Research Article Open Access
Isolation and In vitro Validation of Cardiac Muscle-Specific Promoters in Pigs
Sangsu Shin1,2#, Jinsoo Ahn1,3#, Yeunsu Suh1, Steven J. Moeller1, Seongsoo Hwang1, 4* and Kichoon Lee1,3*
1Department of Animal Sciences, The Ohio State University, Columbus, OH 43210, USA
2Department of Animal Biotechnology, Kyungpook National University, Sangju, Gyeongbuk 37224, Republic of Korea
3The Ohio State University Interdisciplinary Ph.D. Program in Nutrition, The Ohio State University, Columbus, OH 43210, USA
4Animal Biotechnology Division, National Institute of Animal Science, RDA, Wanju-gun, Jeonbuk 55365, Republic of Korea
- *Corresponding Author:
- Seongsoo Hwang
Animal Biotechnology Division, National
Institute of Animal Science, Republic of Korea
Tel: +82-63-238-7253
Fax: +82-63-238-7297
E-mail: hwangss@korea.kr
Kichoon Lee
Department of Animal Sciences
The Ohio State University, Columbus, OH 43210, USA
Tel: 614-688-7963
Fax: 614-292-2929
E-mail: lee.2626@osu.edu
Received date: April 23, 2016; Accepted date: May 10, 2016; Published date: May 15, 2016
Citation: Shin S, Ahn J, Suh Y, Moeller SJ, Hwang S, et al. (2016) Isolation and In vitro Validation of Cardiac Muscle-Specific Promoters in Pigs. Cell Mol Biol 62:123. doi: 10.4172/1165-158X.1000123
Copyright: © 2016 Shin S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Cellular and Molecular Biology
Abstract
Identification of promoter sequences that can drive heart-specific expression of transgenes is essential to investigate gene function in the heart. The aim of this study was to identify heart-specific genes and their promoter sequences that can promote heart-specific transgene expression in pig primary heart cells. Gene expression profiles in the Gene Expression Omnibus (GEO) repository have been integrated and utilized to identify MYH6, TNNI3, and MYBPC3 as common heart-specific genes in the mouse and human. RT-PCR further confirmed heartspecific expression of the genes among various tissues in pigs as well as in mice and humans. Bioinformatics analysis predicted that their promoter sequences contain multiple binding sites for transcription factors involved in cardiogenesis and the promoter sequences were substantially conserved between pigs and humans. In addition, in vitro analysis showed that expression of (i.e., the green fluorescence protein (GFP)) reporter gene under the regulation of promoter sequences of MYH6, TNNI3, and MYBPC3 was detected in primary pig heart cells but not in other primary cells. These findings, along with the physiologic similarities between humans and pigs, suggest that these novel promoters may be valuable candidates for the regulation of heart gene expression in both humans and pigs for biomedical purposes.
Keywords
Pig; Heart; Tissue-specific promoter; MYH6; TNNI3; MYBPC3
Abbreviations
GEO: Gene Expression Omnibus; GFP: Green Fluorescence Protein; MYH6: Myosin Heavy Chain 6; TNNI3: Troponin I Type 3 (Cardiac); MYBPC3: Myosin Binding Protein C; Cardiac; NKX2.5: NK2 Homeobox 5; GATA4: GATA Binding Protein 4; TBX5: T-Box 5; RFP: Red Fluorescence Protein
Introduction
The identification of differentially expressed genes and their promoters, which control gene expression in a tissue-specific manner, is of great importance for the investigation of transgene function and generation of transgenic animals for xenotransplantation. Gene expression profiles generated with cDNA microarrays in the public Gene Expression Omnibus (GEO) repository have been integrated and utilized to verify a large number of differentially expressed genes and discovered novel, tissue-specific genes as described in our previous studies [1,2]. The abundance of mRNA reported in the GEO repository is a useful indicator of tissue-specific patterns of gene expression. In the current study, genes differentially expressed in the heart were investigated for the discovery of novel promoter sequences that can drive heart-specific transgene expression. Selection of promoter sequences is crucial for overcoming a major limitation of cardiac gene transfer, namely ectopic expression of the transgene. Tissue-specific promoters have been widely used to generate transgenic animals in our previous studies. For example, aP2/FABP4 promoter, albumin promoter, and insulin promoter have been used to overexpress DLK1/ Pref-1 in fat, liver, and pancreatic β-cells, respectively, of transgenic mice [3,4]. In the avian species, the RBP7 gene was discovered as a novel adipose tissue-specific gene, and its promoter has been utilized in producing transgenic quail expressing a reporter gene specifically in adipose tissue [5]. For generating cardiac-specific transgenic mice, α-MHC/MYH6 promoter has been used [6-9]; however, its heartspecific gene regulation in pigs has not been investigated.
