Keloid-specific Gene Expression Profiling for Accurate Diagnostic and Therapeutic Applications
Received: 20-Oct-2022 / Manuscript No. DPO-22-77863 / Editor assigned: 21-Oct-2022 / PreQC No. DPO-22-77863 / Reviewed: 04-Nov-2022 / QC No. DPO-22-77863 / Revised: 14-Nov-2022 / Manuscript No. DPO-22-77863 / Accepted Date: 14-Nov-2022 / Published Date: 21-Nov-2022 DOI: 10.4172/ 2476-2024.7.S12.003
Abstract
Scars are a heterogeneous disease including normotrophic scars, hypertrophic scars, and keloids. Of these lesions, keloids are a distinct subtype from any other type of scar. Clinically, it causes pain, itching, or tenderness, causing life discomfort and characteristically irreversible.
Despite various treatment modalities, restoring keloids to normal tissues is difficult, and frequent recurrences have been reported. Therefore, it is essential to identify keloid-specific genes for accurate diagnosis and treatment of keloids. In an effort to find out keloid-specific genes, several studies compared keloids with scar-free normal skin, which leading general scar-related genes to be chosen rather than keloid-specific genes. To select for highly accurate keloid-specific genes and pathways, we compared the transcriptome profile of keloids with those of normotrophic scars and hypertrophic scars, which acquired from formalin-fixed paraffin-embedded human skin samples using high-throughput RNA-sequencing techniques. Differential expression analyses and over-representation analyses revealed that genes related to nervous system process were upregulated in keloids, whereas genes related with immune responses were downregulated in keloids. Additionally, the extracellular matrix related processes were highlighted in both hypertrophic scars and keloids. Finally, we highlight potential keloid-specific biomarkers and expression changes that can be employed for future therapeutics of keloids.
Keywords: Normotrophic scar; Hypertrophic scar; Keloid; RNA sequencing; Scar management
Introduction
A scar is an area of fibrous tissue that replaces normal skin after an injury. Scarring is a biological process of wound healing. Despite tremendous progress in unravelling the science behind wound healing, the mechanisms for elucidating scar formation caused by abnormal wound healing are still unknown [1]. Deep injury to the reticular dermis sometimes results in pathological abnormal fibrous scars such as hypertrophic scars and keloids, which are thought to be caused by abnormal wound healing due to altered gene expression in the wound healing tissue. Hypertrophic scars and keloids consist of numerous inflammatory cells, proliferating fibroblasts, progressive collagen deposition and enhanced angiogenesis [2-4]. And they present distinct clinical, histological, and developmental features. Clinically, hypertrophic scars are confined within the wound boundary, while keloids extend growth beyond the wound borders [5,6]. Histologically, hypertrophic scars have fewer thick collagen fibers than keloids [7,8]. Furthermore, hypertrophic scar tissues are composed of low-density Dermatan Sulfate (DS) proteoglycans, while keloid scar tissues are composed of both low-density DS and Chondroitin Sulfate (CS) proteoglycans, with CS proteoglycans in higher proportion [9,10]. Hypertrophic scars usually develop within two months from injury and tend to regress over time. In contrast, keloids develop and progress after 3 months or more, grow irreversibly for years, and cause medical problems as well as life problems such as itchiness and unusual appearance for patients [11]. Hypertrophic scars tend to respond well to treatment. On the other hand, once formed, keloids are very difficult to restore to normal tissues, and frequent recurrences and exacerbations over a long period of time become another obstacle to treatment [12]. It has not been reported yet what kind of scars would be occurred when the skin is damaged. Therefore, it is essential to understand the pathological factors behind the various scar types and gene expression profiles of pathological scars caused by abnormal wound healing, for the development of efficient treatment strategies for the treatment of abnormal scars.
To date, there have been five reports comparing gene expression patterns in keloid and related scar lesions using whole transcriptome analysis such as cDNA microarray or RNA-seq. Onoufriadis, et al. and Wu, et al. performed RNA-seq on keloids and normal skin tissue [13,14]. Hahn, et al. Chao-Kai, et al. and Matsumoto, et al. compared mRNA expression patterns between keloids and normal skin tissue using cDNA microarrays [15-17]. Although these studies have provided new insights into keloids, there are limitations in that the selected keloid- specific gene expression profile could not be distinguished from a scar- related genes expression profile, since they compared keloids to scar- free normal skin tissue not to reversible scars. Considering different responses of irreversible keloids and reversible hypertrophic scars to the treatment, it is crucial to compare the overall gene expression patterns of irreversible scars such as keloids and reversible scars. In this study, we aimed to compare the overall gene expression patterns of reversible and irreversible scars and provide insight into the molecular and cellular mechanisms behind these lesions. Toward this goal, high-throughput RNA-sequencing techniques using next-generation sequencing technology were employed. By elucidating highly specific genes and related signaling pathways for abnormal fibroproliferative scar lesions, this study can contribute to the development of effective and targeted therapeutic strategies for these lesions.
Materials and Methods
Results
Expression analysis of keloids and related lesions through RNA-seq
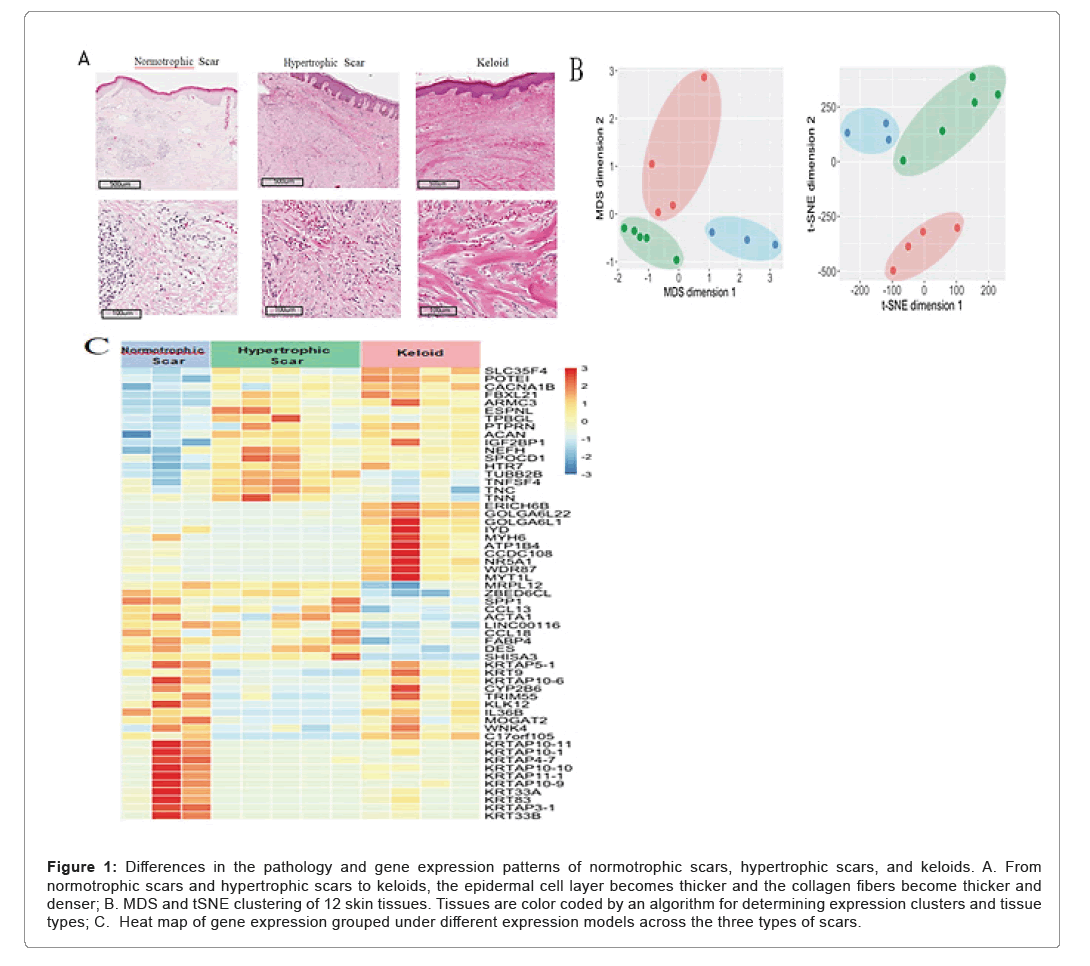
WTo reveal the gene expression characteristics of keloids and related lesions, we collected 4 keloid tissues, 5 hypertrophic scars, and 3 normotrophic scars. The three types of lesions were categorized by characteristic appearance, histological morphology and clinical features [26]. From normotrophic scars and hypertrophic scars to keloids, the epidermal cell layer becomes thicker and the collagen fibers become thicker and denser. Consistent with previous reports, keloids and hypertrophic scars showed abundant vasculature, high mesenchymal cell density, inflammatory cell infiltration, and thickened epidermal cell layer compared to normotrophic scars [27,28]. In particular, thick collagen fibers composed of numerous tightly packed fibrils were observed in keloids, while hypertrophic scars exhibit a nodular structure composed of fibroblasts, small blood vessels, and randomly organized collagen fibers [29].
