Research Article Open Access
Genetic Basis of Naphthalene and Phenanthrene Degradation by Phyllosphere Bacterial Strains Alcaligenes faecalis and Alcaligenes sp. 11SO
| Undugoda LJS, Kannangara S* and Sirisena DM | |
| Department of Botany, Faculty of science, University of Kelaniya, Kelaniya, Sri Lanka | |
| *Corresponding Author : | Kannangara S Department of Botany, Faculty of science University of Kelaniya, Kelaniya, Sri Lanka E-mail: sagarikadpk@kln.ac.lk |
| Received January 11, 2016; Accepted February 10, 2016; Published February 15, 2016 | |
| Citation: Undugoda LJS, Kannangara S, Sirisena DM (2016) Genetic Basis of Naphthalene and Phenanthrene Degradation by Phyllosphere Bacterial Strains Alcaligenes faecalis and Alcaligenes sp. 11SO. J Bioremed Biodeg 7:333. doi:10.4172/2155-6199.1000333 | |
| Copyright: © 2016 Undugoda LJS, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Two bacterial strains, Alcaligenes feacalis and Alcaligenes sp. 11SO isolated from the phyllosphere of four ornamental plant species, Ixora chinensis, Ervatamia divaricata, Hibiscus rosa-sinensis and Amaranthus cruentus found in five highly polluted sites in Sri Lanka, showed a higher level of phenanthrene and naphthalene degradation ability. Both these strains harbor plasmids conferring them resistance to ampicillin. Curing of these strains of their plasmid drastically reduced the ability to degrade the hydrocarbons. Upon transformation of these plasmids into E.coli JM109 enable it to degrade the two hydrocarbons efficiently. Plasmid encoded phenanthrene and naphthalene degradation suggested the presence of required catabolic genes in these plasmids. PCR amplification with degenerate primers and comparison of their nucleotide sequences with Genbank sequences indicated that plasmids of those bacterial strains harbor the genes nahR, nahU involved in naphthalene degradation and phnG for phenanthrene degradation. RFLP and nucleotide sequence comparison of nahU and nahR amplicons revealed that both of these genes in two bacterial strains are homologous. But, phnG gene copies of two bacterial strains exist as two distinct alleles.
| Keywords |
| Plasmid; nahR gene; Phyllosphere bacteria; Phenanthrene; Naphthalene; Alcaligenes sp.11SO |
| Introduction |
| Polyaromatic hydrocarbon (PAH) pollution is a highly concerned environmental problem in the world. Naphthalene and phenanthrene are the highly abundant PAHs in the ambient air due to the vehicular emission, industrial processes and oil refining processes. Naphthalene is the simplest polycyclic aromatic hydrocarbon member of a widespread, well-studied class of environmental pollutants [1,2]. PAHs in the air, deposit on the ground level by the wet deposition and dry deposition and phyllosphere is one of the major exposing surfaces to these PAHs. The microorganisms colonizing the phyllosphere of these polluted areas are able to degrade polyaromatic hydrocarbons. Out of many phyllosphere microorganisms, bacterial strains have been shown to degrade environmental contaminants and their degradation genes are often located on the catabolic plasmids [3]. For example, P. putida NCIB 9816 is a well-characterized bacterium capable of utilizing naphthalene as sole carbon and energy source [4-7]. This trait is conferred in P. putida NCIB 9816-4 by an 81 kb plasmid, pDTG1. That encodes key enzymes in early enzymatic steps in naphthalene degradation [5-8]. The structural genes encoding naphthalene-degrading pathway enzymes in a variety of bacteria are highly conserved [6,9-11]. |
