Research Article Open Access
Isolation of Streptomyces Species from Soil and its Medium Optimization for Microbial Transglutaminase Production by Box-Behnken Design
Gopal Samy B1*, Sujitha S2, Thyagarajan R3 and Jegatheesan K41Liatris Biosciences LLP, Near CSEZ, Kakkanad, Cochin- 682037, Kerala, India
2Department of Biotechnology, St. Michael College of Engg and Tech, Kalayarkoil, Tamil Nadu, India
3Department of Biotechnology, Sathyabama University, Chennai, Tamil Nadu, India
4Center for Research and PG Studies in Botany and Department of Biotechnology Thiagarajar College (Autonomous) Madurai - 625 009, Tamilnadu, India
- *Corresponding Author:
- Gopal Samy
Scientist and CSO, Liatris Biosciences LLP
Near CSEZ, Kakkanad, Cochin- 682037
Kerala, India
Tel: +91 98435 67218
E-mail: gopalsamy2k6@gmail.com
Received: October 09, 2015; Accepted: December 15, 2015; Published date: December 21, 2015
Citation: Samy GB, Sujitha S, Thyagarajan R, Jegatheesan K (2015) Isolation of Streptomyces Species from Soil and its Medium Optimization for Microbial Transglutaminase Production by Box-Behnken Design. J Ecosys Ecograph 6:175. doi:10.4172/2157-7625.1000175
Copyright: © 2015 Samy GB, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Ecosystem & Ecography
Abstract
Transglutaminase (E.C. 2.3.2.13) are a family of enzymes that catalyze the covalent bond formation between open amine groups. They are widely used in food industries and their demand rises daily. Though they are available in mammalian tissues, fish and plants, the complex separation and purification process led to the search of Microbial Transglutaminase (MTGase). Finding a new microbial source of transglutaminase and the medium composition for MTGase production were the purpose of this work. Six Different types of Actinomycetes like strains were isolated from soil sample and two of them named PG03 and PG06 were selected based on their ability to produce 23 mg/ml and 21 mg/ml MTGase enzyme respectively. Strain PG03 was chosen for further studies and it was found to be a Streptomyces species. Standard enzyme production media composition is modified and tested to facilitate the optimized MTGase activity. Strategies like finest nitrogen and carbon source selection, revealing the key ingredients of media by full factorial design and their optimal concentration Box-Behnken design were adopted. At the 95% confidence level, second order polynomial model was applied to fit the research outcome. Under the proposed optimized conditions, the model predicted a transglutaminase yield of 21.7 mg/ml, very closely matching the experimental value of 24.1 mg/ml. The F-test was greater than the table value of 2.82 and the p-value of 0.004 clearly reveals that this regression was statistically significant at the 95% confidence level. Further, the proposed model has the ability to elucidate 48.8% response variation as indicated by the R2 of the regression value.
Keywords
Box-Behnken design; Medium optimization; Microbial transglutaminase; Plackett-Burman design; Streptomyces sp
Introduction
Transglutaminase (TGase) is an enzyme that catalyses an acyl transfer reaction using peptide-bond glutamine residues as acyl donors and several primary amines as acceptors. When the ε-amino groups of the protein-bond lysyl residues are present as acyl receptors, this enzyme transglutaminase is capable of forming intra and intermolecular ε-(γ-Glu)-Lys isopeptide bonds [1]. The covalent crosslinks between a number of proteins and peptides introduced by transglutaminase promote the modification of the food proteins [2]. Therefore transglutaminase catalyzed reactions may be broadly used by food processing industries, for instance, the creation of new product textures, the modification of viscosity, the alteration of emulsifying and foaming properties, and the product nutritional value [3,4].
Insoluble and extensively cross-linked protein polymers essential for the organism to create barriers like blood clots and stable structures like hair and skin are generally produced by transglutaminases. The catalytic reaction is intimately observed with extensive control system as it is irreversible [5]. The reaction catalyzed by Transglutaminase is given as follows:
Peptide-Lysine + Glutamine-Peptide  Peptide- Lysine=Glutamine-Peptide
Peptide- Lysine=Glutamine-Peptide
Industrial Applications
Transglutaminase catalyzed reactions are broadly used by food processing industries in hot dogs, sausages, rationalized steaks for fastening little meat portions into a large one and in improving the quality of low-grade meat like pale, soft, and exudative (PSE) meat. It is also used in making milk and yogurt creamier and noodles stiffer. Transglutaminase is also used to produce some unusual foods.