To confirm the histopathologic classification of the samples, the expression profiles of these collected tissue samples were analyzed by unsupervised clustering using a machine learning-based dimensionality reduction algorithm such as MDS (Multi-Dimensional Scaling) and tSNE (t-Distributed Stochastic Neighbor Embedding). The results showed that these lesions were separated into three clusters, relatively consistent with their diagnoses. The differential genes of these normotrophic Scar (S), Hypertrophic Scar (HS), and Keloid (K) were grouped into 8 classes depending on expression pattern: ○1 UU (Up-Up, S<HS<K), ○2 UF (Up-Flat, S<HS~K), ○3 FD (Flat-Down, S~HS>K), ○4 UD (Up-Down, S<HS>K), ○5 DD (Down-Down, S>HS>K), ○6 DF (Down-Flat, S>HS~K), ○7 FU (Flat-Up, S~HS<K), and ○8 DU (Down-Up, S>HS<K). The genes were separated as upregulated (U), downregulated (D), or not differentially expressed (F). Expression pattern of grouped genes over scar types were also separated in patterns as expected (Figure 1) (Table 1).
Figure 1: Differences in the pathology and gene expression patterns of normotrophic scars, hypertrophic scars, and keloids. A. From normotrophic scars and hypertrophic scars to keloids, the epidermal cell layer becomes thicker and the collagen fibers become thicker and denser; B. MDS and tSNE clustering of 12 skin tissues. Tissues are color coded by an algorithm for determining expression clusters and tissue types; C. Heat map of gene expression grouped under different e xpression models across the three types of scars.
| Type of tissue | Gender and Age | Body part effected |
|---|---|---|
| Normotrophic Scar #1 | F 56 | Skin, Scalp |
| Normotrophic Scar #2 | F 57 | Skin, back |
| Normotrophic Scar #3 | F 35 | Skin, public area |
| Hypertropic Scar #1 | F 55 | Skin, neck |
| Hypertropic Scar #2 | M 69 | Skin, right thigh |
| Hypertropic Scar #3 | M 68 | Skin, post-auricular area |
| Hypertropic Scar #4 | F 81 | Skin, right breast |
| Hypertropic Scar #5 | M 66 | Skin, left cheek |
| Keloid #1 | F 21 | Skin, ear |
| Keloid #2 | F 19 | Skin, ear |
| Keloid #3 | F 53 | Skin, abdomen |
| Keloid #4 | M 15 | Skin, ear |
Table 1: Summary of information on scar tissues used in this study.
Derivation of keloid-specific genes and signaling pathways by comparing gene expression profiles of keloids and reversible scars
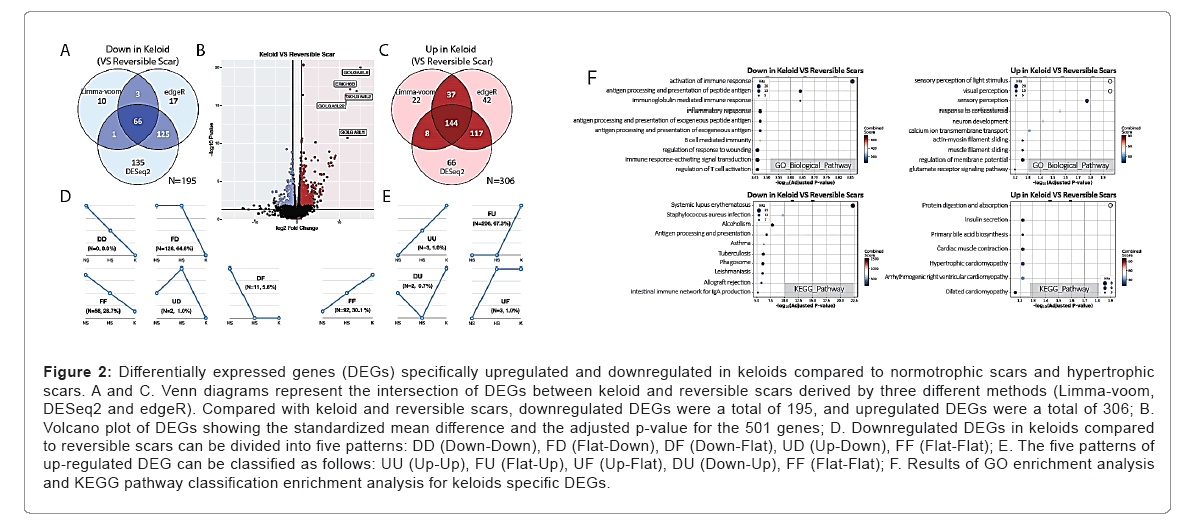
Once keloids were developed, they grow irreversibly for years, compared to normoptophic and hypertrophic scars which regress over time. In an attempt to discover genes specifically regulated in keloids as irreversible scars, we classified normotrophic and hypertrophic scars as one group-reversible scar and compared their gene expression profiles with that of keloids. To identify genes specifically up or downregulated in keloids, we performed mRNA sequencing and differential expression analysis on RNA-Seq data from normotrophic scars, hypertrophic scars, and keloids with following statistical methods. For differential expression analysis, we implemented Ensemble approaches as three widely used methods (Limma-voom, edgeR and DESeq2) to achieve lower false positive DEGs. In this approach, genes were flagged as DEG on at least two different analyses was used for further analysis. Overall, 195 genes were downregulated in keloids compared with reversible scars, whereas 306 genes were upregulated in keloids. Of these genes, golgin A6 family genes and ERICH6B (Glutamate-Rich 6B) genes were identified as the most significantly changed genes as described in volcano plots. It should be noted that the levels of golgin A6 family genes and ERICH6B genes were not only significantly changed, but also exhibited the highest magnitude of fold change among the genes in keloids compared with reversible scars. Golgins are a family of predominantly coiled-coil proteins that are localized to the Golgi apparatus [30]. Anchored to the Golgi membrane, they are predicted to project into the surrounding cytoplasm and are involved in various functions of the Golgi apparatus, such as vesicular traffic through golgin-mediated tethering. Second, golgins are involved in maintenance and positioning of the Golgi apparatus within cells. In addition to acting as tethers, some golgins can sequester various factors at the Golgi membrane, allowing spatiotemporal regulation of downstream cellular functions. However, the specific functions and clinical relevance of the Golgin A6 family has not been elucidated yet. In the case of ERICH6B, there is little information on the structure, function and clinical relevance of the gene, except for its predominant expression in the testis.
Next, up and downregulated 501 DEGs (up: 306, down: 195) was classified as expression changes on normotrophic scars-hypertrophic scars-keloids. Specifically, most of the downregulated DEGs in keloids belonged to the FD (Flat-Down, 64.6%) subgroup, in contrast to very few belonged to DD (Down-Down, 0%) or DF (Down-Flat, 5.6%) subgroup. On the other hand, most of upregulated DEGs in keloids belonged to the FU (Flat-Up, 67.3%) subgroup, while very few belonged to UU (Up-Up, 1.0%) or UF (Up-Flat, 1.0%) subgroup. These results indicate that most of the DEGs exhibited small differences between normotrophic and hypertrophic scars but were higher or lower in keloids.
For better understanding of biological function of genes that exhibited specifically high or low expression in keloids. DEGs are enriched to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. Over-representation analysis on GO Biological Process showed that the genes involved in ‘activation of immune response’, ‘antigen processing and presentation of peptide antigen’ and ‘immunoglobulin mediated immune response’ are significantly downregulated in keloids. These findings suggest that immune regulation and inflammatory mechanisms play important roles in keloid formation. On the other hand, DEGs upregulated in keloids were over-represented in roles of ‘sensory perception’, ‘neuron development’, and ‘muscle filament sliding’. Over-representation analysis on KEGG pathway demonstrated that downregulated DEGs in keloids were enriched in ‘systemic lupus erythematosus’ and ‘antigen processing and presentation’, whereas upregulated DEGs in keloids were enriched in ‘protein digestion and absorption’ and ‘insulin secretion’. Overall, downregulated DEGs in keloids were associated with immune functions, whereas upregulated DEGs in keloids were associated with neuron development and muscle contraction. In keloids, the regulation of T cell proliferation and negative regulation of T cell activation were specifically downregulated, whereas visual/sensory perception was specifically upregulated. In addition, the systemic lupus erythematosus pathway was downregulated, whereas the pathways of protein digestion and absorption were upregulated. (Figure 2) (Tables 2-5).
