Research Article Open Access
Efficient Aerobic Degradation of Various Azo Dyes by a Sphingomonas sp Isolated from Petroleum Sludge
| Liaquat Ali1, Huda Alhassani1, Noushad Karuvantevida2, Muhammad A. Rauf1 and S. Salman Ashraf1* | |
| 1Department of Chemistry, College of Science, UAE University, Al-Ain, UAE | |
| 2Department of Biology, College of Science, UAE University, Al-Ain, UAE | |
| Corresponding Author : | Salman Ashraf Department of Chemistry, College of Science UAE University, Al-Ain, UAE Tel: 971 3 713-6148 E-mail: salman.ashraf@uaeu.ac.ae |
| Received March 31, 2014; Accepted April 24, 2014; Published April 30, 2014 | |
| Citation: Ali L, Alhassani H, Karuvantevida N, Rauf MA, Ashraf SS (2014) Efficient Aerobic Degradation of Various Azo Dyes by a Sphingomonas sp Isolated from Petroleum Sludge. J Bioremed Biodeg 5:223. doi:10.4172/2155-6199.1000223 | |
| Copyright: © 2014 Ali L, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
The extensive discharge of textile dyes in industrial wastewater and the recalcitrant nature of azo dyes have fueled a strong interest in exploring efficient and environmentally friendly approaches for wastewater remediation. In this present study, we report on the isolation and characterization of a Sphingomonas sp. strain from petroleum sludge that was capable of efficiently degrading various classes of dyes. A subsequent detailed analysis using a panel of 12 azo dyes showed that this strain was capable of efficient aerobic degradation of at least 7 diverse azo dyes. Furthermore, these dyes could also be degraded (more than 70% in 24 hours) under static (anaerobic) culture conditions by this Sphingomonas sp. Lastly, HPLC analysis of the metabolites of three different azo dyes showed that depending on the availability of oxygen during the culturing and dye degradation process, and different sets of metabolites could be produced from these dyes. The current study establishes the usefulness of screening petroleum sludge for the isolation of novel bacterial strains for efficient bioremediation and highlights the different degradative pathways that are taken during aerobic and anaerobic bacterial dye degradation processes
| Keywords |
| Dye degradation; Sphingomonas; Azo dyes |
| Introduction |
| Despite increased awareness and governmental regulations, a large amount of organic pollutants are discharged in our water bodies. Sometimes this discharge is accidental (oil spills), but often at times, especially in developing countries, it is deliberate (and criminal). It is well accepted that these organic pollutants pose a direct threat to aquatic and marine life and eventually us. Due to the extensive use of organic dyes by various industries, they have become an integral part of many industrial effluents. Most of these dyes are toxic and potentially carcinogenic in nature and hence their removal from the industrial effluents is a major environmental problem. |