Transglutaminases are found in mammalian tissues, plasma, fish and plants [6]. Eight transglutaminases have been characterized till date and they are Factor XIII (fibrin stabilizing factor), keratinocyte transglutaminase, Tissue transglutaminase, epidermal transglutaminase, Prostate transglutaminase, TGM X, TGM Y and TGM Z [5]. The mammalian enzymes are Ca2+-dependent. However, the relatively small quantities obtained and the complex separation and purification procedures required for these enzymes led to the search for alternative microbiological sources. The first microbial transglutaminase (MTGase) characterized was from an Actinomycetes. Since then, efforts have been made to obtain massive production of this enzyme for commercial applications, especially for the enzymes from Streptomyces.
Streptomyces consists of a vegetative hyphae approximately 0.5 – 2.0 μm in diameter, producing an extensively branched mycelium that rarely fragments. Colonies at the outset are pretty smooth surfaced and soon after build up aerial mycelium weft that looks like powdery, floccose, velvety or granular. They are gram positive but not acidalcohol fast, catalase positive, degrade L-tyrosine, starch, hypoxanthine, gelatin, esculin, casein, adenine and nitrate reduction to nitrite. They are abundantly distributed and in soil as well as composts.
So far, research has been focused on the isolation and screening of microorganisms for transglutaminase activity, and on purifying and characterizing newly found enzymes. The media compositions used to produce microbial transglutaminase from Streptomyces have been almost the same in every work published since Ando et al. [7]. The formulation of the culture media is of grave importance in industrial biotechnology processes as it influences volumetric productivity, product yield and concentration. Reducing medium cost is vital as it impinge on the whole process economics.
The traditional one-at-a-time optimization strategy is simple and constructive in screening and the individual medium component effects are observed on a chart not including complicated statistical analysis.
As the combined effects on the response are not considered, this simple technique often fell short in finding the optimum response section. The lack of understanding about complicated interactions between different factors leads to complications and qualms related with large scale fermentation. Statistically based experimental designs provide an efficient approach to optimization. The Plackett-Burman Design is especially appropriate in determining the most important components in the medium that make up the interactions.
An amalgamation of factors producing an assured optimal response was identified through factorial design along with response surface methodology (RSM). It is a powerful technique for testing multiple process variables because fewer experimental trials are needed compared to the study of one variable at a time. Also, significant interactions between variables can be identified and quantified by this technique.
Taking into account that the soil is a great reservoir of Actinomycetes and that there are few reports concerning optimization of culture medium for transglutaminase production, in this project, the isolation and screening of soil Actinomycetes for transglutaminase production is reported and the nutritional fermentation conditions in order to maximize the transglutaminase yield was studied.
Materials and Methods
Selective isolation and preservation of actinomycetes
The actinomycetes strains used in this study were isolated from soil samples collected from Trichy, Tamil Nadu. Nearly five grams of the soil sample were added to 10 ml sterilized distilled water and the suspension was shaken at 200 rpm for 10 min. Aliquots were inoculated onto Petri dishes containing Rose Bengal Agar and incubated for three days. Actinomycetes-like colonies were streaked onto slants of ISP2 media [8] and checked for purity. Colonies were removed from the agar media and preserved in deep freezer with 10% glycerol solution at -20°C.
MTGase-production screening
The ability to produce transglutaminase was determined by inoculating 1 ml of spore suspension into 250 ml Erlenmeyer flasks containing 50 ml of seed medium. The flasks were incubated for 2 days at 30°C and 200 rpm in a rotational shaker. Aliquots of 15 ml of pre inoculum were transferred to 135 ml of the basal medium [7] in 500 ml Erlenmeyer flasks and cultivated at 30°C for 5 days at 200 rpm. All runs were made in duplicate. After that, the amount of microbial transglutaminase activity was estimated.