Figure 2: Differentially expressed genes (DEGs) specifically upregulated and downregulated in keloids compared to normotrophic scars and hypertrophic scars. A and C. Venn diagrams represent the intersection of DEGs between keloid and reversible scars derived by three different methods (Limma-voom, DESeq2 and edgeR). Compared with keloid and reversible scars, downregulated DEGs were a total of 195, and upregulated DEGs were a total of 306; B. Volcano plot of DEGs showing the standardized mean difference and the adjusted p-value for the 501 genes; D. Downregulated DEGs in keloids compared to reversible scars can be divided into five patterns: DD (Down-Down), FD (Flat-Down), DF (Down-Flat), UD (Up-Down), FF (Flat-Flat); E. The five patterns of up-regulated DEG can be classified as follows: UU (Up-Up), FU (Flat-Up), UF (Up-Flat), DU (Down-Up), FF (Flat-Flat); F. Results of GO enrichment analysis and KEGG pathway classification enrichment analysis for keloids specific DEGs.
| GO ID | Term | Overlap | Adjusted P- value | Genes |
|---|---|---|---|---|
| GO:0002253 | Activation of immune response | 20/487 | 0.000138 | CD86, HLA-DRB5, FCER1G, CTSS, PYCARD, FCGR3A, FCGR2A, C3AR1, HLA-DPB1, CDK1, TLR8, HLA-DRA, CD36, VSIG4, PIK3AP1, FCGR1A, HLA-DRB1, CTSB, C1QC, HLA-DPA1 |
| GO:0048002 | Antigen processing and presentation of peptide antigen | 12/185 | 0.000228 | BCAP31, CD74, HLA-DRB5, FCER1G, HLA-DPB1, HLA-DRA, CD36, FCGR1A, HLA-DRB1, CTSS, PSMA7, HLA-DPA1 |
| GO:0016064 | Immunoglobulin mediated immune response | 5/16 | 0.000228 | CD74, FCER1G, TLR8, FCGR1A, HLA-DRB1 |
| GO:0006954 | Inflammatory response | 16/376 | 0.000337 | IL10, CCL13, CD163, SERPINA1, TNFAIP6, LY86, LYZ, AIF1, PYCARD, C3AR1, SPP1, BDKRB2, TLR8, CCL18, FCGR1A, HLA-DRB1 |
| GO:0002478 | Antigen processing and presentation of exogenous peptide antigen | 11/171 | 0.000337 | CD74, HLA-DRB5, FCER1G, HLA-DPB1, HLA-DRA, CD36, FCGR1A, HLA-DRB1, CTSS, PSMA7, HLA-DPA1 |
| GO:0019884 | Antigen processing and presentation of exogenous antigen | 11/173 | 0.000337 | CD74, HLA-DRB5, FCER1G, HLA-DPB1, HLA-DRA, CD36, FCGR1A, HLA-DRB1, CTSS, PSMA7, HLA-DPA1 |
| GO:0019724 | B-cell mediated immunity | 5/20 | 0.000337 | CD74, FCER1G, TLR8, FCGR1A, HLA-DRB1 |
| GO:1903034 | Regulation of response to wounding | 15/347 | 0.000346 | IL10, FCER1G, SCARA5, CMA1, CAV1, PLA2G7, F3, CTSS, FABP4, ALOX5AP, SPP1, CD36, PIK3AP1, FCGR1A, HLA-DRB1 |
| GO:0002757 | Immune response-activating signal transduction | 17/440 | 0.000346 | CD86, HLA-DRB5, FCER1G, CTSS, FCGR3A, FCGR2A, C3AR1, HLA-DPB1, CDK1, TLR8, HLA-DRA, CD36, PIK3AP1, FCGR1A, HLA-DRB1, CTSB, HLA-DPA1 |
| GO:0050863 | Regulation of T cell activation | 13/259 | 0.000346 | IL10, CD86, CD74, HLA-DRB5, CAV1, AIF1, PYCARD, HLA-DPB1, HLA-DRA, VSIG4, IL7R, HLA-DRB1, HLA-DPA1 |
Table 2: GO Biological Process enrichment result for down regulated DEGs in Keloid VS. Reversible Scars.
| Term | Overlap | Adjusted P-value | Genes |
|---|---|---|---|
| Systemic lupus erythematosus | 25/133 | 3.21E-25 | CD86, HIST1H2BM, HIST1H2BO, FCGR3A, HIST1H3A, HIST1H3H, HIST1H3B, FCGR1A, HIST1H2AB, HLA-DPA1, HIST1H2AM, IL10, HLA-DRB5, HIST1H4L, HIST1H2AJ, HIST1H4A, HIST1H4B, FCGR2A, HIST4H4, HLA-DPB1, HLA-DRA, HIST1H2BH, HIST1H4C, HLA-DRB1, C1QC |
| Staphylococcus aureus infection | 12/68 | 2.37E-12 | IL10, FCGR3A, HLA-DRB5, FCGR2A, C3AR1, HLA-DPB1, HLA-DRA, FPR3, FCGR1A, HLA-DRB1, HLA-DPA1, C1QC |
| Alcoholism | 15/180 | 2.76E-10 | HIST1H2AM, HIST1H2BM, HIST1H2BO, MAOB, HIST1H4L, HIST1H2AJ, HIST1H4A, HIST1H3A, HIST1H4B, HIST4H4, HIST1H2BH, HIST1H3H, HIST1H3B, HIST1H4C, HIST1H2AB |
| Antigen processing and presentation | 10/77 | 3.83E-09 | CD74, HLA-DRB5, HLA-DPB1, HLA-DRA, HSPA2, CTSS, HSPA1B, HLA-DRB1, CTSB, HLA-DPA1 |
| Asthma | 7/31 | 1.62E-08 | IL10, HLA-DRB5, FCER1G, HLA-DPB1, HLA-DRA, HLA-DRB1, HLA-DPA1 |
| Tuberculosis | 13/179 | 2.33E-08 | IL10, CD74, HLA-DRB5, FCER1G, CTSS, FCGR3A, FCGR2A, HLA-DPB1, HLA-DRA, FCGR1A, ATP6V0C, HLA-DRB1, HLA-DPA1 |
| Phagosome | 12/152 | 3.25E-08 | MSR1, HLA-DRB5, FCGR3A, FCGR2A, HLA-DPB1, HLA-DRA, CD36, FCGR1A, ATP6V0C, HLA-DRB1, CTSS, HLA-DPA1 |
| Leishmaniasis | 9/74 | 4.23E-08 | IL10, FCGR3A, HLA-DRB5, FCGR2A, HLA-DPB1, HLA-DRA, FCGR1A, HLA-DRB1, HLA-DPA1 |
| Allograft rejection | 7/38 | 7.35E-08 | CD86, IL10, HLA-DRB5, HLA-DPB1, HLA-DRA, HLA-DRB1, HLA-DPA1 |
| Intestinal immune network for IgA production | 7/48 | 3.95E-07 | CD86, IL10, HLA-DRB5, HLA-DPB1, HLA-DRA, HLA-DRB1, HLA-DPA1 |
Table 3: KEGG Pathway enrichment result for down regulated DEGs in Keloid VS. Reversible Scars.
| GO ID | Term | Overlap | Adjusted P-value | Genes |
|---|---|---|---|---|
| GO:0050953 | sensory perception of light stimulus | 13/198 | 0.0111 | TULP1, RPGR, COL1A1, RP1L1, OCLM, RIMS1, CACNB2, RP1, GRM8, SIX3, CNGB3, PPEF2, GPR179 |
| GO:0007601 | visual perception | 13/195 | 0.0111 | TULP1, RPGR, COL1A1, RP1L1, OCLM, RIMS1, CACNB2, RP1, GRM8, SIX3, CNGB3, PPEF2, GPR179 |
| GO:0007600 | sensory perception | 20/457 | 0.017 | TAS2R60, EYA1, COL11A2, CACNA1D, TULP1, SLC6A3, RPGR, COL1A1, RP1L1, OCLM, CACNB2, RIMS1, RP1, MYO15A, GRM8, SIX3, CNGB3, ASIC3, PPEF2, GPR179 |
| GO:0031960 | response to corticosteroid | 10/140 | 0.0278 | COL1A1, MSTN, UCN2, WNT7B, BCKDHB, PDX1, GRIK4, FOSB, FOS, SOX30 |
| GO:0048666 | neuron development | 9/121 | 0.038 | ALK, ROBO2, TMC1, RP1L1, RP1, TULP1, HOXD10, GLI2, RPGR |
| GO:0070588 | calcium ion transmembrane transport | 9/128 | 0.0487 | TMC1, CACNB2, RYR2, CACNG8, IL1RAPL1, CACNA1B, CACNA1D, ATP2A1, TRPM4 |
| GO:0033275 | actin-myosin filament sliding | 5/38 | 0.0551 | MYH2, ACTN2, MYH8, MYH4, MYH7 |
| GO:0030049 | muscle filament sliding | 5/38 | 0.0551 | MYH2, ACTN2, MYH8, MYH4, MYH7 |
| GO:0042391 | regulation of membrane potential | 13/266 | 0.0551 | RYR2, KCNE2, ACTN2, CACNA1B, CACNA1D, GRIK2, RIMS1, NETO1, GJA5, CHRNE, CNGB3, TRPM4, SHANK1 |
| GO:0007215 | glutamate receptor signaling pathway | 5/40 | 0.0634 | GRM5, GRM8, GRIK4, GRIK2, CDK5R1 |
Table 4: GO Biological Process enrichment result for up regulated DEGs in Keloid VS. Reversible Scars.
| Term | Overlap | Adjusted P-value | Genes |
|---|---|---|---|
| Protein digestion and absorption | 8/90 | 0.0125 | COL1A1, COL27A1, MEP1B, COL22A1, COL7A1, COL11A2, ATP1B4, SLC16A10 |
| Insulin secretion | 6/86 | 0.0601 | RYR2, CCKAR, PDX1, ATP1B4, CACNA1D, TRPM4 |
| Primary bile acid biosynthesis | 3/17 | 0.0601 | CYP46A1, BAAT, CYP7A1 |
| Cardiac muscle contraction | 6/78 | 0.0601 | RYR2, CACNB2, CACNG8, ATP1B4, CACNA1D, MYH7 |
| Hypertrophic Cardiomyopathy (HCM) | 6/85 | 0.0601 | RYR2, CACNB2, CACNG8, ITGA2B, CACNA1D, MYH7 |
| Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC) | 6/72 | 0.0601 | RYR2, CACNB2, CACNG8, ACTN2, ITGA2B, CACNA1D |
| Dilated Cardiomyopathy (DCM) | 6/91 | 0.0685 | RYR2, CACNB2, CACNG8, ITGA2B, CACNA1D, MYH7 |
| Glutamatergic synapse | 6/114 | 0.166 | GRM5, GRM8, GRIK4, CACNA1D, GRIK2, SHANK1 |
| Calcium signaling pathway | 8/188 | 0.166 | RYR2, GRM5, CCKAR, PLCZ1, CACNA1B, CACNA1D, ATP2A1, MYLK3 |
| Taurine and hypotaurine metabolism | 11/02 | 0.2 | GADL1, BAAT |
Table 5: KEGG Pathway enrichment result for up regulated DEGs in Keloid VS. Reversible Scar.