The objectives of the present study were to identify differentially expressed genes in the mouse and human heart, evaluate their expression in pig hearts, analyze their promoter sequences for the regulation of heart-specific expression, and investigate expression of a target gene (green fluorescence protein, GFP) in pig primary heart cells under the regulation of the promoters. The identification of promoters that can drive expression of therapeutic genes, specifically in the heart, will direct the generation of transgenic pigs for biomedical applications including transgenic pigs as animal models for human heart diseases and xenotransplantation [10,11].
Materials and Methods
General information
Our study protocol and standard operating procedures for the treatments of the animals used were reviewed and approved by the Institutional Animal Care and Use Committee of the Ohio State University.
Data mining from the GEO datasets
The procedures for data mining and sources of microarray data obtained from the GEO DataSets (GDS) in the NCBI web site were described in our previous report [1]. Two sources of DataSets: GDS3142 for the mouse containing a total of 70 expression profiles for 22 tissues with an average replication of three for individuals for each tissue, and GDS596 for the human containing expression profiles for 79 tissues with duplication for each tissue, were used in the present study. Among the various tissues in each GDS source, the expression profiles for seven and six tissues for the mouse and human, respectively, were compared for selecting heart-specific genes. Heart-specific genes were determined as described previously [1]: i) gene expression values for each tissue were averaged, ii) the averaged values of heart tissue were divided by an average value for all other tissues, iii) the averaged results were sorted in descending order representing a higher value with a greater heartspecific expression, and iv) highly ranked genes in both the human and mouse were selected for further statistical analysis.
Tissue sampling, RNA extraction, cDNA synthesis, and RTPCR
Human RNAs from the kidney, liver, lung, heart, and muscle were purchased from Agilent Technologies (Santa Clara, CA, USA) and human adipose RNA from Clontech Laboratories (Mountain View, CA, USA). Mouse and pig RNAs were extracted from the tissues of 3-monthold, ICR (CD-1) outbred line (Harlan Laboratories, Indianapolis, IN) and 120-day-old Landrace, respectively. Trizol reagent (Invitrogen, Carlsbad, CA, USA) was used for extracting total RNA from adipose tissue, muscle, heart, lung, liver, kidney, spleen, and intestine of mice and pigs according to the manufacturer’s instructions. The quality of extracted RNA was assessed via gel electrophoresis, and quantity of total RNA was measured with a Nanodrop spectrophotometer (NanoDrop Technologies, Wilmington, DE). Reverse transcription for the generation of cDNA was performed with the following conditions, 65°C for 5 min, 37°C for 52 min and 70°C for 15 min, using 0.5μg of total RNA and Moloney murine leukemia virus reverse transcriptase (Invitrogen). For the semi-quantitative PCR of TNNI3, MYBPC3, and MYH6 expression in several tissues of mice, humans, and pigs, primers for each gene listed in Supplementary Table 1 were used. Conditions for the PCR using AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA) were 95°C for 10 min and 25, 27, 29, or 31 cycles of 94°C for 15 s, 58°C for 40 s, 72°C for 30 s, and 82°C for 32 s to get a linear amplification of PCR products. The PCR products after 29 cycles were separated in a 1% agarose gel, and images of gels were captured by a geldocumentation system (Alpha Inotech Corporation, San Leandro, CA). The cyclophilin (CYC) gene was used as a housekeeping control for an equal amount of cDNA used for PCR reaction.