MTGase Estimation
Aliquots of 1 ml of culture medium were taken and after centrifugation the enzyme amount was detected in the supernatant. Equal volumes (50 μl) of supernatant and transglutaminase reaction mix containing transglutaminase assay buffer with both donor and acceptor substrate and 1 M DTT, is taken in a 96 well plate, mixed and incubated for 2 hrs. at 37°C. After incubation, 50 μl of stop solution was added and mixed by pipetting and centrifuged at rpm for 15 min to pellet the precipitate formed. 100 μl of supernatant was transferred to a new 96 well plate containing hydroxamate standards and the colour developed was read at 525 nm. From the hydroxamate standard curve, the absorbance value is converted into nmole of hydroxamate which is then converted to mg/ml of hydroxamate using the molecular weight of hydroxamate. This value is used to represent the amount of transglutaminase produced in the entire article.
Identification of Species
The selected strain was identified by Gram's staining and confirmed by culturing in Starch-Casein Agar plates.
Screening of optimal carbon and nitrogen sources
The effects of different sources of nitrogen and carbon on the TGase production were scrutinized with classical one at time approach. The nitrogen sources and corresponding concentrations tested were: 2% peptone with 0.2% yeast extract; 2% peptone; 2% corn steep liquor (CSL); and 2% casein. The carbon sources were: 2% potato starch with 0.2% glucose; 2% molasses; 2% sucrose with 0.2% glucose; 2% glycerol; and 2% soluble starch. In the investigation of the nitrogen sources, growth was carried out in the medium containing: 0.2% KH2PO4; 0.1% MgSO4.7H2O; 2% soybean flour; 2% potato starch; 0.2% glucose. In the process of screening carbon sources, fermentation was carried out using the medium containing: 0.2% KH2PO4; 0.1% MgSO4.7H2O; 2% soybean flour and 2% peptone.
Elucidation of significant components by Plackett-Burman Method
10 ml of medium was prepared with the following components as shown in the Table 1 in distilled water. The mediums are then autoclaved and after cooling inoculated with 1 ml of strain. Incubation was done in a shaker at 37°C at 200 rpm for 72 hrs. After incubation the enzyme produced were estimated by transglutaminase assay method provided earlier and are analyzed using F-test [9].
| Trial | Variable (%) | Yield (mg/ml) | ||||||
|---|---|---|---|---|---|---|---|---|
| KH2PO4 | MgSO4.7H2O | Glucose | Peptone | Yeast Extract | Sucrose | Molasses | ||
| 1 | 0.3 | 0.15 | 3.0 | 1.0 | 0.2 | 1.0 | 1.0 | 25.8 |
| 2 | 0.1 | 0.15 | 3.0 | 3.0 | 0.2 | 3.0 | 1.0 | 40.3 |
| 3 | 0.1 | 0.05 | 3.0 | 3.0 | 0.2 | 1.0 | 3.0 | 37.4 |
| 4 | 0.3 | 0.05 | 1.0 | 3.0 | 0.2 | 3.0 | 1.0 | 46.9 |
| 5 | 0.1 | 0.15 | 1.0 | 1.0 | 0.2 | 3.0 | 3.0 | 52.8 |
| 6 | 0.3 | 0.05 | 3.0 | 1.0 | 0.2 | 3.0 | 3.0 | 55.3 |
| 7 | 0.3 | 0.15 | 1.0 | 3.0 | 0.2 | 1.0 | 3.0 | 82.4 |
| 8 | 0.1 | 0.05 | 1.0 | 1.0 | 0.2 | 1.0 | 1.0 | 33.3 |
Table 1: Plackett-Burman design.
Optimization of key ingredient concentration using RSM
The concentrations of key ingredients identified as a result of Plackett-Burman method were optimized using Response Surface Methodology (RSM), especially by Box-Behnken Design. The experiment was designed by NCSS 2007 software. The experimental design including the variables and concentration levels were shown in the following Table 1. The experimental results were analyzed using the StatistXL software and NCSS 2007.
