Methanotrophs: The Natural Way to Tackle Greenhouse Effect
Received: 22-Jan-2018 / Accepted Date: 15-Feb-2018 / Published Date: 19-Feb-2018 DOI: 10.4172/2155-6199.1000432
Abstract
Methane is considered as an important greenhouse gas which produced from a wide range of anthropogenic and natural sources. It plays a key role in global warming. Capturing and disposal of methane is technically both costly and problematic. A low-cost alternate to the conventional methods is the microbial oxidation of methane. Methanotrophs are the type of bacteria that aerobically oxidize methane as a source of energy through their key enzyme, monooxygenase (MMO), especially the soluble MMO, it is noteworthy in its broad substrate specificity. This exceptional capability, i.e., catalyzing reactions of environmental importance, has owned methanotrophs attention from applied microbiologists and biotechnologist. From about 30 years, it is observed that copper (Cu) is playing an important role in the physiology and activity of methanotrophs, but the discovery of how Cu collect by these cells, more importantly what is the role Cu playing in CH4 oxidation by the particulate CH4 monooxygenase and how Cu affects the ability of methanotrophs to oxidize different substrates has been made. In this review we summarize the morphology, phylogeny, ecology, and possible applications of methanotrophs to address the global as well as regional issues, along with the role of gene expression regulation by Cu and how it affects the cell activity of methane-oxidizing bacteria. Our focus was on two main aspects of potential value and application of methanotrophs in environmental bioremediation, namely physiology along with working of methanotrophs and methane removal from atmosphere.
Keywords: Methanotrophs; Mono-oxygenase (MMO); Methane; Greenhouse effect; Methanobactins
Introduction
Methane (CH4) is a potent greenhouse gas and its concentration in the atmosphere is since forever on an up rise with an approximate 1% increase per year [1]. It is a major contributor to direct radiative forcing from all the long lasting greenhouse gases (GHGs) accountable for about 20% of it (IPCC, 2001). Industrial revolution has led to the amplified CH4 concentration from 0.7 to 1.8 p.p.m.v. due to increased anthropogenic inputs [2]. This increased anthropogenic emission of greenhouse gases is what has led to the globally hyped phenomena known as “The Greenhouse Effect”. This occurrence can potentially direct towards hazardous changes in climate namely temperature fluctuations leading to disastrous outcomes.
GHGs, chiefly CH4 reduction can lead to stabilization of the effect resulting in optimization of global temperatures. However, curbing these gases is difficult i.e., capturing and disposal of methane is technically costly and problematic. A low-cost alternate to the conventional methods is the microbial oxidation of methane [3,4].
Methanotrophs aka Methane-oxidizing bacteria, are a major group of bacteria primarily responsible for running the naturally present global methane cycle. Thus, methanotrophs consist of one of the major groups of free-living microorganisms that metabolically playing role in determining the nature of the environment. Although it is self-evident that methanotrophs play a role in maintaining low levels of methane in the atmosphere but to do so they require certain conditions and environmental factors [5].
Phylogenetic and physiological diversity of methanotrophs should be considered of prime focus while working with them since they are newly discovered vague assets of bioremediation at the moment. Some other important features to be considered are how methanotrophs can be used as a pollutant degrader by removing greenhouse gas and Cu role in methanotrophic regulating activities by shedding light on mechanism for its uptake used by methanotrophs [6].
The above mentioned aspect are examined in this review, and an attempt is made to give a contemporary view of the environmental significance of these microorganisms and to evaluate the potential for their industrial exploitation.
Diversity of Methanogens: A Taxonomic and Phylogenetic overview
Methanotrophic bacteria have been a center of attention since their discovery by Kaserer and Sohngen in the start of 20th century. In 1906, Bacillus methanicum was the first methane oxidizing bacteria to be isolated but it was not until Whittenbury and his colleague’s isolation and characterization of over a 100 new methanotrophs that the current basis of classification of these bacteria was established [7].