| Removal of dyes from wastewaters using various methods has been suggested, such as coagulation, adsorption, advanced oxidation (AOPs) and the membrane processes [1-6]. All these approaches have some advantages and disadvantages as well as limitations. Besides these, traditional physical techniques like adsorption on activated carbon, ultrafiltration, reverse osmosis, coagulation by chemical agents and ion exchange on synthetic adsorbent resins have also been used by various groups [7-9]. However in many cases, these techniques are either very costly or economically unfeasible or may have technical constraints. |
| The versatility and adaptability of microbes for hazardous waste degradation is gaining a lot of attention lately. Literature survey shows the promise of efficient biodegradation of various classes of dyes using microorganisms, mainly due to the low cost, ability to produce less sludge and environmental compatibility [10]. In this regard, various microorganisms (e.g. Bacillus subtilis, Phanerochaete chrysosporium, Aeromonas hydrophila, Penicillium sp., Klebsiella pneumoniae, Proteus mirabilis and Pseudomonas cepacia) have been isolated and have been shown to be very promising for degrading different dyes [11-14]. |
| In this paper, we present results on the isolation of twelve strains of dye degrading bacteria from petroleum sludge and characterized their abilities to degrade various types of organic dyes in aqueous solutions. Since most biodegradation of azo dyes are generally carried out under anaerobic conditions, part of the novelty of this work is that our strains were very active (in dye degradation) under aerobic (shaking) conditions. Additionally, they could also degrade dyes under anaerobic conditions. Analyses of degradation products of different dyes suggest that different degradation products are generated by our Sphingomonas strain depending on the availability of oxygen during the degradation process. |
| Materials and Methods |
| Toluidine Blue, Amido Black, Crystal Ponceau 6R, Trypan Blue, Methyl Blue, Orange G, Acid Red 40, Reactive Black 5 and Ponceau BS were obtained from Sigma-Aldrich. Eriochrome Black T, Thiazole Yellow G, Naphthol Green B and Congo Red were purchased from Fluka chemical and Malachite Green from BDH. Drimarene Black CL BGR, Drimarene Red CL 4BN and Drimarene Yellow CL 4R were obtained from Clariant chemicals. The chemical structures, dye class and the λmax of all dyes are listed in Table 1. Media for culturing was obtained from Sigma-Aldrich. Nutrient Broth composition (Sigma) was as follows: 1 g/L D(+)-glucose, 15 g/L peptone, 6 g/L sodium chloride, 3 g/L yeast extract, 3 g/L, final pH 7.5 ± 0.2 (25°C). All organic solvents (HPLC grade Acetonitrile and Methanol) were from Fisher Scientific (UK). All chemical used in this work were of analytical grade and used without further purification. |
| Bacterial strain identification and phylogenetic analysis |
| The 12 bacterial strains (H1-H12) were identified using partial 16S rRNA sequencing of the crude DNA on a 3500 Genetic Analyzer, Applied Biosystems, USA. The obtained DNA sequences were compiled in FASTA format and analyzed using BLAST (blastn) through NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The phylogenetic analyses were performed using the online site http://www.phylogeny.fr [15,16] which produced the phylogenetic tree. |