Keloids and hypertrophic specific upregulated DEGs are predominantly associated with remodeling of extracellular matrix structural tissue
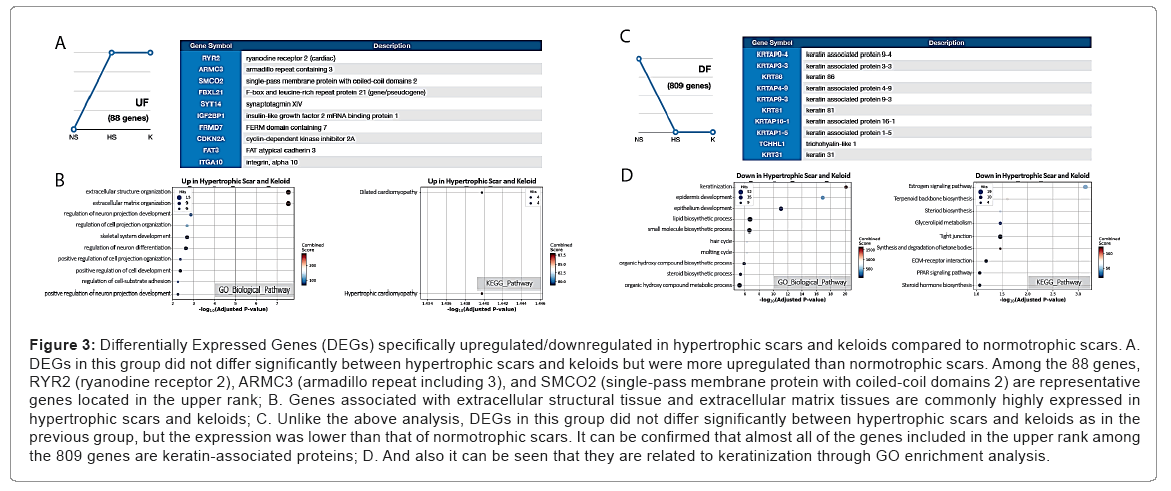
Hypertrophic scars and keloids have a commonality in that they are present in abnormally high levels in fibroproliferative lesions. To gain insight into the mechanisms behind this abnormality, we attempted to identify DEGs that are specifically upregulated in hypertrophic scars and keloids. Our study revealed 88 upregulated genes in these exaggerated scars compared to normotrophic scars, including RYR2 (ryanodine receptor 2), ARMC3 (armadillo repeat including 3), and SMCO2 (single-pass membrane protein with coiled-coil domains 2) as representative genes with larger fold changes. Particularly, these 88 DEGs were highly over-represented on ‘extracellular structure/matrix organization’ among GO Biological Process terms. Overall, these results are in consistently implies that the clinical observation of excessive ECM depositions are represented in both hypertrophic scars and keloids (Tables 6 and 7).
| GO ID | Term | Overlap | Adjusted P-value | Genes |
|---|---|---|---|---|
| GO:0043062 | Extracellular structure organization | 15/360 | 2.97E-08 | ITGB1, POSTN, OLFML2B, TGFB3, PLOD2, NID2, GDF5, ADAMTS4, ACAN, MMP14, ADAMTS14, BMP1, ITGA10, SH3PXD2B, EMILIN1 |
| GO:0030198 | Extracellular matrix organization | 15/359 | 2.97E-08 | ITGB1, POSTN, OLFML2B, TGFB3, PLOD2, NID2, GDF5, ADAMTS4, ACAN, MMP14, ADAMTS14, BMP1, ITGA10, SH3PXD2B, EMILIN1 |
| GO:0010975 | Regulation of neuron projection development | 9/275 | 0.00139 | PTPRD, NTRK1, ITGB1, FRMD7, MAP1B, NCS1, NEFL, FAT3, EPHA3 |
| GO:0001501 | Skeletal system development | 7/162 | 0.00206 | ADAMTS4, ACAN, POSTN, BMP1, SH3PXD2B, ALPL, PAPSS2 |
| GO:0031344 | Regulation of cell projection organization | 10/391 | 0.00206 | PTPRD, NTRK1, ITGB1, FRMD7, TGFB3, MAP1B, NCS1, NEFL, FAT3, EPHA3 |
| GO:0045664 | Regulation of neuron differentiation | 10/405 | 0.00233 | PTPRD, NTRK1, ITGB1, FRMD7, MAP1B, NCS1, NEFL, FAT3, GDF5, EPHA3 |
| GO:0031346 | Positive regulation of cell projection organization | 7/195 | 0.00427 | PTPRD, NTRK1, ITGB1, TGFB3, MAP1B, NEFL, EPHA3 |
| GO:0010720 | Positive regulation of cell development | 9/358 | 0.00428 | PTPRD, NTRK1, ITGB1, TGFB3, MAP1B, SNAI1, NEFL, GDF5, EPHA3 |
| GO:0010976 | Positive regulation of neuron projection development | 6/144 | 0.00562 | PTPRD, NTRK1, ITGB1, MAP1B, NEFL, EPHA3 |
| GO:0045666 | Positive regulation of neuron differentiation | 7/217 | 0.00562 | PTPRD, NTRK1, ITGB1, MAP1B, NEFL, GDF5, EPHA3 |
Table 6: GO Biological Process enrichment result for UF pattern of DEGs.
| Term | Overlap | Adjusted P-value | Genes |
|---|---|---|---|
| Hypertrophic Cardiomyopathy (HCM) | 4/85 | 0.0363 | ITGB1, RYR2, TGFB3, ITGA10 |
| Dilated Cardiomyopathy (DCM) | 4/91 | 0.0363 | ITGB1, RYR2, TGFB3, ITGA10 |
| Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC) | 3/72 | 0.137 | ITGB1, RYR2, ITGA10 |
| Amyotrophic Lateral Sclerosis (ALS) | 2/51 | 0.476 | NEFL, NEFH |
| Pathogenic Escherichia coli infection | 2/55 | 0.476 | ITGB1, TUBB3 |
| Sulfur metabolism | 9/01 | 0.476 | PAPSS2 |
| Leishmaniasis | 2/74 | 0.476 | ITGB1, TGFB3 |
| Pancreatic cancer | 2/75 | 0.476 | TGFB3, CDKN2A |
| Chronic myeloid leukemia | 2/76 | 0.476 | TGFB3, CDKN2A |
| Axon guidance | 3/181 | 0.476 | SEMA6B, ITGB1, EPHA3 |
Table 7: KEGG Pathway enrichment result for UF pattern of DEGs.
Keloids and hypertrophic scar specific downregulated DEGs are associated with keratinization, epidermal development, and the estrogen signaling pathway
Compared to upregulated genes, we identified 809 downregulated genes in exaggerated scars such as hypertrophic scars and keloids. Over representation analysis on GO Biological Process terms found most represented terms as ‘keratinization’ and ‘epidermal development’. Also, in KEGG pathways, ‘estrogen signaling pathway’ was the most represented term (Figure 3) (Tables 8 and 9).