Classification
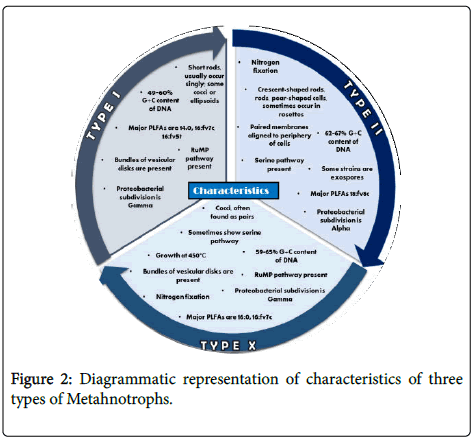
The classification of methanotrophs according to researchers is as so that these bacteria are separated into five groups (proposed genera) on the basis of resting stage formed, morphological differences, structure of intracytoplasmic membranes and some other physiological characteristics. Study shows that, methanotrophs have 14 recognize genera, which then categorized into 3 main types i.e., type I, type II and type X, based on the above mentioned basis i.e., morphology, structure of membrane and some other physiological characteristics ( Figure 1). The discovery of diverse types helped us to further study of methane-utilizing bacteria and has led to the belief that natural environment is more suitable for them [5,7].
Figure 1: All known methanotrophs showing phylogenetic link created with the help of 16sRNA gene sequence using MEGA4 [8].
Types: Type I is the type of methanotrophs that dominants in environments where methane is present in a limited quantity and relatively high levels of combined nitrogen and copper are present. Type II bacteria on the other hand favors environment with high concentration of methane, as well as the environment where dissolved oxygen level is low, and restrictive combined concentration of nitrogen and/or copper. Type I strains can also be characterized on the basis of having other apart from condition characteristics i.e., they have intracytoplasmic membranes throughout the cell in the form vascular disk present in bundles, for carbon absorption they use ribulose monophosphate (RuMP) pathway. They also have phospholipid fatty acids which consist of carbon length of 14 and 16. They can be characterized in a similar comparative way i.e., they too have an intracytoplasmic membranes but unlike Type I it is aligned along the periphery of the cell, with the help of serine pathway they assimilate carbons and they have phospholipid fatty acids with 18 carbon length [ 5,9].
Type X: The best of both strains: Type X strains are a combination of Type I and II strains i.e., they have phospholipid fatty acids of 16 carbon, a RuMP pathway along with possessing ribulose-1,5- bisphosphate, and ability to grow at a temperatures which is higher than Type I or II strains [5]. Type X methanotrophs though are quite similar to Type I they are distinguished from type I methanotrophs on the basis that they have enzyme of serine pathway which are lower in levels named as ribulosebisphosphate carboxylase. This enzyme is present in the Calvin Benson cycle. Type X have a DNA, with a distinctive property that it contains G1C content with high moles percentage as compared to type I methanotrophs. Type X have the property that they can grow at higher temperature which isn’t present in any other type [10,11].
Proteobacteria defined types: The main difference between the types is the pathway they utilize. For the assimilation of carbon type I methanotrophs utilize RuMP pathway, as they are Gammaproteo bacteria (Figure 2). As type II methanotrophs uses Serine pathway for carbon absorption, so they are Alphaproteo bacteria [12].
Ecology
Majority of methane-oxidixzing species of bacteria isolated from a wide range of environments suggest that they are mostly aerobic and obligate in nature. However, it cannot be established as a fact due to two main reasons. Firstly, for a fact methane is continuously generated in anaerobic environments, and there is good evidence for its anaerobic oxidation linked which is linked to sulfate reduction by uncharacterized microorganisms present in sediments. Secondly, there is no surety as to if this is the true reflection of relative abundance or just an artifact due to isolation procedures. A thing for ecology for sure is that aerobic environment e.g., soils, surface layers of sediments and natural waters where methane is diffusing, have diversity in aerobic methanotrophs population [13,14].
MMO at Work
Methane monooxygenase (MMO), all known aerobic methanotrophs the first step in oxidation is the methane converstion into methanol by using MMO [15]. In the second step the methanol further oxidized into formaldehyde, after that it has two options, first is to convert into biomass and other is the further oxidation into formate and then into CO2.
Two iso-enzymes of MMO is known: soluble MMO (sMMO), found only as subset of known methanotrophs, and also membrane bound (or particulate) MMO (pMMO). It is located in specialized internal membrane structures, called ICMs [15,16]. sMMO and pMMO both have mixed function of oxidation, that is one atom from O2 goes to methanol and the other to water, involving the input of 2 electrons and 2 protons. sMMO utilizes NADH, but it is still unknown that what is the physiological electron donor to the MMO [15].
pMMO vs sMMo
pMMO is found in almost all known methanotrophs, it also shows more affinity towards methane when it is compared to cells which are expressing sMMO (Figure 3). Further studies show that cells shows higher growth yield which are using pMMO for growth, which signifies that oxidation of methane is more effectual by pMMO [17].