Analysis of binding sites of cardiac muscle-specific transcription factors
The DNA sequences of 5 kb upstream regions from the start codon of three genes were obtained from the human, mouse, and pig genome browser at the UCSC Genome Bioinformatics site (http://genome.ucsc. edu). With these DNA sequences, the binding sites of transcription factors were predicted using the MatInspector program (Genomatix Software GmbH, Munich, Germany). Among the transcription factors, the cardiac muscle-specific transcription factors, NKX2.5, GATA4, and TBX5, were selected and marked on the sequences to select the possible promoter regions.
Cloning of pig cardiac muscle-specific promoters
Among the 5 kb sequences, 1.2 kb, 2 kb, and 3.6 kb of the 5’ upstream region of TNNI3, MYBPC3, and MYH6 genes, respectively, were selected for promoters, as the promoter regions were conserved among the species and had multiple binding sites of cardiac musclespecific transcription factors. The pig genomic DNA was extracted from the muscle tissues by using the Gentra Puregene Tissue Kit (Qiagen, Venlo, Netherlands) and following the manufacturer’s instruction, and then used as a template for PCR. To clone the promoters, each promoter was amplified from the pig genomic DNA by PCR with each primer set of TNNI3-pro, MYBPC3-pro, and MYH6-pro that are identified in Supplementary Table 1. Each PCR product was directly cloned into the pGlow-TOPO TA cloning vector (Invitrogen) for further studies.
| Species | Rank | Gene | Expression levels of the genes in tissues | Folda | P value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spleen | Muscle | Liver | Lung | Kidney | Adipose | Heart | |||||
| Mouse | 1 | PLN | 74 ± 1 | 112 ± 8 | 75 ± 8 | 185 ± 8 | 102 ± 3 | 94 ± 9 | 14565 ± 615 | 136.2 | <0.0001 |
| 2 | MYH6 | 70 ± 2 | 83 ± 2 | 73 ± 2 | 839 ± 124 | 71 ± 2 | 73 ± 3 | 19317 ± 1430 | 95.8 | <0.0001 | |
| 3 | TNNI3 | 94 ± 2 | 100 ± 5 | 86 ± 4 | 374 ± 26 | 86 ± 2 | 89 ± 4 | 12016 ± 156 | 87 | <0.0001 | |
| 4 | NPPB | 84 ± 4 | 102 ± 2 | 119 ± 5 | 97 ± 5 | 98 ± 3 | 121 ± 6 | 7339 ± 629 | 70.9 | <0.0001 | |
| 5 | FHL2 | 158 ± 9 | 161 ± 2 | 140 ± 5 | 173 ± 6 | 279 ± 7 | 146 ± 7 | 8829 ± 429 | 50.2 | <0.0001 | |
| 6 | MYH7 | 90 ± 5 | 1133 ± 157 | 104 ± 5 | 313 ± 33 | 90 ± 2 | 90 ± 6 | 13749 ± 161 | 45.3 | <0.0001 | |
| 7 | MYL2 | 81 ± 3 | 2634 ± 315 | 84 ± 4 | 82 ± 8 | 80 ± 7 | 84 ± 3 | 22237 ± 370 | 43.8 | <0.0001 | |
| 8 | MYBPC3 | 109 ± 2 | 128 ± 4 | 116 ± 13 | 166 ± 6 | 115 ± 2 | 122 ± 5 | 5316 ± 204 | 42.2 | <0.0001 | |
| 9 | NPPA | 85 ± 3 | 83 ± 3 | 88 ± 7 | 261 ± 19 | 77 ± 4 | 80 ± 2 | 4431 ± 361 | 39.5 | <0.0001 | |
| 10 | RYR2 | 72 ± 2 | 74 ± 3 | 69 ± 3 | 128 ± 6 | 70 ± 1 | 68 ± 2 | 2889 ± 51 | 36.1 | <0.0001 | |
| Human | 1 | TNNI3 | NA | 4884 ± 1696 | 2165 ± 265 | 1877 ± 143 | 2056 ± 181 | 1655 ± 390 | 381987 ± 30441 | 151.2 | <0.0001 |