Result
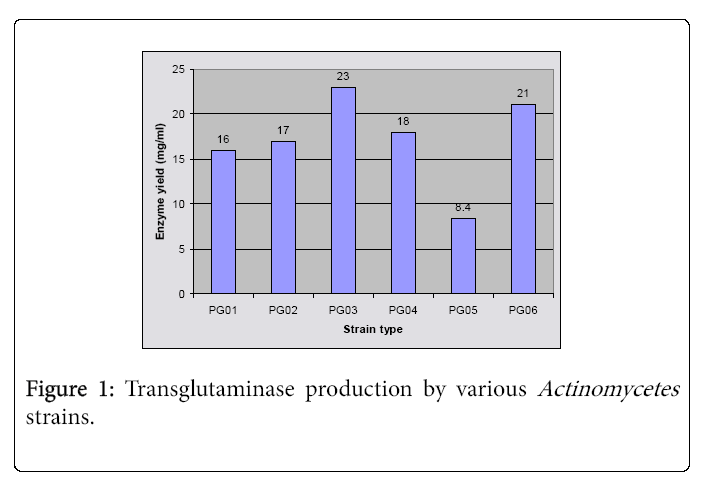
The strategy involved in this study, comprising selective isolation conditions for actinomycetes from soil sample from Trichy, was successful for the recovery of around 6 actinomycete pure cultures. These isolates were investigated for transglutaminase production and the results (Figure 1) showed that more enzyme was produced by the strains PG03 (23 mg/ml) and PG06 (21 mg/ml). The former strain was chosen to continue the studies and was taxonomically identified as Starch and casein hydrolyzing, Gram positive Streptomyces sp.
The results of the effects of the carbon and nitrogen sources on the microbial transglutaminase production by Streptomyces sp. PG03 are shown in the Figure 2. Of all nitrogen sources investigated, the most promising was peptone and a mixture of peptone with yeast extract where the microbial transglutaminase production was around 27.4 mg/ml and 27 mg/ml respectively. Hence both peptone and yeast extract were taken for further studies. For the carbon source the best results were obtained with a mixture of glucose with sucrose (17 mg/ml) and molasses (18.3 mg/ml).
Since the outcome for both carbon supplies were not drastically diverse, glucose, sucrose and molasses were chosen to continue the study. The most important nutrient factors were screened by Plackett- Burman Design as described earlier in Table 1. The experimental design and the results of Plackett-Burman design observations are presented in Table 1. Transglutaminase production varied from 25.8 mg/ml to 82.4 mg/ml with different combinations of the components in the media. The F-test for the design was given in the Table 2.
| Variable | KH2PO4 | MgSO4 .7H2O | Glucose | Peptone | Yeast Extract | Sucrose | Molasses |
|---|---|---|---|---|---|---|---|
| ∑(H) | 208.4 | 201.3 | 158.8 | 205.0 | 160.9 | 193.3 | 190.5 |
| ∑(L) | 163.8 | 170.9 | 213.4 | 167.2 | 211.3 | 178.9 | 181.7 |
| ∑(H) - ∑(L) | 44.61 | 30.4 | -54.6 | 37.8 | -50.4 | 14.4 | 8.8 |
| Effect | 11.15 | 7.6 | -13.65 | 9.45 | -12.6 | 3.6 | 2.2 |
| Mean Square | 248.75 | 115.52 | 372.64 | 178.60 | 317.52 | 25.92 | 9.68 |
| F-test | 0.78 | 0.36 | 1.17 | 0.5625 | 1.0 | 0.081 | 0.03 |
Table 2: F-test for Plackett-Burman design.
Response Surface Methodology, especially Box-Behnken design was chosen to optimize the key ingredients selected in the media. The experimental design and results are shown in the Table 3. The quadratic model calculated using NCSS for maximum microbial transglutaminase yield after eliminating the statistically insignificant terms was:
Y = 26.94444 - 9.8625x1 - 222.75x3 + 3.341667x4 + 2.882292x12 + 1137.917x32.