| The NAH7 naphthalene catabolic plasmid in Pseudomonas putida G7 has been well characterized [6,8,12,13]. Analysis of the NAH7 plasmid [13] showed that nah operons are divided into two clusters. The genes of upper operon have genes encoding enzymes that convert naphthalene to salicylate and the lower operon has genes encoding enzymes that convert salicylate to tricarboxylic acid. |
| Isolated a phenanthrene degrading bacterium [14], Alcaligenes faecalis AFK2 which was able to degrade phenanthrene but it was unable to degrade naphthalene. The phenanthrene degradation genes of Alcaligenes faecalis AFK2 have since been sequenced and are unique with regard to both the gene organization and sequence similarity of genes when compared to other published sequences. Furthermore, the phn genes of Burkholderia sp. strain RP007 encode the enzymes for phenanthrene degradation [15]. |
| However in Sri Lanka there are no such recorded studies. So that the present study was carried out with an attempt to investigate the presence of nahR and nahU genes involved in naphthalene degradation and phnG gene required for phenanthrene degradation in Alcaligenes feacalis and Alcaligenes sp.11SO, phenanthrene and naphthalene degrading two bacterial strains isolated from the phyllosphere of some ornamental plants (Ixora chinensis, Ervatamia dervaticata, Hibiscus rosasinensis and Amaranthus cruentus). |
| Methods and Materials |
| Sampling sites |
| Leaves of four plant species, Ixora chinensis, Ervatamia dervaticata, Hibiscus rosasinensis and Amaranthus cruentus from five polluted sites in Sri Lanka, Colombo fort, Orugodawattha, Maradana, Panchikawattha and Sapugaskanda were collected to isolate PAH degrading phyllosphere bacteria. Meemure an isolated less polluted remote village was selected as control site. |
| Isolation of phyllosphere bacteria |
| Each leaf sample (4 g) was washed with 100 ml of phosphate buffer and then shaken at 200 rev/min for one hour. Then the diluted samples were directly added to the modified mineral salt agar plates. Plates were then incubated at room temperature (28°C-30°C) for five days. Bacterial colonies were streaked on PAH added Bacto-Bushnell Haas medium to select PAH utilizing bacteria. |
| Selection of efficient PAH degrading bacteria |
| The best PAH degrading bacterial strains were selected based on the results obtained from the colorimetric and HPLC methods indicated below. |
| Colorimetric assay |
| Each bacterial strain was inoculated into Bacto Bushnell-Haas broth incorporated with PAH compound (1%v/v) and Methylene blue (2%v/v), the redox indicator and incubated at room temperature (28°C-30°C) with constant shaking at 180 rev/min, for 14 days with a control without bacterial inoculation. From broth culture 5 ml sample was centrifuged at 6000 rev/min for five minutes. The recovered supernatant was assayed spectrophotometrically by measuring absorbance at 609 nm for the residual hydrocarbon. Six replicates were done for each bacterial strain and PAH degradation percentage was determined using the following equation [16]. |
| HPLC determination of PAH degradation |