| 2 | SLC4A3 | NA | 921 ± 241 | 198 ± 45 | 124 ± 12 | 231 ± 33 | 130 ± 18 | 19470 ± 4416 | 60.7 | <0.0001 | |
| 3 | TNNT2 | NA | 4233 ± 2167 | 1086 ± 354 | 462 ± 36 | 1562 ± 144 | 1136 ± 471 | 98686 ± 15502 | 58.2 | <0.0001 | |
| 4 | ACTC1 | NA | 17644 ± 750 | 3124 ± 1120 | 2256 ± 804 | 4831 ± 1210 | 1888 ± 555 | 290000 ± 22478 | 48.8 | <0.0001 | |
| 5 | MYBPC3 | NA | 4812 ± 2986 | 2474 ± 1636 | 525 ± 52 | 3212 ± 1836 | 613 ± 69 | 76667 ± 1990 | 32.9 | <0.0001 | |
| 6 | NDRG4 | NA | 1036 ± 219 | 355 ± 9 | 1433 ± 22 | 529 ± 158 | 286 ± 33 | 21423 ± 566 | 29.5 | <0.0001 | |
| 7 | MYH6 | NA | 10985 ± 2647 | 287 ± 48 | 531 ± 155 | 176 ± 27 | 127 ± 70 | 56135 ± 3208 | 23.2 | <0.0001 | |
| 8 | FHL2 | NA | 9967 ± 2088 | 1490 ± 912 | 2704 ± 72 | 4484 ± 1475 | 5450 ± 88 | 108298 ± 1964 | 22.5 | <0.0001 | |
| 9 | ITGB1BP3 | NA | 11047 ± 839 | 1730 ± 582 | 1252 ± 467 | 913 ± 207 | 1573 ± 770 | 63148 ± 6895 | 19.1 | <0.0001 | |
| 10 | PTGDS | NA | 8975 ± 978 | 7032 ± 289 | 23824 ± 3615 | 6307 ± 783 | 2037 ± 201 | 154356 ± 25658 | 16 | <0.0001 | |
Table 1: Comparison of the gene expression levels in various tissues of mouse and human.
Cell culture and transfection into pig primary cells
For cell culture, the tissues of fat, muscle, heart, liver, lung, kidney, and spleen were collected from 28-day-old Landrace. The tissues were washed several times with PBS containing 100 U/ml penicillin and 100 μg/ml streptomycin. To dissociate the cells, tissues were incubated with Hank’s Balanced Salt Solution (HBSS) (Gibco, Grand Island, NY, USA) containing 5 mg/ml of collagenase, Type II (Gibco) for the heart and 3.2 mg/ml of collagenase, Type I (Gibco) for the rest of the tissues at 37°C for 30 - 60 min by shaking the tubes several times. Then, the collagenase was inactivated by adding fetal bovine serum (FBS, 10% final, Gibco LOT1584266), and the cells were filtrated through the cell strainer (70 μm diameter; BD Bioscience, Franklin Lakes, NJ, USA). The cells that passed the cell strainer were collected by centrifugation and then seeded into 6-well cell culture plates (Greiner Bio-One North America, Inc., Monroe, NC, USA) coated with collagen or gelatin depending on cell type. The cell culture medium was based on Dulbecco Modified Eagle Medium (DMEM) (Gibco) supplemented with 10% FBS, 100 U/ ml penicillin and 100 μg/ml streptomycin. When the cells reached 90% confluency, the cells were co-transfected with a pGlow vector containing each promoter and pLKO.1-puro-CMV-TagRFP (Sigma) as a positive control by using Lipofectamine® 2000 (Invitrogen). After two days, cell pictures were taken under a fluorescent microscope (AXIO-Vert. A1 equipped with an AxioCam MRc5 camera; Carl Zeiss Microscopy, Thornwood, NY, USA) and lysed to collect protein samples.