Mthanotroph “The Degrader”
In the prevailing twenty five years, study shows that methaneoxidizing bacteria have the ability to degrade wide ranges of halogenated hydrocarbons. Methanotrophic enrichments are capable of degrading priority pollutants e.g., chlorinated hydrocarbons [18], in US and various other countries they are present in aquifers, landfills, wastewaters, and waste disposal sites [19,20]. Methanotrophs make for a good pollutant degrader given the abilities that methanotrophs are ubiquitous and the rate of degradation of pollutant with the help of methanotrophs have magnitude greater than that have witnessed by other cells having monooxygenases [21].
pMMO: The main degrader MMO
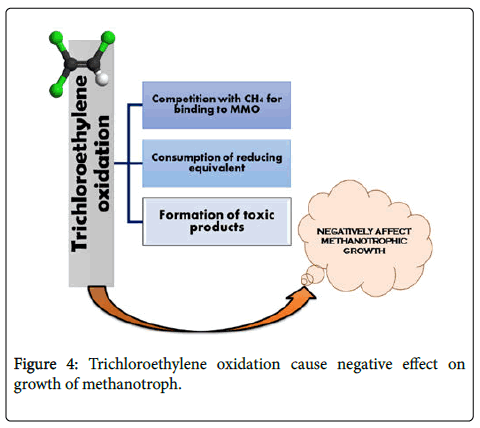
sMMO was initially thought of as the MMO responsible for degredation given it’s broader substrate range. However, in a study by DiSpirito et al. [22], it is stated that pMMO could indeed degrade trichloroethylene shattering the old belief that it cannot degrade chlorinated solvents [22]. On a side note it was noted that increasing Cu availability led to increased degradation of trichloroethylene [ 23,24].
Primarily owing to mRNA levels and enzymatic activity which is present in different environments, it is largely difficult to measure prevalent form of MMO expressed in in-situ communities [25]. Using competitive RT-PCR assays cultures from land resources like soil slurries and groundwater from a mixture of two gases i.e., chloroethane/chloroethene plume were observed they showed only pMMO instead of sMMO concluding that on the site degradation of chlorinated hydrocarbons are done by the pMMO expressing cells [ 26].
Further lab studies show that survival chance of pMMO expressing cells is increases if chlorinated ethenes are present and also they have the ability of degradation of these compounds in higher concentration because of pMMO specificity of CH4 and pollutant transformation to more toxic products in slow time [27,28] (Figure 4).
Unique Cu-Specific Uptake Systems Present in Methanotrophs
Copper is considered to be the key factor in controlling Methanotrophic activity along with being of central importance in the physiology and metabolism of methanotrophs. From past decade the activity of methane-oxidizing bacteria is popular, particularly from the perspective of sMMO and pMMO activities [29-31]. Methanotophs respond differently toward the diverse concentration of Cu. Organic acids such as humic acid can decreased the bioavailabity of Cu especially in subsurface environments leading to different response of methanotrophs [32].
Copper is important because pMMO is copper dependent i.e., the first step in methanotrophic metabolism pathway depends on it, while on the other hand copper determines the MMO type that is during copper starved conditions sMMO are produced. Few methanotrophs strain possess copper mediated switches between MMO’s helping in transcription repression of sMMO gene, which resulting in the enhancing of pMMO expression, as well as formation of the intracytoplasmic membrane. This copper switch mechanism is yet to be clarified [15,33].
Methanobactins: The copper binding molecules
Recent studies have suggested that Methanobactin (Mb) plays important role in copper uptake by methanotrophs. They are basically copper-binding molecules with low-molecular mass, and are secreted by many methanotrophic bacteria, specifically M. trichosporium OB3b. Methanotrophs fulfill their copper requirement by the secretion of mb. They bind either Cu+ without changing their oxidation state or Cu 2+ which is later reduced to Cu+ [34,35]. Choi et al. [36] suggested that Cu-mb work as a vehicle which directs the electrons from the in vivo reductant toward the dinuclear Cu site, with subsequent transfer to the diiron site for CH4 oxidation [36] (Figure 5).