| Dye decolorization |
| A loopful of bacteria culture from glycerol stock was inoculated in a 50 mL sterile tube containing 15 ml nutrient broth and incubated at 37°C under shaking condition (200 rpm) for 24 h. For sampling, all dyes from stock solution (2000ppm) were sterilized by filtering through 0.45 μm Nylon filter, before being added at 20 ppm in 15 mL nutrient broth and 0.15 mL freshly grown (overnight) bacterial culture, incubated for 24 h while shaking. One ml of sample was withdrawn at different time intervals and each time it was centrifuged at 13,000 rpm for 10 minutes to separate the bacterial cell mass. Decolorization of dye was determined by measuring the absorbance of the cleared supernatant at the absorbance maxima of the respective dyes. All experiments were performed in triplicate. Nitrogen purging (blowing Nitrogen gas over the surface of the culture to displace the air and immediately tightly closing the tube cap) followed by static incubation were used to obtain anaerobic conditions in the experiments. Percentage decoloration was calculated as follows: |
| % Decoloration= [(Ai – Af) / Ai] x 100 (1) |
| Where Ai is the initial absorbance and Af is the final absorbance value. |
| Optimization |
| The effect of pH, concentration and carbon sources were studied and optimized. The decolorization of dye was observed at pH 5, 7, 9 and 11. Nutrient broth of pH 7 was used at various concentrations of dye. The effect of carbon and nitrogen sources was examined by using different nutrient media such as Nutrient Broth, LB and 2xYT. All experiments were carried out as mentioned above. |
| HPLC and UV analysis for decolorization |
| High performance liquid chromatography (HPLC) was carried out as previously described [17]. Briefly, an Agilent PH 1100 liquid chromatography system, (Agilent, USA) with an Agilent Zorbax SBC18 column 150 mm x 4.6 mm packed with 5 μm particle size, coupled to a diode array detector was used (Agilent, USA). The mobile phase consisted of solution A (0.1 M ammonium formate (pH 6.7) and solution B (1:1 acetonitrile/methanol) and gradient from 0% B to 80% B in 40 minutes and a flow rate of 1 mL/min was used to obtain the chromatographs. Percentage of dye decolorization was analyzed by using Epoch microplate reader from BioTek (USA). Sample from all tubes were examined at different times (0h, 2h, 4h, 6h, 8h and 24h) during the period of 24h. During all time intervals, each sample was centrifuged and the supernatant was used for absorbance measurements at λmax values of the dyes. |
| Result and Discussion |
| In initial screening we were interested in identifying bacterial strains that would be most efficient at degrading various classes of dyes. Therefore, we tested our 12 bacterial strains, isolated from petroleum sludge, on twelve different dyes under aerobic conditions. The screening results are summed up in Table 2. All dyes showed reasonable degradation when exposed to various bacterial strains, except H5 strain which failed to show any decolorization of the dyes under investigation. Additional experiments with H5 showed that it could in fact degrade various dyes, but only under anaerobic conditions. Based on the results from this initial screening, isolate H8 showed most promising results in degrading various classes of dyes and was chosen for further study. |