| Trial | Glucose (%) | KH2PO4 (%) | MgSO4.7H2O (%) | Peptone (%) | Yield (mg/ml) | |
|---|---|---|---|---|---|---|
| Predicted | Experimental | |||||
| 1 | 1.0 | 0.1 | 0.10 | 2.0 | 15.7 | 13.3 |
| 2 | 3.0 | 0.1 | 0.10 | 2.0 | 19.1 | 17.1 |
| 3 | 1.0 | 0.3 | 0.10 | 2.0 | 15.7 | 19.1 |
| 4 | 3.0 | 0.3 | 0.10 | 2.0 | 19.1 | 21.6 |
| 5 | 2.0 | 0.2 | 0.05 | 1.0 | 13.8 | 11.2 |
| 6 | 2.0 | 0.2 | 0.15 | 1.0 | 14.3 | 15.8 |
| 7 | 2.0 | 0.2 | 0.05 | 3.0 | 20.5 | 20.0 |
| 8 | 2.0 | 0.2 | 0.15 | 3.0 | 21.0 | 19.5 |
| 9 | 2.0 | 0.2 | 0.10 | 2.0 | 14.5 | 16.2 |
| 10 | 1.0 | 0.2 | 0.10 | 1.0 | 12.4 | 11.6 |
| 11 | 3.0 | 0.2 | 0.10 | 1.0 | 15.7 | 15.8 |
| 12 | 1.0 | 0.2 | 0.10 | 3.0 | 19.1 | 17.9 |
| 13 | 3.0 | 0.2 | 0.10 | 3.0 | 22.4 | 19.1 |
| 14 | 2.0 | 0.1 | 0.05 | 2.0 | 17.1 | 15.4 |
| 15 | 2.0 | 0.3 | 0.05 | 2.0 | 17.1 | 17.9 |
| 16 | 2.0 | 0.1 | 0.15 | 2.0 | 17.6 | 20.0 |
| 17 | 2.0 | 0.3 | 0.15 | 2.0 | 17.6 | 15.4 |
| 18 | 2.0 | 0.2 | 0.10 | 2.0 | 14.5 | 16.2 |
| 19 | 1.0 | 0.2 | 0.05 | 2.0 | 18.3 | 20.0 |
| 20 | 3.0 | 0.2 | 0.05 | 2.0 | 21.7 | 24.1 |
| 21 | 1.0 | 0.2 | 0.15 | 2.0 | 18.8 | 18.3 |
| 22 | 3.0 | 0.2 | 0.15 | 2.0 | 22.2 | 22.5 |
| 23 | 2.0 | 0.1 | 0.10 | 1.0 | 11.2 | 08.7 |
| 24 | 2.0 | 0.3 | 0.10 | 1.0 | 11.2 | 10.8 |
| 25 | 2.0 | 0.1 | 0.10 | 3.0 | 17.9 | 20.4 |
| 26 | 2.0 | 0.3 | 0.10 | 3.0 | 17.9 | 17.1 |
| 27 | 2.0 | 0.2 | 0.10 | 2.0 | 14.5 | 16.2 |
Table 3: Response surface methodology.
The ANOVA obtained by StatistXL (Table 4) proved that the model was momentous. The F-test was greater than the table value of 2.82 [10] and the p-value of 0.004 clearly reveals that this regression was statistically significant at the 95% confidence level [11]. Further, the R2 of the regression obtained was 0.488 indicating that 48.8% of the response variation can be elucidated by the model.
| Source of Variation | Sum of Squares. | Degrees of Freedom | Mean Squares | Ftest | p-value |
|---|---|---|---|---|---|
| Regression | 172.118 | 4 | 43.030 | 5.253 | 0.004 |
| Residual | 180.222 | 22 | 8.192 | ||
| Total | 352.341 | 26 |
Table 4: Analysis of variance for Box-Behnken design.
The response surfaces were selected with the probable mishmash to visualize the simultaneous effects of Glucose, KH2PO4, MgSO4.7H2O and peptone on microbial transglutaminase production pattern.
Discussion
A Streptomyces sp. strain from soil sample was secluded as a potential producer of transglutaminase enzyme. It was as an extracellular calcium independent enzyme producer making it much more appealing and sensible for commercial applications [2].
Different compounds were examined with classical one at a time strategy to pick the best carbon and nitrogen sources for transglutaminase production. When peptone (27 mg/ml) and a mixture of peptone with yeast extract (27.4 mg/ml) were used as nitrogen sources and molasses (18.3 mg/ml) and a mixture of glucose and sucrose (17 mg/ml) as carbon sources, superior results were achieved. The results for both the carbon and nitrogen sources were not significantly different and hence all of them (Peptone, yeast extract, molasses, sucrose and glucose) were selected the Plackett-Burman design.