| Each bacterial strain was inoculated into Bacto Bushnell-Haas broth incorporated with PAH (phenanthrene and naphthalene) compound (100 ppm). Then it was incubated at room temperature (28°C-30°C) with constant shaking at 180 rev/min, for 14 days with a control without bacterial inoculation. |
| Residual PAH in the culture was extracted with hexane and acetone containing mixture. Then extract was analyzed by high performance liquid chromatography (HPLC) equipped with UV detector. Analytical column (250 mm long, 4.6 mm diameter) was packed totally porous spherical C-18 material (packed size, 5μm). Acetonitrile-water mixture (75: 25) was used as mobile phase for PAHs at a flow rate of 1.0 ml min-1. Sample (20 μL) was injected into column through sample loop. UV - detector was set at 254 nm for compound detection. The Chromeleon chromatography software was used for quantification of PAHs throughout the experiment. Finally percentage of degradation was determined. |
| Isolation of catabolic plasmid and confirmation of plasmid encoded PAH degradation |
| Catabolic plasmids of Alcaligenes faecalis and Alcaligenes sp.11SO were isolated according to the method of [17] and each plasmid was transformed into the E.coli JM109. PAH degrading ability of transformants were tested using colorimetric and HPLC method to determine the plasmid based PAH degradation. Plasmid based PAH degradation was confirmed by curing the plasmid of bacterial cells using acridine orange [18] and then testing their PAH degradation ability by colorimetric and HPLC methods. |
| PCR amplification of nahR, nahU, phnG and phnAc regions in catabolic plasmid |
| Plasmid isolation, gel electrophoresis, transformation and amplification of DNA by PCR were performed by standard procedures [17]. The conserved regions of G7-nahR (P. putida G7), nahR (P. stutzeri AN10) and NCIB-nahR (P. putida NCIB 9816-4) (GenBank accession no.s M22723, AF039534 and AF491307, respectively) have been used to design the degenerate primers for PCR amplifying nahR genes. The conserved regions of pND6-nahU (P. putida ND6) (GenBank accession no AAP44249) genes has been used to design the degenerate primers for amplifying nahU gene and the conserved region of GZ38-phnG (P.putida G7) (GenBank accession no AF112137) has been used to design the degenerate primers of PCR for amplifying phnG genes. The conserved region of Burkholderia sp. strain RP007 (phnAc (AF061751) and Alcaligenes faecalis AFK2 (phnAc (AB024945) have been used to design phnAc primers. Sequence of these primers are, nahR-F, 5«-CGCGAATTCATGGAACTGCRTGAYCTGGA-3«,nahR-R,5«- CGCGAATTCTCAATCMGWAAACAGSTCRAAC-3«. nahU-F, 5<<- GGAGACATCATATGCAAAATTCTACTTCTGCTCTGAA- 3’,nahU-R, 5’-CCAATCTCGAGGGCCGCTTGCGCGC-3’, phnAc-F, 5<<- TTCGAGCTGGAATGTGAGC-3’, phnAc-R 5<<- AATAACCGGCGATTCCAAAC- 3’, phnG-F, 5<<- GGAAGGATCCGAATTCATTAAAGAGGAGAAATTAACTATGCACGCAGATACCGCGACCCGCCAGCACTGGATGTCCG- 3’, phnG-R, 5<<- TGGTTGGGATCCCGAGCCATGGTTATTAATGGTGATGGTGATGGTGTGCGTTGTCTCCGCGAACCATCGTAAAG AGTCGACCC A-3’. |
| Plasmid DNA templates of Alcaligenes faecalis and Alcaligenes sp.11SO were amplified using above primers. Then PCR products were visualized and their size estimation was done by gel electrophoresis. |
| Sequencing of PCR products |
| Automated sequencing was carried out using applied Biosystems automated sequencer (ABI3730XL) at Macrogen, Seoul, Korea. Purified PCR products representing nahR, nahU, and phnG gene segments were sequenced directly using appropriate primers. |