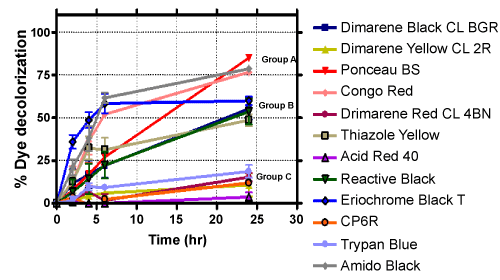
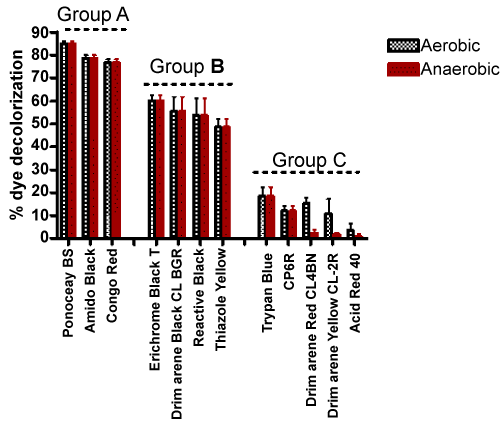
| Among the many dyes used in various applications, azo dyes are most extensively used in industries because they are easy to synthesize and are thus cost effective. Unfortunately, most azo dyes are toxic, carcinogenic and mutagenic in nature [18]. The azo bonds present in these compounds are resistant to breakdown, which can result in the accumulation of these molecules in the environment. However, as has been reported previously and shown in Table 2, they can be degraded by specific strains of bacteria under aerobic and/or anaerobic conditions. Subsequent experiments were carried out with additional azo dyes with strain H8, under aerobic conditions. The % decoloration observed with a set of twelve diverse azo dyes are presented in Table 3. Based on the % decolorization achieved, these azo dyes were divided into three main groups. The results are shown in Figure 1. Dyes belonging to group A showed maximum decoloration, whereas, group C dyes showed minimum decoloration. An interesting part of this investigation is that, when same dyes were examined under static (anaerobic) conditions, they showed almost the same amount of decolorization as compared to shaking (aerobic) condition - the comparative results are shown in Figure 2. At the end of second screening, three azo dyes namely, Amido Black, Congo Red and Ponceau BS (all from group A) were chosen for further analysis as they showed more than 75% decoloration. |
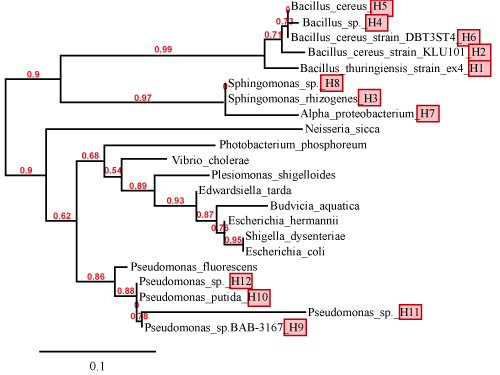
| Partial sequencing of the 16S rRNA gene showed that the 12 bacterial strains belonged to four different bacterial genuses: Bacillus (H1, H2, H4, H5 and H6), Sphingomonas (H3 and H8), Alphaprotobacterium (H7) and Pseudomonas (H9, H10, H11 and H12). Table 4 shows the individual bacterial strains, their GenBank accession number, and their identity (based on 16S rRNA sequence similarity to published sequences). The phylogenetic analysis of these twelve strains (and a few representative bacteria) is also shown in Figure 3. It is interesting to note that of the four genera of bacterial strains that we isolated from petroleum sludge, three of them (Bacillus, Sphingomonas, and Pseudomonas) have been previously shown to be able to degrade various classes of organic pollutants [19,20]. For example, Ding and colleagues have shown that a Sphingomonas strain isolated from Fe(OH)3 enriched microbial electrochemical reactor was able to efficiently degrade Acid Orange 7 dye under anaerobic conditions [20]. |
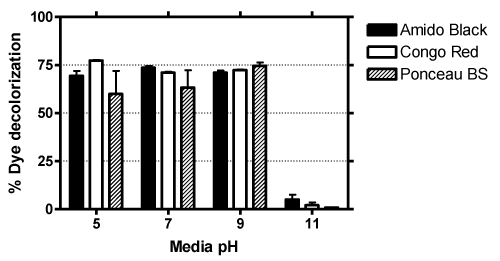