Figure 3: Differentially Expressed Genes (DEGs) specifically upregulated/downregulated in hypertrophic scars and keloids compared to normotrophic scars. A. DEGs in this group did not differ significantly between hypertrophic scars and keloids but were more upregulated than normotrophic scars. Among the 88 genes, RYR2 (ryanodine receptor 2), ARMC3 (armadillo repeat including 3), and SMCO2 (single-pass membrane protein with coiled-coil domains 2) are representative genes located in the upper rank; B. Genes associated with extracellular structural tissue and extracellular matrix tissues are commonly highly expressed in hypertrophic scars and keloids; C. Unlike the above analysis, DEGs in this group did not differ significantly between hypertrophic scars and keloids as in the previous group, but the expression was lower than that of normotrophic scars. It can be confirmed that almost all of the genes included in the upper rank among the 809 genes are keratin-associated proteins; D. And also it can be seen that they are related to keratinization through GO enrichment analysis.
| GO ID | Term | Overlap | Adjusted P-value | Genes |
|---|---|---|---|---|
| GO:0031424 | Keratinization | 26/47 | 7.28E-21 | SPRR4, SPRR2E, SPRR2F, SPRR2G, TCHH, PPL, EVPL, LCE1A, LCE1B, LCE3D, CASP14, CDH3, SFN, SPRR2A, SPRR2B, TGM3, CNFN, LCE1F, LOR, LCE1C, LCE1D, LCE2B, KRT17, LCE6A, SPRR1A, IVL |
| GO:0008544 | Epidermis development | 35/120 | 1.19E-17 | COL17A1, SPRR2E, SPRR2F, SPRR2G, CALML5, BNC1, KLK5, HOXC13, SOX21, EVPL, CST6, KLK7, KRT83, NTF4, CASP14, SOX9, SPRR2A, SPRR2B, DSP, WNT10A, LAMB3, KDF1, KRT34, KRT31, EDAR, ALDH3A2, LCE2B, DCT, KRT17, FABP5, KRT16, KRT15, KRT14, DNASE1L2, SPRR1A |
| GO:0060429 | Epithelium development | 42/256 | 8.96E-12 | SPRR2E, RET, COL17A1, SPRR2F, SPRR2G, FOXC1, CALML5, SDC4, BNC1, KLK5, HOXC13, SOX21, EVPL, CST6, KLK7, KRT83, NTF4, CASP14, CECR2, EPCAM, SOX9, SPRR2A, SPRR2B, DSP, TFAP2B, WNT10A, LAMB3, KDF1, KRT34, KRT31, LCE2B, EDAR, ALDH3A2, BMP2, KRT17, FABP5, DCT, KRT16, KRT15, KRT14, DNASE1L2, SPRR1A |
| GO:0008610 | Lipid biosynthetic process | 52/491 | 1.96E-07 | SLC44A5, PECR, TECR, MSMO1, TFCP2L1, PLB1, TM7SF2, FADS2, FADS6, SPTLC3, GGT1, GGT2, CERS4, DGAT2, EDN2, ELOVL4, HMGCS1, ACSL1, ELOVL3, CYP39A1, MVD, FAR2, SLC27A2, CFTR, CDS1, ACSS2, MVK, FAXDC2, INSIG1, ABHD5, HMGCR, CYP3A4, HSD17B8, AGPAT3, HSD11B1, HSD17B2, PIP5K1B, SPTSSB, SDR42E1, FDPS, PLA2G2D, GK, SRD5A1, DHCR24, ALOX15B, FABP5, GPAM, FASN, PNPLA3, DHCR7, DEGS2, LPIN3 |
| GO:0044283 | Small molecule biosynthetic process | 45/394 | 2.34E-07 | CDS1, SLC44A5, PECR, ACSS2, MVK, GLS2, FAXDC2, INSIG1, TECR, GPT, MSMO1, HMGCR, CYP3A4, HSD17B8, TM7SF2, FADS2, IMPA2, FADS6, KYNU, GGT1, GGT2, FDPS, EDN2, ELOVL4, PKLR, HMGCS1, GPT2, ELOVL3, BBOX1, DHCR24, UPB1, CYP4F12, ALOX15B, CYP39A1, GATM, BDH1, FABP5, FASN, PADI3, MVD, DHCR7, SLC27A2, CFTR, LPIN3, BCAT2 |
| GO:0042633 | Hair cycle | 9/15 | 5.09E-07 | KRT33B, KRTAP4-9, KRTAP4-8, KRTAP4-7, PPP1R13L, KRT16, KRT14, MPZL3, KRT83 |
| GO:0042303 | Molting cycle | 9/15 | 5.09E-07 | KRT33B, KRTAP4-9, KRTAP4-8, KRTAP4-7, PPP1R13L, KRT16, KRT14, MPZL3, KRT83 |
| GO:1901617 | Organic hydroxy compound biosynthetic process | 26/166 | 1.24E-06 | CDS1, SLC44A5, PMEL, OCA2, MVK, INSIG1, TYRP1, MSMO1, HMGCR, CYP3A4, TM7SF2, IMPA2, FDPS, GPR37, HMGCS1, BBOX1, DHCR24, TYR, CYP39A1, DCT, FABP5, MVD, DHCR7, SLC27A2, CFTR, LPIN3 |
| GO:0006694 | Steroid biosynthetic process | 20/108 | 3.94E-06 | SDR42E1, FDPS, MVK, HMGCS1, SRD5A1, INSIG1, MSMO1, DHCR24, TFCP2L1, HMGCR, CYP3A4, HSD17B8, TM7SF2, CYP39A1, HSD11B1, HSD17B2, MVD, DHCR7, SLC27A2, CFTR |
| GO:1901615 | Organic hydroxy compound metabolic process | 47/476 | 5.41E-06 | CDS1, SLC44A5, OCA2, PECR, CEBPA, PMEL, ACSS2, MVK, FOXE1, INSIG1, TYRP1, GDPD2, MSMO1, HMGCR, CYP3A4, PLB1, TM7SF2, SDR16C5, PPIP5K1, IMPA2, AWAT2, LRAT, FDPS, PLA2G2D, GPR37, DGAT2, GK, HMGCS1, SORD, BBOX1, DHCR24, TYR, CYP4F12, CYP39A1, ALDH3A2, AKR1B10, FABP5, SOAT1, DCT, PLCH1, MVD, DHCR7, DEGS2, SLC27A2, GPLD1, CFTR, LPIN3 |
Table 8: GO Biological Process enrichment result for DF pattern of DEGs.
| Term | Overlap | Adjusted P-value | Genes |
|---|---|---|---|
| Estrogen signaling pathway | 19/137 | 0.000726 | CALML5, TGFA, KRT23, KRT34, CALML3, KRT31, ESR1, KRT33B, KRT33A, KRT18, KRT39, KRT17, KRT16, KRT26, KRT15, KRT36, KRT14, PGR, FKBP5 |
| Terpenoid backbone biosynthesis | 6/22 | 0.0246 | FDPS, MVK, HMGCS1, MVD, HMGCR, ACAT2 |
| Steroid biosynthesis | 5/19 | 0.0346 | SOAT1, DHCR24, MSMO1, DHCR7, TM7SF2 |
| Glycerolipid metabolism | 9/61 | 0.0346 | ALDH3A2, DGAT2, AKR1B10, GK, GPAM, PNPLA3, PNLIPRP3, AGPAT3, LPIN3 |
| Tight junction | 17/170 | 0.0346 | MAP3K1, F11R, PRKCZ, CD1A, TIAM1, SLC9A3R1, EPB41L4B, OCLN, CLDN3, MARVELD2, PPP2R2C, CLDN16, EZR, CFTR, LLGL2, TJP2, CRB3 |
| Synthesis and degradation of ketone bodies | 10/04 | 0.0346 | BDH1, HMGCS1, OXCT2, ACAT2 |
| ECM-receptor interaction | 10/82 | 0.0653 | SDC4, LAMB3, LAMC3, ITGB4, ITGA2, LAMB4, COL4A6, COL4A5, ITGB6, GP6 |
| PPAR signaling pathway | 9/74 | 0.0868 | FADS2, GK, FABP5, ACSL1, FABP7, ME1, SLC27A6, ACSBG1, SLC27A2 |
| Steroid hormone biosynthesis | 8/60 | 0.0868 | SULT2B1, HSD11B1, SULT1E1, SRD5A1, HSD17B2, UGT1A4, CYP3A4, HSD17B8 |
| Pyruvate metabolism | 6/39 | 0.118 | ALDH3A2, ACSS2, PC, PKLR, ME1, ACAT2 |
Table 9: KEGG Pathway enrichment result for DF pattern of DEGs.
Hypertrophic scar specific genes are associated with epidermal cell differentiation and the lipid catabolic process
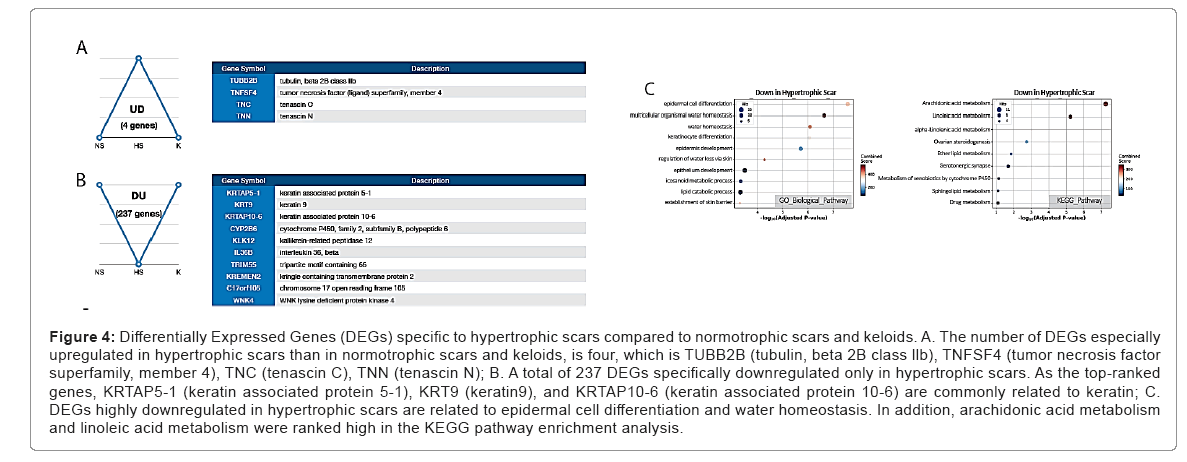
Although hypertrophic scars are reversible, the degree of fibrosis is more severe and prolonged compared to that of normotrophic scars. Therefore, we attempted to find hypertrophic scar-specific genes to distinguish them from both keloids and normotrophic scars. Only four DEGs specifically were upregulated in hypertrophic scars, including TUBB2B (tubulin, beta 2B class 2 B), TNFSF4 (tumor necrosis factor superfamily, member 4), TNC (tenascin C), and TNN (tenascin N). In contrast, a total of 237 DEGs was specifically downregulated in hypertrophic scars. Among the mostly changed genes in fold changes, KRTAP5-1 (keratin associated protein 5-1), KRT9 (keratin9), and KRTAP10-6 (keratin associated protein 10-6) are commonly related to keratin. Over-representation analysis results on GO Biological processes revealed that these hypertrophic scar specific expressions were associated with ‘epidermal cell differentiation’, ‘epidermal development’, and ‘water homeostasis. Conversely, over-represented KEGG pathway terms were ‘arachidonic acid metabolism’, ‘linoleic acid metabolism’ (Figure 4) (Tables 10 and 11).