| Restriction digestion of PCR products |
| PCR amplicons of nahR, nahU and phnG genes were digested using HindIII restriction enzyme to determine their RFLP patterns. |
| Computer analysis |
| Nucleotide sequences of nahR, nahU and phnG gene segments were aligned with gene sequences of Genbank using BLAST [19]. |
| Results and Discussion |
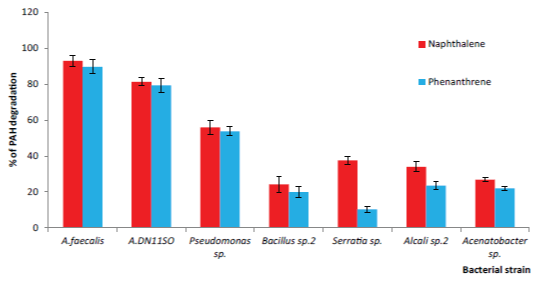
| PAH degrading phyllosphere bacterial population of the leaf samples collected from five highly polluted sites were much higher compared to that of less polluted control site. Colorimetric test and HPLC analysis results revealed that out of isolated 38 bacterial strains twenty could degrade one or both of tested two PAH compounds, naphthalene and phenanthrene. Eight of these PAH degraders belong to five genera Alcaligenes, Pseudomonas, Serratia, Bacillus and Acenatobacter which showed relatively higher degradation ability of these chemicals (Figure 1). Furthermore, A.feacalis (KT356811) had the highest naphthalene (92.9%) and phenanthrene (89.6%) degradation ability (Figure 1). Alcaligenes sp.11SO (KT356809) also had higher naphthalene (81.32%) and phenanthrene (79.24%) degradation ability compare to other bacterial strains. According to the literature [20] most of the naphthalene degraders were Pseudomonas sp. and Alcaligenes sp. were the predominant phenanthrene degraders. But the present investigation showed significantly high efficiencies of the two isolated Alcaligenes sp. in degrading both naphthalene and phenanthrene. |
| These two bacterial strains harbor an approximately 23 kb plasmid. Upon transformation of these plasmids into E.coli JM109 strain, it’s PAH degradation ability was similar to that of original organism. Further, after curing of plasmids, the two Alcaligenes sp. lost their PAH degradation ability. These results revealed that PAH degradation ability of Alcaligenes faecalis and Alcaligenes sp.11SO was a plasmid encoded character. Therefore, these plasmids should harbor naphthalene and phenanthrene catabolic genes nah and phn respectively. |
| nahR gene fragment analysis |
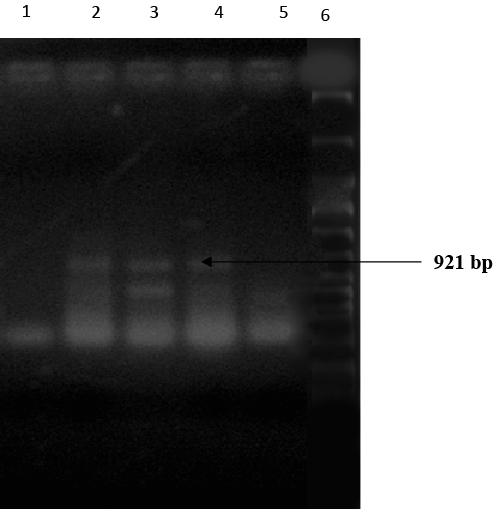
| Expected 921 bp nahR gene fragment [21] was observed in gel electrophoresis of PCR amplicons of nahR genes of Alcaligenes faecalis and Alcaligenes sp.11SO (Figure 2). Therefore, both of these Alcaligenes strains may harbor nahR gene on their catabolic plasmids. |
| According to the literature, nahR gene was predominant in the plasmids of Pseudomonas strains. For example Pseudomonas putida G7 haboured nahR gene on their NAH7 plasmid [22,23] P. putida NCIB 9816- 4, harboured nahR gene in their plasmid pDTG1 [6,7]. The two phyllosphere bacterial strains, Alcaligenes faecalis and Alcaligenes sp.11SO were the best AH degraders having plasmid born nahR genes responsible for naphthalene degradation. |
| PCR amplicons of nahR gene were sequenced and their nucleotide sequences were compared with the nucleotide sequences of nahR genes deposited in Genbank. Plasmid- haboured nahR gene of Alcaligenes sp.11SO had 79% sequence similarity to the nahR gene located on the pDTG1 plasmid of P. putida. Nucleotide sequence of nahR amplicon of Alcaligenes faecalis is almost close (88% sequence similarity) to the nahR gene of plasmid pND6 in Pseudomonas sp. nahR genes of two Alcaligens sp. were similar to the nahR gene located on the plasmid of Pseudomonas sp. Close sequence relationship of Alcaligenes nahR gene with the nahR genes of Pseudomonas sp. suggest that this gene may have transferred from Pseudomonas sp. to Alcaligenes sp. by conjugation at some stage of evolutionary process. |