| The growth of bacteria depends on the pH of culture media, which has been previously shown to directly affect dye decoloration [21]. Thus the ability of Sphingomonas strain H8 to degrade dyes at different range of pH 5, 9 and 11 (control was pH 7) was carried out. Figure 4 shows that all dyes were best degraded in 5 to 9 pH, but no significant amount of decolorization was observed in alkaline media (pH 11), which is most likely due to the inability of bacteria to grow sufficiently in highly acid or alkaline media [22]. |
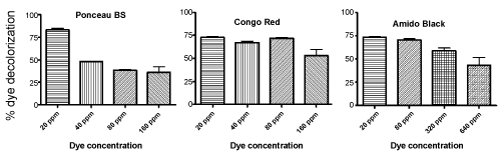
| It is well known that the dye concentration has a direct effect on the decolorization rate as well [23]. Therefore, Sphingomonas strain H8 was incubated with dyes of varying concentrations to see how this effected the decolorization of the tested dyes. It was found that decolorization, in general, increased with decreasing dye concentration (Figure 5). Ponceau BS, Congo Red, and Amido Black all showed almost 75% decoloration after 24 h when the concentration of these dyes were 20 ppm. Furthermore, Sphingomonas strain H8 was able to decolorize Congo Red quite efficiently even at much higher concentrations of up to 160 ppm (more than 50% decolorization in 24 hours). Amido Black could also be decolorized quite well by the Sphingomonas strain at concentrations up to 80 ppm (>60% in 24 hours). However, Ponceau BS decolorization seemed to be very sensitive toward dye concentration, with significantly reduced decolorization at concentrations higher than 20 ppm (Figure 5). Such dependence of dye concentration on bacteriamediated decolorization has been reported in the literature for other dyes as well [24,25]. |
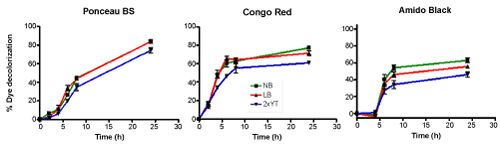
| Azodyes can be poor carbon sources, thus to biodegrade such dyes, pure culture with supplement of carbon or nitrogen sources are required [26]. Therefore, to examine the effect of supplement on dye decolorization, three media cultures were used and the results are shown in Figure 6. Although our Sphingomonas strain was able to decolorize the three chosen dyes in all media, best decolorization was observed with Nutrient Broth (NB). Luria Broth (LB) gave almost similar results as Nutrient Broth, with 2xYT showing statistically significantly lesser decolorization for all three dyes. Interestingly we noticed that both Congo Red and Amido Black reached maximum decolorization within 10 hours after which % decolorization became constant, most likely due to nutrient depletion [27]. |
| Although majority of the published literature on microbial azo dye degradation report their studies under anaerobic conditions, bacterial azo dye degradation can proceed under both aerobic and anaerobic conditions, or in mixed-batch mode [28]. It has been shown that the first step in azo dye degradation under both aerobic and anaerobic conditions proceeds via the reductive cleavage of the azo bond, leading to subsequent generation of aromatic amines [29].Under anaerobic conditions, and depending on the ring substituents and microbial strains, these amines are further converted to various metabolites such as 1-amino-2-naphthol, sulfanilic acid and nitroaniline [30]. However, if the degradation is being carried out under aerobic conditions, then microbial oxygenases can work on the aromatic amines to produce mono and dihydroxyaromatic