Application of Methanotrophs in Greenhouse Gas Removal
Global warming poses a fundamental threat to planet Earth, and emission of greenhouse gases play a key role in this. From the decade questions like: What’s the best way to reduce anthropogenic emission of these gases? How CH4 emitting from landfills and agricultural soil can be prevented? Have been asked.
CH4 sink
Approximately 10-90% of CH4 produced from the methanogenic activity is consumed by methanotrophs even before it can be released into the environment, making methanotrophs key proprietor in degradation of CH4 from atmosphere for e.g., acidic forest soil express pMMO activity for Methanotrophic oxidation of CH4. Reduction of free CH4 with the help of methanotrophs is now attracting attention from the researchers since this activity broadly occurs in landfills which covers soil thus making landfills natural CH4 sinks [37-39].
Engineered strategies for methane removal
Various strategies can be adopted considering stimulation of Methanotrophic activity for CH4 reduction in atmosphere. ‘Biofilters or Biocovers’ are the well-known engineered system involved in this activity (Figure 6).
Biocovers are typically organic matter, where the permeable material, may it be compost, sewage sludge or wood chips, cover the surface area of landfill. These materials have the characteristic of effective gas transportation for both CH4 coming out from landfills and the atmospheric oxygen, along with the ability of water retention for the Methanotrophic activity [40,41].
Strain discovery (pMMO2)
Knief and Dunfield isolated two strains of methanotrophs which helps in CH4 reduction by the time period of 3 months, these strains however are unable to grow in atmospheric concentration of CH4 and require special conditions. Surprisingly, a new strain of Methylocystis has been discovered which has the ability to grow at 10 p.p.m.v. CH4. Further studies of this strain shows that this cell has two pMMO isozymes, pMMO1 and pMMO2 leading to the discovery that Oxidation of CH4 by pMMO2 is more effective than pMMO1 [42,43].
Biotechnology Meets Methanotrophs
There are only few methane based products present commercially but, none of them involves methanotrophs strains which are metabolically engineered. The reason behind this is the limited study of methanotrophs and studies only being done during the production process for biopolymers, vitamins, antibiotics, single cell protein (SCP) as well as carboxylic acids [16,44].
However, the studies that have been done show that poly-β- hydroxybutyrate (PHB), which is a biopolymer produced by methanotrophs have biomedical applications, can be used as biodegradable food packaging, and encapsulation of agrochemicals on a large scale [45].
Biotechnology play an important role in methanotrophic bioremediation by the conversion of methane into SCP at industrial scale by Methanotrophic cultivation. This concept is not more than 3 decades old, but remarkably enhancement is only now in rapid expansion of genetic and metabolic knowledge [46,47].
Unfortunately limited studies are done to make methanotrophs useful for the purpose of biocatalysts in the lipid or fuel production with Bio-GTL process, mainly because there is no suitable strain is produced or discovered [48].
Scientists of 21st century are now using biotechnology for enhancing Methanotrophic bacteria effectively, that is controlling of methane emission, bioremediation, biobleaching and well known Methanotrophic based biofilters for landfill methane reduction [41,49].
MB an asset for biotechnologist
Many species of methanotrophs, which expresses pMMO activities, produce Methanobactin (mb), when there is high demand of copper, as it binds and reduce copper at high affinity. This high binding affinity catch interest of biotechnologists, for using them in biotechnological application including controlling of copper homeostasis in Wilson’s disease patient by working as a therapeutic agent [50].
Interestingly, mb can also bind other metals like binding and reduction of both trivalent gold and bivalent mercury ions, their mechanism of binding is very much similar to that of copper ion binding. This can lead to the using of methanotrophs in the field of biobleaching with mining and environmental remediation. Gold nanoparticles and uniform copper can also be produced by Methanobactin [36,49].
