, van der Heidjen I, Rossi F, Levin AS, Costa SF (2009) Multiplex PCR for rapid detection of genes encoding oxacillinases and metallo-beta-lactamases in carbapenem-resistant Acinetobacter spp. J Med Microbiol 58: 1522-1524.
- Mostachio AK, Levin AS, Rizek C, Rossi F, Zerbini J, et al. (2012) High prevalence of OXA-143 and alteration of outer membrane proteins in carbapenem-resistant Acinetobacter spp. isolates in Brazil. Int J Antimicrob Agents 39: 396-401.
- Bradford PA, Bratu S, Urban C, Visalli M, Mariano N, et al. (2004) Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin Infect Dis 39: 55-60.
- Durmaz R, Otlu B, Koksal F, Hosoglu S, Ozturk R, et al. (2009) The optimization of a rapid pulsed-field gel electrophoresis protocol for the typing of Acinetobacterbaumannii, Escherichia coli and Klebsiella spp. Jpn J Infect Dis 62: 372-377.
- Pournaras S, Protonotariou E, Voulgari E, Kristo I, Dimitroulia E, et al. (2009) Clonal spread of KPC-2 carbapenemase-producing Klebsiellapneumoniae strains in Greece. J AntimicrobChemother 64: 348-352.
- Tascini C, Tagliaferri E, Giani T, Leonildi A, Flammini S, et al. (2013) Synergistic activity of colistin plus rifampin against colistin-resistant KPC-producing Klebsiellapneumoniae. Antimicrob Agents Chemother 57: 3990-3993.
- Pillai SK, Moellering RC, Eliopoulos GM (2005) Antibiotics in laboratory medicine. (5thedn.) Lorian V, Lippincott Williams and Wilkins, Philadelphia, USA.
- Hickman RA, Hughes D, Cars T, Malmberg C, Cars O (2014) Cell-wall-inhibiting antibiotic combinations with activity against multidrug-resistant Klebsiellapneumoniae and Escherichia coli. ClinMicrobiol Infect 20: O267-273.
- Yoon J, Urban C, Terzian C, Mariano N, Rahal JJ (2004) In vitro double and triple synergistic activities of Polymyxin B, imipenem, and rifampin against multidrug-resistant Acinetobacterbaumannii. Antimicrob Agents Chemother 48: 753-757.
- Nakamura A, Hosoda M, Kato T, Yamada Y, Itoh M, et al. (2000) Combined effects of meropenem and aminoglycosides on Pseudomonas aeruginosa in vitro. J AntimicrobChemother 46: 901-904.
- Lim TP, Lee W, Tan TY, Sasikala S, Teo J, et al. (2011) Effective antibiotics in combination against extreme drug-resistant Pseudomonas aeruginosa with decreased susceptibility to polymyxin B. PLoS One 6: e28177.
- Kmeid JG, Youssef MM, Kanafani ZA, Kanj SS (2013) Combination therapy for Gram-negative bacteria: what is the evidence? Expert Rev Anti Infect Ther 11: 1355-1362.
- Bergen PJ, Forrest A, Bulitta JB, Tsuji BT, Sidjabat HE, et al. (2011) Clinically relevant plasma concentrations of colistin in combination with imipenem enhance pharmacodynamic activity against multidrug-resistant Pseudomonas aeruginosa at multiple inocula. Antimicrob Agents Chemother 55: 5134-5142.
- Bergen PJ, Tsuji BT, Bulitta JB, Forrest A, Jacob J, et al. (2011) Synergistic killing of multidrug-resistant Pseudomonas aeruginosa at multiple inocula by colistin combined with doripenem in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother 55:5685-5695.
- Zhang Y, Chen F, Sun E, Ma R, Qu C, et al. (2013) In vitro antibacterial activity of combinations of fosfomycin, minocycline and polymyxin B on pandrug-resistant Acinetobacterbaumannii. Experimental Therapeutic Medic 5:1737-1739.
- Martinez-Martinez L, Rodriguez G, Pascual A, Suárez AI, Perea EJ (1996) In vitro activity of antimicrobial agent combinations against multidrug resistant Acinetobacterbaumannii. J AntimicrobChemother 38:1107-1108.
- Grosso F, Carvalho KR, Quinteira S, Ramos A, Carvalho-Assef AP, et al. (2011) OXA-23-producing Acinetobacterbaumannii: a new hotspot of diversity in Rio de Janeiro? J AntimicrobChemother 66: 62-65.
- Garnacho-Montero J, Amaya-Villar R, Gutiérrez-Pizarraya A, Espejo-Gutiérrez de Tena E, Artero-González ML, et al. (2013) Clinical efficacy and safety of the combination of colistin plus vancomycin for the treatment of severe infections caused by carbapenem-resistant Acinetobacterbaumannii. Chemotherapy 59: 225-231.
- Tan TY, Lim TP, Lee WH, Sasikala S, Hsu LY, et al. (2011) In vitro antibiotic synergy in extensively drug-resistant Acinetobacterbaumannii: the effect of testing by time-kill, checkerboard, and Etest methods. Antimicrob Agents Chemother 55: 436-438.
- Tängdén T, Hickman RA, Forsberg P, Lagerbäck P, Giske CG, et al. (2014) Evaluation of double- and triple-antibiotic combinations for VIM- and NDM-producing Klebsiellapneumoniae by in vitro time-kill experiments. Antimicrob Agents Chemother 58: 1757-1762.
- Kádár B, Kocsis B, Tóth Á, Damjanova I, Szász M, et al. (2013) Synergistic antibiotic combinations for colistin-resistant Klebsiellapneumoniae. ActaMicrobiolImmunol Hung 60: 201-209.
- Tascini C, Tagliaferri E, Giani T, Leonildi A, Flammini S, et al. (2013) Synergistic activity of colistin plus rifampin against colistin-resistant KPC-producing Klebsiellapneumoniae. Antimicrob Agents Chemother 57: 3990-3993.
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 | Table 4 | Table 5 |
Figures at a glance
 |
| Figure 1 |
Relevant Topics
- Advanced Therapies
- Chicken Pox
- Ciprofloxacin
- Colon Infection
- Conjunctivitis
- Herpes Virus
- HIV and AIDS Research
- Human Papilloma Virus
- Infection
- Infection in Blood
- Infections Prevention
- Infectious Diseases in Children
- Influenza
- Liver Diseases
- Respiratory Tract Infections
- T Cell Lymphomatic Virus
- Treatment for Infectious Diseases
- Viral Encephalitis
- Yeast Infection
Recommended Journals
Article Tools
Article Usage
- Total views: 18891
- [From(publication date):
April-2015 - Aug 18, 2025] - Breakdown by view type
- HTML page views : 13781
- PDF downloads : 5110