| RFLP pattern obtained with HindIII digestion of nahR amplicon of Alcaligenes faecalis plasmid was similar to that of Alcaligenes sp.11SO plasmid. It revealed, nahR gene was homologous to each other and same gene type exists in these PAH degrading phyllosphere bacterial population. |
| nahU gene fragment analysis |
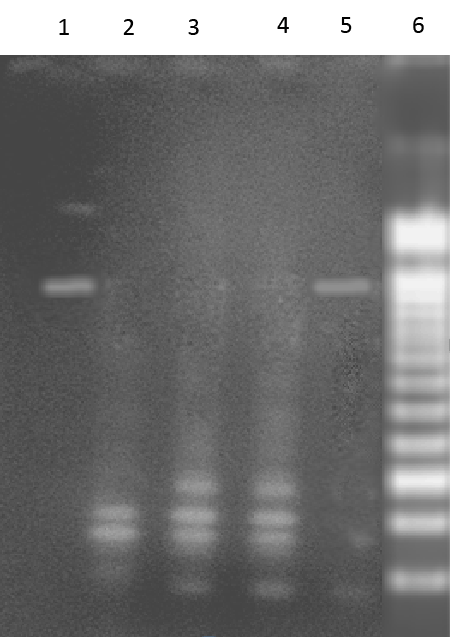
| Expected 712 bp nahU [20] gene fragment was observed in gel electrophoresis of PCR amplicons of both Alcaligenes faecalis and Alcaligenes sp.11SO. Therefore, both of these Alcaligenes strains harbor nahU gene on their catabolic plasmids. Presence of similar RFLP patterns in HindIII digest of these amplicons (Figure 3) indicating that both Alcaligenes sp. harbor similar nahU genes. nahU gene is an isofunctional gene located on the outside of the lower pathway operon [20]. |
| phnG gene fragment analysis |
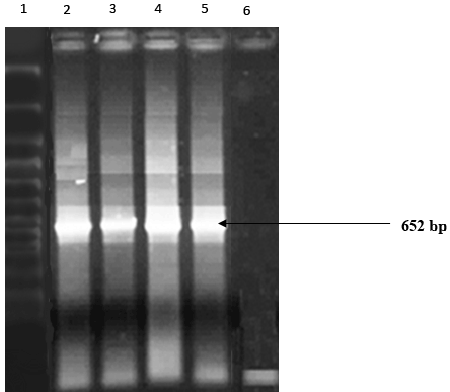
| Expected 652 bp [15] amplicons of phnG observed in PCR amplicons of two Alcaligenes sp. (Figure 4). Therefore, these two Alcaligenes spp. harbored phnG gene on their catabolic plasmids. Failures to amplify plasmid templates with phnAc-specific primers suggest the absence of phnAc gene on these catabolic plasmids. |
| All phnG amplicons of two bacterial strains were digested with HindIII enzyme. RFLP pattern of HindIII digest with phnG amplicon in Alcaligenes faecalis was different from that of Alcaligenes sp.11SO (Figure 5). Thus, phnG gene exists as two different alleles in these two strains enabling them to degrade phenanthrene. |
| The phyllosphere bacterial strains, Alcaligenes feacalis and Alcaligenes sp.11SO are different PAH degraders. Their phenanthrene and naphthalene degradation ability is a plasmid encoded character. Catabolic plasmids of these two strains harbored naphthalene specific nahR and nahU genes. The nahR and nahU genes of these two Alcaligenes sp. are homologous to each other. Since naphthalene is a simplest compound, it is easy to degrade to survive under harsh conditions [24,25]. Therefore, it may have limited allele types. But phenanthrene specific isofunctional gene phnG exists as two different allele types in the two species of Alcaligenes. Phenanthrene is a complex compound. Phenanthrene degradation ability of them was lower than naphthalene degradation. Therefore, they should have several variations in their genes to survive under harsh conditions. Therefore they may have different allele types. Although, these catabolic plasmids lack the gene phnAc, their ability to degrade phenanthrene suggests its chromosomal location in these strains. The ability of degradation of two chemicals (PAHs) by one organism is very important when applying them in to the bioremediation. Because, they can survive very well under the harsh conditions created by PAHs. |
References
|
Figures at a glance
 |
 |
 |
 |
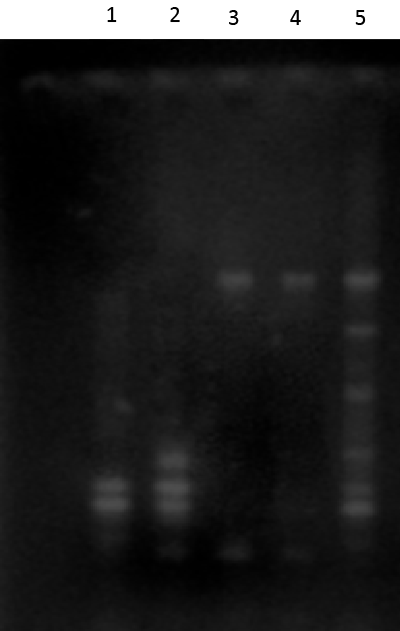 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 | Figure 5 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 12691
- [From(publication date):
March-2016 - Sep 03, 2025] - Breakdown by view type
- HTML page views : 11554
- PDF downloads : 1137