Figure 4: Differentially Expressed Genes (DEGs) specific to hypertrophic scars compared to normotrophic scars and keloids. A. The number of DEGs especially upregulated in hypertrophic scars than in normotrophic scars and keloids, is four, which is TUBB2B (tubulin, beta 2B class llb), TNFSF4 (tumor necrosis factor superfamily, member 4), TNC (tenascin C), TNN (tenascin N); B. A total of 237 DEGs specifically downregulated only in hypertrophic scars. As the top-ranked genes, KRTAP5-1 (keratin associated protein 5-1), KRT9 (keratin9), and KRTAP10-6 (keratin associated protein 10-6) are commonly related to keratin; C. DEGs highly downregulated in hypertrophic scars are related to epidermal cell differentiation and water homeostasis. In addition, arachidonic acid metabolism and linoleic acid metabolism were ranked high in the KEGG pathw ay enrichment analysis.
| GO ID | Term | Overlap | Adjusted P-value | Genes |
|---|---|---|---|---|
| GO:0009913 | Epidermal cell differentiation | 13/76 | 2.68E-08 | CERS3, CSTA, CDSN, SPINK5, OVOL1, POU2F3, FOXN1, TGM1, SCEL, FA2H, ACER1, IRF6, TP63 |
| GO:0050891 | Multicellular organismal water homeostasis | 9/32 | 2.27E-07 | CDH1, CYP4F2, SCNN1B, SCNN1A, ALOX12B, ABCA12, CLDN1, ALOXE3, TP63 |
| GO:0030104 | Water homeostasis | 9/38 | 8.28E-07 | CDH1, CYP4F2, SCNN1B, SCNN1A, ALOX12B, ABCA12, CLDN1, ALOXE3, TP63 |
| GO:0030216 | Keratinocyte differentiation | 10/54 | 8.88E-07 | TGM1, CERS3, SCEL, ACER1, CSTA, CDSN, IRF6, POU2F3, FOXN1, TP63 |
| GO:0008544 | Epidermis development | 13/120 | 1.94E-06 | TFAP2C, CDSN, GRHL3, KRT5, FOXN1, POU2F3, KRT9, ACER1, SCEL, GJB5, TGM5, SPRR2D, ZNF750 |
| GO:0033561 | Regulation of water loss via skin | 6/19 | 5.36E-05 | CDH1, ALOX12B, ABCA12, CLDN1, ALOXE3, TP63 |
| GO:0060429 | Epithelium development | 15/256 | 0.000325 | TFAP2C, CDSN, GRHL3, KRT5, GATA3, FOXN1, POU2F3, KRT9, ACER1, SCEL, GJB5, TGM5, FGFR3, SPRR2D, ZNF750 |
| GO:0006690 | Icosanoid metabolic process | 9/88 | 0.000473 | CYP2J2;CYP2C9;PLA2G4F;GGT6;PLA2G4B;CYP4F2;CYP4F3;ALOX12B;ALOXE3 |
| GO:0016042 | Lipid catabolic process | 14/232 | 0.000488 | PLA2G2F, PLA2G4F, PLA2G4D, PLA2G4E, CYP4F2, PLA2G4B, CYP4F3, SMPD3, ACER1, LIPH, LIPM, LIPK, PLCH2, PNPLA1 |
| GO:0061436 | Establishment of skin barrier | 5/16 | 0.000507 | ALOX12B, ABCA12, CLDN1, ALOXE3, TP63 |
Table 10: GO Biological Process enrichment result for DU pattern of DEGs.
| Term | Overlap | Adjusted P-value | Genes |
|---|---|---|---|
| Arachidonic acid metabolism | 11/63 | 5.66E-08 | PLA2G2F, CYP2J2, CYP2C9, PLA2G4F, CYP2B6, PLA2G4D, PLA2G4E, PLA2G4B, CYP4F2, CYP4F3, ALOX12B |
| Linoleic acid metabolism | 7/29 | 5.78E-06 | PLA2G2F, CYP2J2, CYP2C9, PLA2G4F, PLA2G4D, PLA2G4E, PLA2G4B |
| alpha-Linolenic acid metabolism | 5/25 | 0.00101 | PLA2G2F, PLA2G4F, PLA2G4D, PLA2G4E, PLA2G4B |
| Ovarian steroidogenesis | 6/49 | 0.00183 | CYP2J2, PLA2G4F, PLA2G4D, STAR, PLA2G4E, PLA2G4B |
| Ether lipid metabolism | 5/47 | 0.0141 | PLA2G2F, PLA2G4F, PLA2G4D, PLA2G4E, PLA2G4B |
| Serotonergic synapse | 7/113 | 0.0205 | CYP2J2, CYP2C9, PLA2G4F, PLA2G4D, PLA2G4E, PLA2G4B, ALOX12B |
| Metabolism of xenobiotics by cytochrome P450 | 5/74 | 0.0715 | ALDH3B2, CYP2C9, CYP2B6, GSTA3, UGT1A6 |
| Sphingolipid metabolism | 4/47 | 0.0787 | SMPD3, CERS3, ACER1, SGPP2 |
| Drug metabolism | 6/108 | 0.0801 | ALDH3B2, CYP2C9, CYP2B6, GSTA3, XDH, UGT1A6 |
| VEGF signaling pathway | 4/59 | 0.162 | PLA2G4F, PLA2G4D, PLA2G4E, PLA2G4B |
Table 11: KEGG Pathway enrichment result for DU pattern of DEGs.