Western blot analysis
Western blotting was performed as described in our previous report [4]. In detail, proteins isolated from cell lysates were separated on 10% SDS-PAGE and then electrophoretically transferred onto polyvinylidene difluoride (PVDF) membranes (Amersham Biosciences Hybond-P; Amersham Biosciences, Piscataway, NJ). After blocking for 30 min in 4% nonfat dry milk in 1× Tris-buffered saline (TBS) containing 0.05% Tween-20 (TBST), the membranes were incubated overnight at 4°C with a primary antibody specifically binding to amino acids 3-17 of GFP (Sigma-Aldrich, St Louis, MO) and to full-length RFP (ThermoFisher Scientific, Waltham, MA) in 4% nonfat dry milk. The membranes were washed with 1×TBST and incubated with the corresponding HRP-conjugated secondary antibody for 1 h at room temperature. After washing with 1×TBST, Amersham ECL plus Western Blotting Detection Reagents (GE Healthcare Biosciences, Pittsburgh, PA) was applied on the membrane, and the blots were developed onto Hyperfilm (GE Healthcare Biosciences).
Statistical analysis
A comparison of multiple means of mRNA expression in mouse and human tissues was conducted by using one-way ANOVA followed by a Fisher’s protected least significant difference test. Densitometry of the Western blots was analyzed with the Image J software (NIH). The differences of GFP and RFP protein expression were evaluated by using one-way ANOVA followed by Tukey’s post hoc test at P<0.05. All statistical analyses were performed using SAS 9.4 software (SAS Institute, Inc., Cary, NC).
Results
Microarray analysis of heart-specific genes in mice and humans
By analyzing the microarray data of mouse and human genes in various tissues obtained from the GEO Datasets, the ten most highly expressed genes in either mice or humans were collected and listed by fold changes (Table 1). Among the ten genes, MYH6, TNNI3, MYBPC3, and FHL2 were present in both lists. Following ranks in the human, the top three expressed genes (TNNI3, MYBPC3, and MYH6) were selected as candidate genes commonly expressed in the heart (Table 1). In the mouse, expression of TNNI3, MYBPC3, and MYH6 genes were 87.0, 42.2, and 95.8 folds greater in the heart, respectively, than the average of other tissues. In the human, the three genes expressed 151.2, 32.9, and 23.2 folds greater in the heart, respectively.
Confirmation of the heart-specific expression and selection of pig cardiac muscle-specific genes
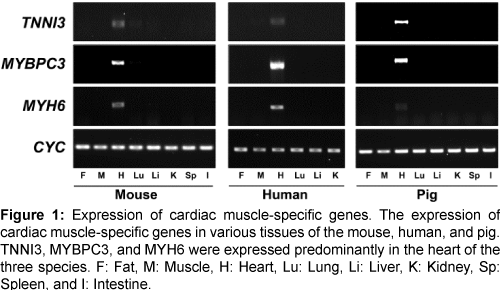
The expression of the three genes (TNNI3, MYBPC3, and MYH6) was confirmed in various tissues of the mouse, human, and pig by reverse transcription-PCR (RT-PCR) (Figure 1). All three genes were highly expressed in heart tissues of each of the three species and did not, or were barely expressed, in other tissues. Therefore, our comparative analysis of microarray and RT-PCR led to identification of common, heart-specific genes in the mouse, human, and pig.