Variations in the concentration of molasses and sucrose in Plackett- Burman Design did not affect microbial transglutaminase production significantly. Considering this and the cost parameter, the concentration of sucrose is maintained at low level and molasses was not at all added. Conversely, KH2PO4, MgSO4.7H2O, Glucose and peptone affected the enzyme production to a greater extent. These four components were further investigated using Box-Behnken Design in a broader concentration range.
The results from the Box-Behnken Design showed that the concentrations of the four key ingredients were: 3% Glucose, 0.2% KH2PO4, 0.05% MgSO4.7H2O and 2% peptone. The maximum yield of microbial transglutaminase predicted with the model was 21.7 mg/ml whereas the experimental value was 24.1 mg/ml proving that the model was ample to forecast the optimization of transglutaminase production by Streptomyces sp PG03.
Conclusion
The fermentation media for microbial transglutaminase production have been modified and optimized via statistical methods like Plackett- Burman Design and Box-Behnken Design. The medium optimization not only resulted in an increased microbial transglutaminase production, but also reduced the constituent costs and an improvement in repeatability. Besides, the factors responsible for better activity were found and are imperative for further studies.
Acknowledgement
The authors wish to acknowledge the Management of St. Michael College of Engineering & Technology and the Faculty of Biotechnology, Sathyabama University, for their continuous support and encouragement in carrying out such a study.
Conflict of Interest Disclosure
The authors declare that they have no conflict of interest.
References
- Soares LHB, Assmann F, Ayub MAZ (2003) Purification and properties of a transglutaminase produced by a Bacillus circulans strain isolated from the Amazon environment. Biotechnology and Applied Biochemistry 37: 295-299.
- Yokoyama K, Nio N, Kikuchi Y (2004) Properties and application of microbial transglutaminase. Applied Microbiology and Biotechnology 64: 447-454.
- Kwan SW, EasaAM (2003) Comparing physical properties of retort-resistant glucono-δ-lactone tofu treated with commercial transglutaminase enzyme or low levels of glucose. Lebensmittel-Wissenschaft und-Technologie 36: 643-646.
- Zhu Y, Rinzema A, Tramper J, Bol J (1995) Microbial transglutaminase - areview of its production and application in food processing. Applied Microbiology and Biotechnology, 44: 277-282.
- Griffin M, Casadio R, Bergamini CM (2002) Transglutaminases: nature's biological glues. Biochem J 368: 377-396.
- Pasternack R, Dorsch S, Otterbach JT, Robenek IR,Wolf S, et al. (1998) Bacterial protransglutaminase from Streptoverticilliummobaraense, purification, characterization and sequence of the zymogen. European Journal of Biochemistry 257: 570-576.
- Ando H, Adachi M, Umeda K, Matsura A, Onaka M, et al. (1989) Purification and characteristics of a novel transglutaminase derived from microorganisms. Agricultural and Biological Chemistry 53: 2613-2617.
- Küster E, Williams ST (1964) Selection of media for isolation of streptomycetes. Nature 202: 928-929.
- Nelson LS (1982)Technical Aids, Jour Qual Tech 14: 99-100.
- Walpole RL, Meyers RH, Meyers SL (1998) Probability and Statistics for Engineers and Scientists. Prentice Hall International Inc New Jersey.
- Gopal SB, Sujitha S, Thyagarajan R, Jegatheesan K (2012) Isolation of Streptomyces sp. from soil and its medium optimization for microbial transglutaminase production by box-behnken design, Journal of Biotech & Biomaterial 2: 83.
Relevant Topics
- Aquatic Ecosystems
- Biodiversity
- Conservation Biology
- Coral Reef Ecology
- Distribution Aggregation
- Ecology and Migration of Animal
- Ecosystem Service
- Ecosystem-Level Measuring
- Endangered Species
- Environmental Tourism
- Forest Biome
- Lake Circulation
- Leaf Morphology
- Marine Conservation
- Marine Ecosystems
- Phytoplankton Abundance
- Population Dyanamics
- Semiarid Ecosystem Soil Properties
- Spatial Distribution
- Species Composition
- Species Rarity
- Sustainability Dynamics
- Sustainable Forest Management
- Tropical Aquaculture
- Tropical Ecosystems
Recommended Journals
Article Tools
Article Usage
- Total views: 12617
- [From(publication date):
March-2016 - Aug 23, 2025] - Breakdown by view type
- HTML page views : 11456
- PDF downloads : 1161