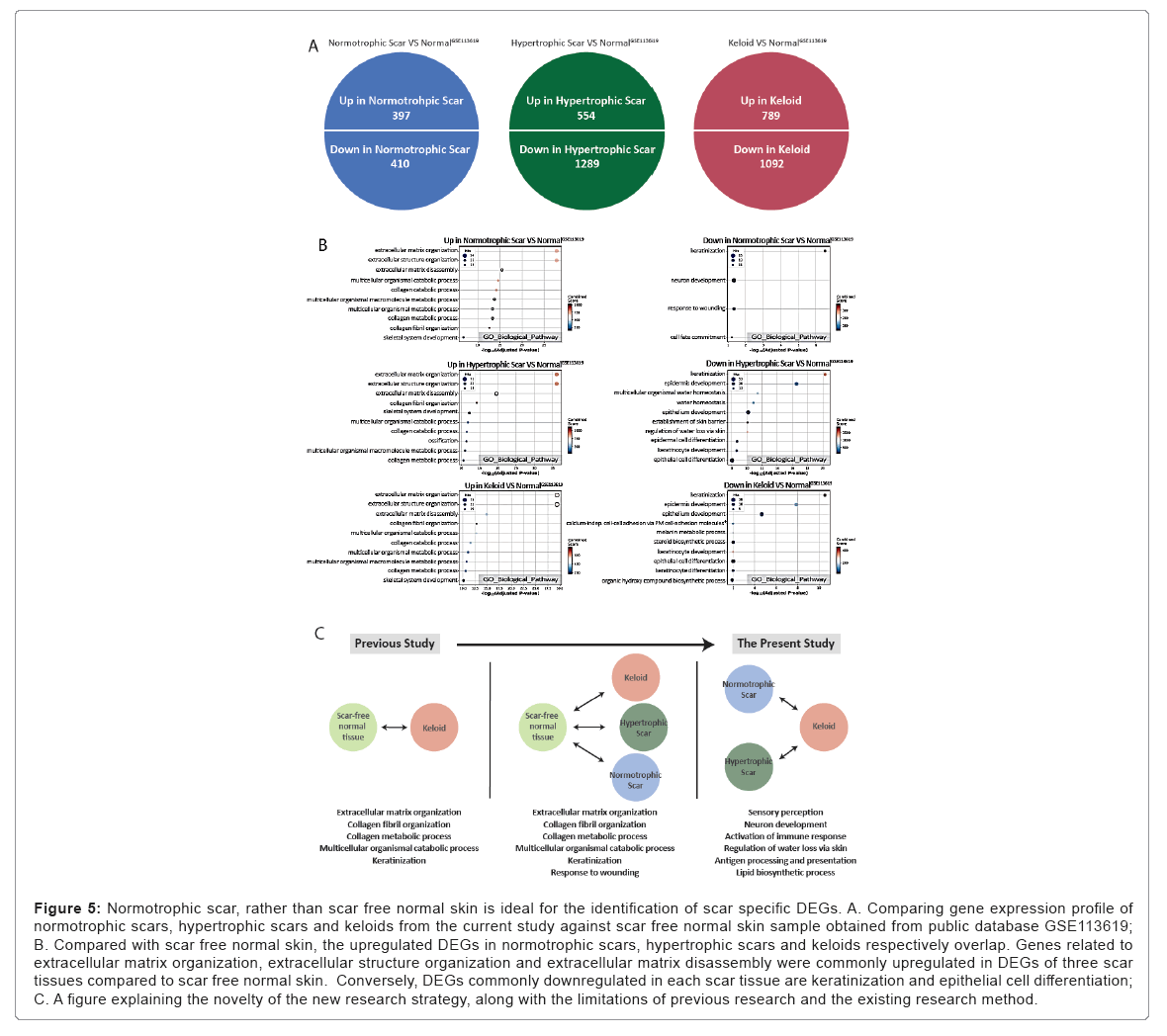
Scar-free normal skin is not a proper control group for determining keloid-specific genes
Previous studies have used scar-free normal skin as a control for comparison with keloids. Since keloids are a possible outcome of wound healing, it is uncertain if it is appropriate to compare keloids with unwounded skin tissue. This limitation suggests that documented studies comparing keloids with scar-free normal skin as a control group are including expression profiles of overall scars rather than keloid- specific characteristics. To overcome this issue, we hypothesized that comparing expression profiles between types of scar tissues would find scar subtype specific features better than comparison between keloid and scar-free normal tissue. For scar-free normal comparisons, we imported publicly available normal skin tissue data from GEO (ID: GSE113619). We implemted the same Ensemble approaches on differential expression analysis and identified DEGs. According to over representation analyses with those DEGs, expression profile changes of keloids, hypertrophic scars, and normotrophic scars than normal skin present very similar shared results especially in GO Biological Processes. Specifically, the genes for ‘extracellular matrix/structure organization’ and ‘extracellular matrix assembly’ were upregulated in all three types of scars compared to normal skin. In addition, the genes for ‘keratinization’, ‘epidermal development’, and ‘epithelial cell differentiation’ were downregulated in all three types of scars. These results imply previous studies that directly compared keloids with scar-free normal skin were more likely to discover general scar- specific features rather than keloid-specific features. Therefore, keloid- specific features can be found better by comparing keloids with other reversible scars instead of with scar free normal skin. From these scar tissue type based comparisons, the keloid-specific genes obtained from this improved comparison were related to sensory perception, neuron development, activation of the immune response, and the lipid biosynthetic process (Figure 5) (Tables 12-17).
Figure 5: Normotrophic scar, rather than scar free normal skin is ideal for the identification of scar specific DEGs. A. Comparing gene expression profile of normotrophic scars, hypertrophic scars and keloids from the current study against scar free normal skin sample obtained from public database GSE113619; B. Compared with scar free normal skin, the upregulated DEGs in normotrophic scars, hypertrophic scars and keloids respectively overlap. Genes related to extracellular matrix organization, extracellular structure organization and extracellular matrix disassembly were commonly upregulated in DEGs of three scar tissues compared to scar free normal skin. Conversely, DEGs commonly downregulated in each scar tissue are keratinization and epithelial cell differentiation; C. A figure explaining the novelty of the new research strategy, along with the limitations of previous research and the existing research method.
| GO ID | Term | Overlap | Adjusted P-value | Genes |
|---|---|---|---|---|
| GO:0030198 | Extracellular matrix organization | 54/359 | 1.44E-28 | COL14A1, SERPINE1, COL12A1, CYR61, LOXL2, COMP, EFEMP2, ADAMTS2, IBSP, COL10A1, POSTN, EGFL6, MMP1, MMP2, MMP3, FGG, BGN, ADAMTS20, MMP11, VCAN, SFRP2, COL2A1, MMP13, COL4A1, COL6A2, PXDN, COL6A1, COL6A3, MATN3, ENG, FBN2, FOXC2, LAMA2, COL11A1, KLK4, LTBP3, NID2, ACAN, ADAMTS14, ABI3BP, SPP1, TGFB3, FN1, SULF1, SULF2, DSPP, COL1A1, COL1A2, COL5A1, ITGA10, COL5A2, MFAP2, COL20A1, FBN1 |
| GO:0043062 | Extracellular structure organization | 54/360 | 1.44E-28 | COL14A1, SERPINE1, COL12A1, CYR61, LOXL2, COMP, EFEMP2, ADAMTS2, IBSP, COL10A1, POSTN, EGFL6, MMP1, MMP2, MMP3, FGG, BGN, ADAMTS20, MMP11, VCAN, SFRP2, COL2A1, MMP13, COL4A1, COL6A2, PXDN, COL6A1, COL6A3, MATN3, ENG, FBN2, FOXC2, LAMA2, COL11A1, KLK4, LTBP3, NID2, ACAN, ADAMTS14, ABI3BP, SPP1, TGFB3, FN1, SULF1, SULF2, DSPP, COL1A1, COL1A2, COL5A1, ITGA10, COL5A2, MFAP2, COL20A1, FBN1 |
| GO:0022617 | Extracellular matrix disassembly | 25/116 | 3.48E-16 | FBN2, COL14A1, COL11A1, COL12A1, KLK4, ACAN, SPP1, COL10A1, MMP1, MMP2, MMP3, FN1, COL1A1, MMP11, COL1A2, COL2A1, MMP13, COL5A1, COL4A1, COL6A2, COL5A2, COL6A1, COL6A3, FBN1, ENG |
| GO:0044243 | Multicellular organismal catabolic process | 21/80 | 2.35E-15 | ACE, COL14A1, MMP1, MMP2, COL11A1, COL12A1, MMP3, COL1A1, MMP11, ADAMTS2, ADAMTS14, MMP13, COL2A1, COL1A2, COL5A1, COL4A1, COL6A2, COL6A1, COL5A2, COL10A1, COL6A3 |
| GO:0030574 | Collagen catabolic process | 20/74 | 6.36E-15 | COL14A1, MMP1, MMP2, COL11A1, COL12A1, MMP3, COL1A1, MMP11, ADAMTS2, ADAMTS14, MMP13, COL2A1, COL1A2, COL5A1, COL4A1, COL6A2, COL6A1, COL5A2, COL10A1, COL6A3 |
| GO:0044259 | Multicellular organismal macromolecule metabolic process | 21/89 | 1.72E-14 | ACE, COL14A1, MMP1, MMP2, COL11A1, COL12A1, MMP3, COL1A1, MMP11, ADAMTS2, ADAMTS14, MMP13, COL2A1, COL1A2, COL5A1, COL4A1, COL6A2, COL6A1, COL5A2, COL10A1, COL6A3 |
| GO:0044236 | Multicellular organismal metabolic process | 21/94 | 4.69E-14 | ACE, COL14A1, MMP1, MMP2, COL11A1, COL12A1, MMP3, COL1A1, MMP11, ADAMTS2, ADAMTS14, MMP13, COL2A1, COL1A2, COL5A1, COL4A1, COL6A2, COL6A1, COL5A2, COL10A1, COL6A3 |
| GO:0032963 | Collagen metabolic process | 20/83 | 4.69E-14 | COL14A1, MMP1, MMP2, COL11A1, COL12A1, MMP3, COL1A1, MMP11, ADAMTS2, ADAMTS14, MMP13, COL2A1, COL1A2, COL5A1, COL4A1, COL6A2, COL6A1, COL5A2, COL10A1, COL6A3 |
| GO:0030199 | Collagen fibril organization | 15/41 | 2.55E-13 | FOXC2, COL14A1, COL11A1, COL12A1, LOXL2, COL1A1, ACAN, MMP11, ADAMTS14, ADAMTS2, COL1A2, COL2A1, SFRP2, COL5A1, COL5A2 |
| GO:0001501 | Skeletal system development | 19/162 | 1.49E-07 | POSTN, COL12A1, IGF2, AEBP1, SULF1, PAPSS2, PAX1, SULF2, DSPP, COMP, COL1A1, ACAN, COL1A2, COL2A1, COL5A2, COL10A1, ARSE, MATN3, FBN1 |
Table 12: GO Biological Process enrichment result for up regulated DEGs in Normotrophic Scar VS. Normal (GSE113619).