Figure 1: Expression of cardiac muscle-specific genes. The expression of cardiac muscle-specific genes in various tissues of the mouse, human, and pig. TNNI3, MYBPC3, and MYH6 were expressed predominantly in the heart of the three species. F: Fat, M: Muscle, H: Heart, Lu: Lung, Li: Liver, K: Kidney, Sp: Spleen, and I: Intestine.
Promoter analysis
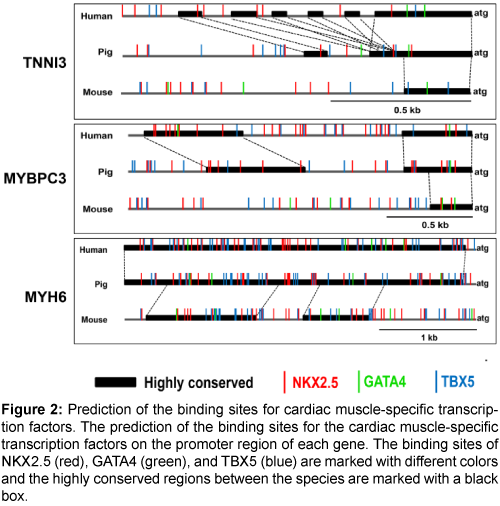
Next, the binding sites of the cardiac muscle-specific transcription factors, NKX2.5, GATA4, and TBX5, were predicted (Figure 2). There were multiple transcription factor binding sites for NKX2.5, GATA4, and TBX5 on the promoter regions of the three genes, and the promoter regions were more highly conserved between pigs and humans than between pigs and mice (Supplementary Figure 1). Therefore, the promoters of the three genes could be valuable candidates for a cardiacmuscle specific promoter.
Figure 2: Prediction of the binding sites for cardiac muscle-specific transcription factors. The prediction of the binding sites for the cardiac muscle-specific transcription factors on the promoter region of each gene. The binding sites of NKX2.5 (red), GATA4 (green), and TBX5 (blue) are marked with different colors and the highly conserved regions between the species are marked with a black box.
The expression of green fluorescence protein (GFP) by cardiac muscle-specific promoters in pig cardiac muscle cells
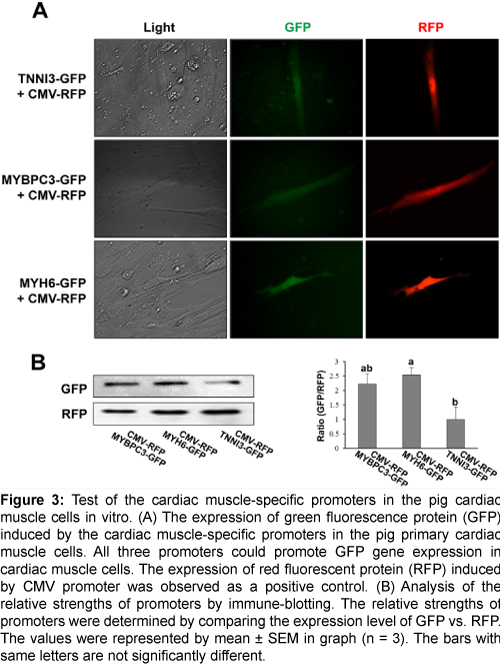
To confirm the capability as a cardiac muscle-specific promoter, the above three candidate promoters were tested in vitro with various pig tissues. The 1.2 kb, 2 kb, and 3.6 kb of the 5’ upstream region of TNNI3, MYBPC3, and MYH6 genes, respectively, were selected for a promoter and inserted into the pGlow vector to test the cardiac muscle-specific expression. Each vector was co-transfected with CMV-RFP vector which could express red fluorescent protein initiated by CMV promoter into various pig primary cells including the heart, fat, kidney, lung, liver, spleen, and muscle. The expression of GFP was observed only in the heart primary cells, while the RFP was observed in all primary cells (Figure 3A, and Supplementary Figures 2A-2F); suggesting that all three promoters can initiate the expression of a gene specifically in the heart. Western blot analysis showed that GFP expression was stimulated under the regulation of MYH6, MYBPC3, and TNNI3 promoters, and the strength of the promoters may be in the descending order of MYH6, MYBPC3, and TNNI3 (Figure 3B). Collectively, all three promoters are able to initiate the gene expression in a cardiac cell-specific manner in vitro.