compounds [31]. These compounds can be very different than those produced under anaerobic degradation conditions, and include metabolites such as 3-aminobenzenesulphonates, 2-aminonapthyl sulfonate, and hydroxysalicylic acids [32,33]. Interestingly, one of the metabolites, 1-amino-2-naphthol, has been reported to be produced under both aerobic and anaerobic conditions [30,32]. Although interesting and useful, most of the above mentioned studies have been carried out on different strains and different dyes, and different degradation conditions (aerobic and anaerobic). In this manuscript, we present preliminary results from our systematic degradation study using a same set of three dyes with one specific strain (Sphingomonas sp) and understand aerobic and anaerobic conditions. |
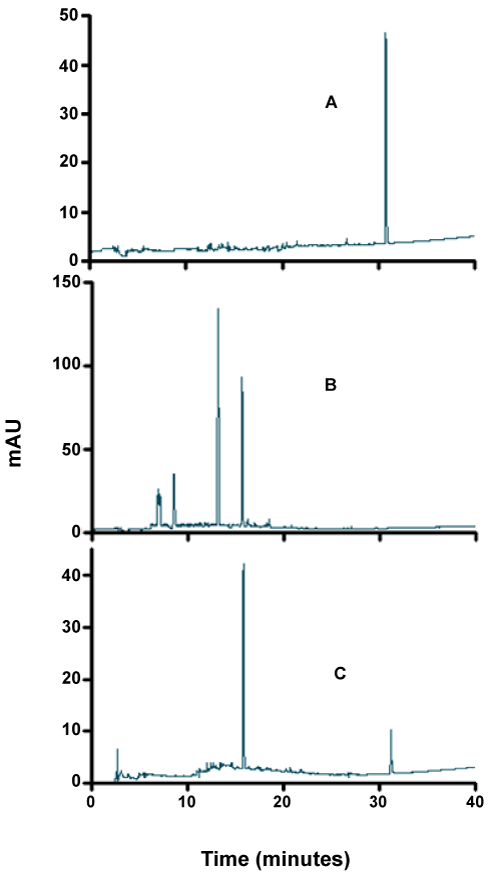
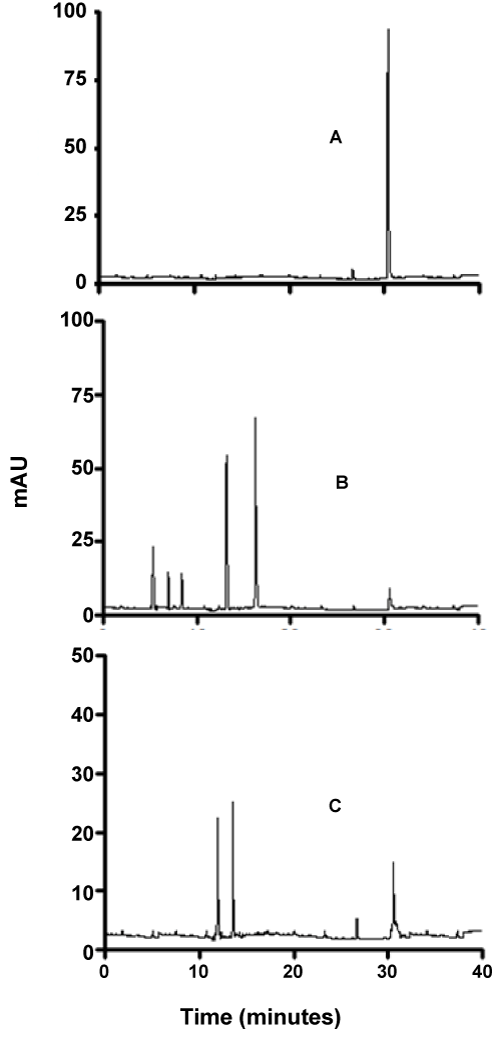
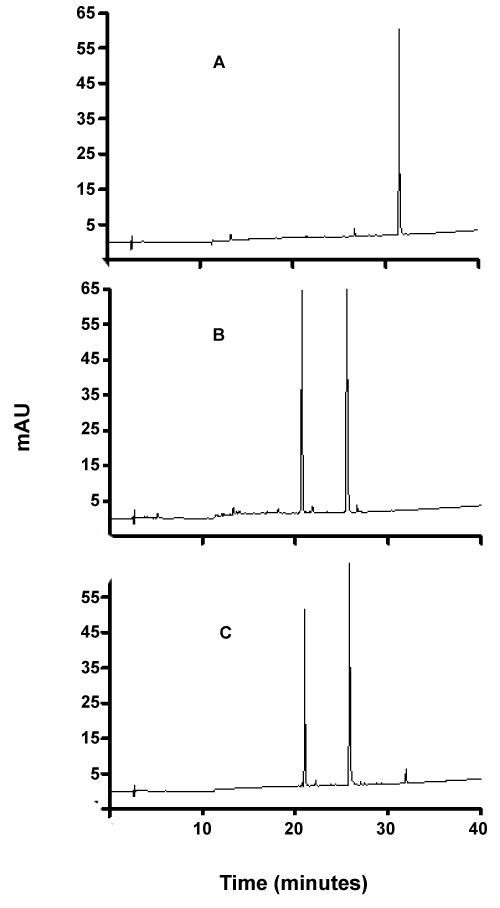
| As can be seen from figures 7-9, although the dyes seemed to be degraded equally well in both aerobic and anaerobic conditions, there appears to be very significant differences in the way these dyes are broken down. For example, the chromatograms of Ponceau BS (Figure 7) show four major peaks under aerobic but only one major peak under anaerobic condition. Similarly, Congo Red degradation chromatograms show 5 peaks in aerobic and 2 peaks in anaerobic condition (Figure 8). Surprisingly, Amido Black chromatograms showed the apparently same metabolites under both aerobic and anaerobic conditions (Figure 9). These results show that the degradation pathway for Ponceau BS and Congo Red are probably very different under aerobic and anaerobic conditions based on their HPLC profile, but Amido Black may proceed via a common scheme under both aerobic and anaerobic conditions. Future studies are planned to identify these metabolites that are produced under different degradation conditions. |
| Conclusion |
| In summary, we report here that petroleum sludge is a rich source of microbes that could be used to degrade various classes of dyes, including diverse azo dyes, which are normally recalcitrant to degradation. Furthermore, we identified one of the strains as Sphingomonas sp which could degrade up to 7 different azo dyes, including three (Amido Black, Congo Red, and Ponceau BS) very efficiently under both aerobic and anaerobic conditions. Lastly, preliminary HPLC analyses showed that depending on the presence or absence of oxygen, very different metabolic products can be produced, the identities of which will be reported in a future publication. |
| Acknowledgements |
| The authors graciously acknowledge the generous funding support from UAEU to SSA and MAR (UAEU/NRF Research Grant Program 27/11/2). |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 | Table 4 |
Figures at a glance
 |
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 | Figure 5 |
 |
 |
 |
 |
| Figure 6 | Figure 7 | Figure 8 | Figure 9 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 14637
- [From(publication date):
June-2014 - Aug 24, 2025] - Breakdown by view type
- HTML page views : 10006
- PDF downloads : 4631