| GO ID | Term | Overlap | Adjusted P-value | Genes |
|---|---|---|---|---|
| GO:0031424 | Keratinization | 12/47 | 3.08E-07 | LCE1A, LCE2B, LCE1B, LCE4A, LCE6A, LCE1E, EVPL, LCE1F, LCE2D, LOR, LCE1C, LCE1D |
| GO:0030182 | Neuron differentiation | 15/222 | 0.0461 | BCL11B, WNT3A, WNT7B, WNT9B, WNT7A, RORA, WNT16, GDF7, NTF4, NGRN, ID4, FGFR3, WNT3, HES5, WNT4 |
| GO:0009611 | Response to wounding | 13/167 | 0.0461 | SERPINB2, PLLP, TGFA, NMNAT3, KLK8, POU2F3, EREG, RTN4RL1, CYP1A1, BCL2, TSPO, FGFR3, MATN4 |
| GO:0045165 | Cell fate commitment | 11/134 | 0.0608 | WNT3A, WNT7B, BCL2, WNT9B, WNT7A, WNT16, FGFR3, WNT3, HES5, WNT4, GDF7 |
| GO:0060429 | Epithelium development | 15/256 | 0.131 | CALML5, SDC4, WNT7B, KDF1, KLK5, FOXJ1, EVPL, POU2F3, POU3F3, LCE2B, NTF4, FGFR3, WNT3, HES5, WNT4 |
| GO:0035588 | G-protein coupled purinergic receptor signaling pathway | 4/17 | 0.131 | ADORA2A, P2RY2, P2RY1, ADORA1 |
| GO:0061098 | Positive regulation of protein tyrosine kinase activity | 5/32 | 0.131 | WNT3A, DOK7, ADORA1, TGFA, EREG |
| GO:0014014 | Negative regulation of gliogenesis | 5/32 | 0.131 | ID4, TSPO, ASCL2, FGFR3, HES5 |
| GO:0022010 | Central nervous system myelination | 9/03 | 0.131 | ID4, FGFR3, HES5 |
| GO:0060428 | Lung epithelium development | 9/03 | 0.131 | WNT7B, FOXJ1, FGFR3 |
Table 13: GO Biological Process enrichment result for down regulated DEGs in Normotrophic Scar VS Normal (GSE113619).
| GO ID | Term | Overlap | Adjusted P-value | Genes |
|---|---|---|---|---|
| GO:0030198 | Extracellular matrix organization | 71/359 | 8.08E-37 | OLFML2B, COL14A1, ELN, SERPINE1, COL12A1, TNC, SCX, PLOD2, PLOD1, CYR61, LOXL2, COMP, ADAMTS4, EFEMP2, ADAMTS2, IBSP, SH3PXD2B, COL10A1, POSTN, EGFL6, MMP2, BGN, HSPG2, ADAMTS20, MMP11, VCAN, SFRP2, COL2A1, MMP13, COL4A1, COL6A2, PXDN, COL6A1, COL8A1, COL6A3, MATN3, ENG, FBN2, FOXC2, LAMA2, COL11A1, LAMA4, KLK4, LTBP3, NID2, ACAN, ADAMTS14, ABI3BP, SPP1, TGFB1, COL24A1, TGFB3, LUM, FN1, LAMB1, SULF1, GDF5, SULF2, DSPP, COL1A1, COL3A1, COL1A2, BMP1, COL5A1, WT1, ITGA10, COL5A2, ITGA11, MFAP2, TLL2, FBN1 |
| GO:0043062 | Extracellular structure organization | 71/360 | 8.08E-37 | OLFML2B, COL14A1, ELN, SERPINE1, COL12A1, TNC, SCX, PLOD2, PLOD1, CYR61, LOXL2, COMP, ADAMTS4, EFEMP2, ADAMTS2, IBSP, SH3PXD2B, COL10A1, POSTN, EGFL6, MMP2, BGN, HSPG2, ADAMTS20, MMP11, VCAN, SFRP2, COL2A1, MMP13, COL4A1, COL6A2, PXDN, COL6A1, COL8A1, COL6A3, MATN3, ENG, FBN2, FOXC2, LAMA2, COL11A1, LAMA4, KLK4, LTBP3, NID2, ACAN, ADAMTS14, ABI3BP, SPP1, TGFB1, COL24A1, TGFB3, LUM, FN1, LAMB1, SULF1, GDF5, SULF2, DSPP, COL1A1, COL3A1, COL1A2, BMP1, COL5A1, WT1, ITGA10, COL5A2, ITGA11, MFAP2, TLL2, FBN1 |
| GO:0022617 | Extracellular matrix disassembly | 32/116 | 2.68E-20 | FBN2, COL14A1, ELN, COL11A1, COL12A1, KLK4, ADAMTS4, ACAN, SH3PXD2B, SPP1, COL10A1, MMP2, FN1, LAMB1, HSPG2, COL1A1, MMP11, COL3A1, COL1A2, BMP1, COL2A1, MMP13, COL5A1, COL4A1, COL6A2, COL5A2, COL6A1, COL8A1, COL6A3, TLL2, FBN1, ENG |
| GO:0030199 | Collagen fibril organization | 18/41 | 5.93E-15 | FOXC2, COL14A1, LUM, COL11A1, COL12A1, SCX, LOXL2, COL1A1, ACAN, MMP11, ADAMTS14, ADAMTS2, COL3A1, COL1A2, COL2A1, SFRP2, COL5A1, COL5A2 |
| GO:0001501 | Skeletal system development | 29/162 | 6.02E-13 | HOXD1, COL12A1, SCX, AEBP1, PHEX, PAPSS2, COMP, ADAMTS4, ACAN, SH3PXD2B, COL10A1, ARSE, POSTN, DMRT2, IGF2, SULF1, PAX1, SULF2, DSPP, COL1A1, COL3A1, COL1A2, BMP1, COL2A1, COL5A2, CDH11, ALPL, MATN3, FBN1 |
| GO:0044243 | Multicellular organismal catabolic process | 21/80 | 1.50E-12 | ACE, COL14A1, MMP2, COL11A1, COL12A1, COL1A1, MMP11, ADAMTS2, ADAMTS14, COL3A1, MMP13, COL2A1, COL1A2, COL5A1, COL4A1, COL6A2, COL6A1, COL5A2, COL10A1, COL8A1, COL6A3 |
| GO:0030574 | Collagen catabolic process | 20/74 | 3.19E-12 | COL14A1, MMP2, COL11A1, COL12A1, COL1A1, MMP11, ADAMTS2, ADAMTS14, COL3A1, MMP13, COL2A1, COL1A2, COL5A1, COL4A1, COL6A2, COL6A1, COL5A2, COL10A1, COL8A1, COL6A3 |
| GO:0001503 | Ossification | 24/116 | 3.93E-12 | FOXC2, ECM1, TGFB1, TGFB3, MMP2, BMP8A, SCX, CHRDL2, HSPG2, RUNX2, CTGF, DSPP, COL1A1, IGSF10, MN1, BMP1, COL2A1, MMP13, CDH11, GNAS, SPP1, ALPL, COL10A1, CTHRC1 |
| GO:0044259 | Multicellular organismal macromolecule metabolic process | 21/89 | 1.02E-11 | ACE, COL14A1, MMP2, COL11A1, COL12A1, COL1A1, MMP11, ADAMTS2, ADAMTS14, COL3A1, MMP13, COL2A1, COL1A2, COL5A1, COL4A1, COL6A2, COL6A1, COL5A2, COL10A1, COL8A1, COL6A3 |
| GO:0032963 | Collagen metabolic process | 20/83 | 2.46E-11 | COL14A1, MMP2, COL11A1, COL12A1, COL1A1, MMP11, ADAMTS2, ADAMTS14, COL3A1, MMP13, COL2A1, COL1A2, COL5A1, COL4A1, COL6A2, COL6A1, COL5A2, COL10A1, COL8A1, COL6A3 |
Table 14: GO Biological Process enrichment result for up regulated DEGs in Hypertrophic Scar VS. Normal (GSE113619).
| GO ID | Term | Overlap | Adjusted P-value | Genes |
|---|---|---|---|---|
| GO:0031424 | Keratinization | 30/47 | 4.70E-21 | SPRR2E, ABCA12, PPL, EVPL, LCE2D, TGM1, LCE1A, LCE1B, LCE3D, CASP14, LCE5A, SFN, TGM3, SPRR2D, KRT2, CNFN, LCE1E, LCE1F, LOR, LCE1C, LCE3E, LCE1D, LCE2B, LCE2C, LCE2A, KRT17, LCE4A, LCE6A, SPRR1B, IVL |
| GO:0008544 | Epidermis development | 42/120 | 2.90E-17 | COL17A1, SPRR2E, CALML5, BNC1, KLK5, HOXC13, SOX21, EVPL, CST6, KLK7, NTF4, CASP14, SCEL, ALX4, TGM5, SPRR2D, DSP, TFAP2C, WNT10A, CDSN, LAMB3, KRT2, KDF1, GRHL3, KRT5, KRT31, FOXN1, POU2F3, KRT9, EDAR, LCE2B, ACER1, ELF3, DCT, KRT17, KRT16, KRT15, GJB5, KRT14, DNASE1L2, SPRR1B, ZNF750 |
| GO:0050891 | Multicellular organismal water homeostasis | 19/32 | 3.93E-12 | FLG, CYP4F2, KRT1, KDF1, ALOX12B, ALOX12, ABCA12, CLDN1, CYP4F12, FLG2, SCNN1G, CLDN4, CDH1, SCNN1B, TMEM79, SCNN1A, SFN, ALOXE3, TP63 |
| GO:0030104 | Water homeostasis | 20/38 | 1.32E-11 |