Figure 3: Test of the cardiac muscle-specific promoters in the pig cardiac muscle cells in vitro. (A) The expression of green fluorescence protein (GFP) induced by the cardiac muscle-specific promoters in the pig primary cardiac muscle cells. All three promoters could promote GFP gene expression in cardiac muscle cells. The expression of red fluorescent protein (RFP) induced by CMV promoter was observed as a positive control. (B) Analysis of the relative strengths of promoters by immune-blotting. The relative strengths of promoters were determined by comparing the expression level of GFP vs. RFP. The values were represented by mean ± SEM in graph (n = 3). The bars with same letters are not significantly different.
Discussion
In this study, we cloned three heart-specific promoters from the pig genome and confirmed the availability of the promoters in vitro, assessing whether they could restrict the foreign gene expression to the pig heart. As previously shown in other studies, comparing the gene expression profile in many tissues and several species was a useful approach to select the candidate promoters showing tissue-specificity [1]. Among the top 10 genes expressed abundantly in heart tissue, three genes, TNNI3, MYBPC3, and MYH6, were nominated in both the mouse and human. The specific expression of the genes was tested by RT-PCR, and all three genes were detected specifically in heart tissues of three species including the mouse, human, and pig. Three promoters were selected based on a well-received concept that promoters of tissue-specific genes have cis-acting elements on which tissue-specific transcription factors bind to drive gene expression in a specific tissue. There are three well-known transcription factors (NKX2.5, GATA4, and TBX5) that are called the core cardiac transcription factors in the heart. These transcription factors are involved in heart development, cardiomyocyte proliferation and differentiation, and heart function (reviewed by Accornero et al. [6]). In the present study, there were many putative binding sites for those transcription factors on the candidate promoters. The ratio of binding site numbers of each transcription factor was similar in all promoters and the binding sites of NKX2.5 and TBX5 were more abundant than GATA4 (ratio = 8:1). Of note, a large part of the promoter region was conserved between the pig and human, especially the MYH6 promoter, which was more highly conserved in the pig and human than the other promoters even though its size was larger than the other promoters. In the human TNNI3 promoter, there were several repetitive satellite sequences corresponding to a site on the pig promoter. All results of the present study support the three promoters as good candidates for the heart-specific promoter in future studies.
The heart-specific promoter may be a useful tool to study gene function for specifically over-expressing or knocking-out genes in the heart. The specific expression of GFP driven by three promoters was tested in various tissues of pigs in vitro, and it was confirmed that all three promoters have the capability to express GFP specifically in primary heart muscle cells. Interestingly, the expression of GFP driven by these heart-specific promoters was not observed in the skeletal muscle cells. This finding suggests that three promoters could regulate the expression of genes strictly in heart muscle cells; a finding that will need to be confirmed in vivo in future studies. In conclusion, three heart-specific promoters were identified from the pig by analysing the gene expression profiles deposited in GEO. These promoters successfully drove the expression of the GFP gene specifically in heart muscle cells in vitro. Therefore, these promoters might be useful for driving expression of target genes in the heart in vivo for biomedical applications such as developing human heart disease models and heart xenograft.
References
- Song Y., Ahn J., Suh Y., Davis M.E. & Lee K. Identification of novel tissue-specific genes by analysis of microarray databases: a human and mouse model. PLoS ONE 2013, 8:e64483.
- Zhang J., Ahn J., Suh Y., Hwang S., Davis M.E. & Lee K. Identification of CTLA2A, DEFB29, WFDC15B, SERPINA1F and MUP19 as novel tissue-specific secretory factors in mouse. PLoS ONE 2015, 10:e0124962.
- Lee K., Villena J.A., Moon Y.S., Kim K.H., Lee S., Kang C. & Sul H.S. Inhibition of adipogenesis and development of glucose intolerance by soluble preadipocyte factor-1 (Pref-1). J. Clin. Invest. 2003, 111:453-461.
- Wang Y., Lee K., Moon Y.S., Ahmadian M., Kim K.H., Roder K., Kang C. & Sul H.S. Overexpression of Pref-1 in pancreatic islet beta-cells in mice causes hyperinsulinemia with increased islet mass and insulin secretion. Biochem. Biophys. Res. Commun. 2015, 461:630-635.
- Ahn J., Shin S., Suh Y., Park J.Y., Hwang S. & Lee K. Identification of the avian RBP7 gene as a new adipose-specific gene and RBP7 promoter-driven GFP expression in adipose tissue of transgenic quail. PLoS ONE 2015, 10:e0124768.
- Accornero F., van Berlo J.H., Correll R.N., Elrod J.W., Sargent M.A., York A., Rabinowitz J.E., Leask A. & Molkentin J.D. Genetic analysis of connective tissue growth factor as an effector of transforming growth factor beta signaling and cardiac remodeling. Mol. Cell. Biol. 2015, 35:2154-2164.
- Liu X., Wang X., Bi Y., Bu P. & Zhang M. The histone demethylase PHF8 represses cardiac hypertrophy upon pressure overload. Exp. Cell Res. 2015, 335:123-134.
- Subramaniam A., Jones W.K., Gulick J., Wert S., Neumann J. & Robbins J. Tissue-specific regulation of the alpha-myosin heavy chain gene promoter in transgenic mice. J. Biol. Chem. 1991, 266:24613-24620.
- Zieseniss A., Hesse A.R., Jatho A., Krull S., Holscher M., Vogel S. & Katschinski D.M. Cardiomyocyte-specific transgenic expression of Prolyl-4-Hydroxylase Domain 3 impairs the myocardial response to ischemia. Cell. Physiol. Biochem. 2015, 36:843-851.
- Li X., Suh Y., Kim E., Moeller S.J. & Lee K. Alternative splicing and developmental and hormonal regulation of porcine comparative gene identification-58 (CGI-58) mRNA. J. Anim. Sci. 2012, 90:4346-4354.
- McCulley D.J. & Black B.L. Transcription factor pathways and congenital heart disease. Curr. Top. Dev. Biol. 2012, 100:253-277.
Relevant Topics
- Biomolecular Structure and Function
- Cell Biology Junctions
- Cell Biology Techniques
- Cell Cycle
- Cell Death: Apoptosis
- Cell Regeneration
- Cell synthesis
- Cellular and Molecular Biology
- Cellular Biology
- Cellular DNA Studies
- Cellular Dynamics
- Cellular Signalling
- Gene Expression and Regulation
- Methods and Techniques in Molecular Biology
- Molecular Biochemistry
- Molecular Biotechnology
- Molecular Cell
- Molecular Genetics
- Stem Cell Biology
Recommended Journals
- Air and Water Borne Diseases
- Cellular and Molecular Biology
- Archives of Parasitology
- Archives of Science
- Biochemistry & Physiology: Open Acess
- Clinical Pharmacology & Biopharmaceutics
- Diagnostic Pathology: Open Access
- Diagnostic Pathology: Open Access
- Epidemiology: Open Access
- Epidemiology: Open Access
- Journal of Biochemistry and Cell Biology
- Journal of Cell Biology & Immunology
- Journal Cell Science and Apoptosis
Article Tools
Article Usage
- Total views: 12655
- [From(publication date):
May-2016 - Jul 16, 2025] - Breakdown by view type
- HTML page views : 11586
- PDF downloads : 1069