Conclusions and Future Prospects
In this review, we try our best to summarize current knowledge on morphology and application of methanotrophs in environmental bioremediation, mainly for removal of methane from atmosphere. These interesting microorganisms were merely discovered not more than a century ago and attracting great interest. There are plenty more possibilities then discussed in the literature above to make methaneoxidizing bacteria become important and universal microorganism industries. Role of methanotrophs in biogeochemical carbon cycle and also in controlling of global climate change can’t be neglected. There is still too much to discover about these organisms like how can unique Methanotrophic structures such as acidophilic, thermoacidophilic, and nitrite-utilizing methanotrophs be best used for the purpose of pollutant degradation? How prevalent are these types of methanotrophs? Can they be with little effort stimulated in situ? Answers to all the above issues will not only guides us to the use of methanotrophs for pollutant degradation but will also help act as a gateway to other interesting issues in methanotrophy. We believe that mighty progress in basic research, together with novel and cuttingedge biotechnological methods eventually will enable the engineering applications of methanotrophs to be realized.
Acknowledgment
I am highly grateful to my teacher and mentor Dr. Bushra Uzair, for helping and guiding me in writing this review article.
References
- Lelieveld J, Crutzen P, Brühl C (1993) Climate effects of atmospheric methane. Chemosphere 26: 739-768.
- Etheridge DM, Steele L, Francey RJ, Langenfelds RL (1998) Atmospheric methane between 1000 AD and present: Evidence of anthropogenic emissions and climatic variability. Journal of Geophysical Research: Atmospheres 103: 15979-15993.
- Brosius LS (2010) Investigating controls over methane production and bubbling from interior Alaskan lakes using stable isotopes and radiocarbon ages.
- Reeburgh WS (2007) Oceanic methane biogeochemistry. Chemical Reviews 107: 486-513.
- Hanson RS, Hanson TE (1996) Methanotrophic bacteria. Microbiological Reviews 60: 439-471.
- Conrad R (1996) Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiological Reviews 60: 609-640.
- Knief C (2015) Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Frontiers in Microbiology 6: 1346.
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24: 1596-1599.
- Bowman JP, McCammon SA, Skerrat JH (1997) Methylosphaera hansonii gen. nov., sp. nov., a psychrophilic, group I methanotroph from Antarctic marine-salinity, meromictic lakes. Microbiology 143:Â 1451-1459.
- Whittenbury R (1981) The interrelationship of autotrophy and methylotrophy as seen in Methylococcus capsulatus (Bath). Microbial growth on C1 compounds. Heyden, London, UK, pp: 181-190.
- Alvarez-Cohen L, McCarty PL, Boulygina E, Hanson RS, Brusseau GA, et al. (1992) Characterization of a methane-utilizing bacterium from a bacterial consortium that rapidly degrades trichloroethylene and chloroform. Am Soc Microbiol 58: 1886-1893
- Hanson RL, Hanson RL, Imperatore G, Venkat Narayan KM, Roumain J, et al. (2001) Family and genetic studies of indices of insulin sensitivity and insulin secretion in Pima Indians. Diabetes/Metabolism Research and Reviews 17: 296-303.
- Bezrukova LV, Nikolenko Y, Nesterov AI, Galchenko VF, Ivanov MV, et al. (1983) Comparative serological analysis of methanotrophic bacteria. Mikrobiologiya 52: 800-805.
- De Angelis MA (1989) Studies of microbial methane oxidation in deep-sea hydrothermal vent environments. University of Washington, Washington, USA.
- Semrau JD, DiSpirito AA, Yoon S (2010) Methanotrophs and copper. FEMS Microbiology Reviews 34: 496-531.
- Anthony C (1982) Biochemistry of methylotrophs. Academic Press, Massachusetts, USA.
- Leak DJ, Dalton H (1986) Growth yields of methanotrophs 2. A theoretical analysis. Applied Microbiology and Biotechnology 23: 477-481.
- Wilson JT, Wilson BH (1985) Biotransformation of trichloroethylene in soil. Applied and Environmental Microbiology 49: 242.
- Westrick JJ, Mello JW, Thomas RF (1984) The groundwater supply survey. American Water Works Association 76: 52-59.
- Semprini L (1997) In situ transformation of halogenated aliphatic compounds under anaerobic conditions. Subsurface Restoration, Ann Arbor Press Inc., Chelsea, Michigan, USA, pp: 429-450.
- Oldenhuis R, Vink RL, Janssen DB, Witholt B (1989) Degradation of chlorinated aliphatic hydrocarbons by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Applied and Environmental Microbiology 55: 2819-2826.
- DiSpirito AA, Gulledge J, Shiemke AK, Murrell JC, Lidstrom ME, et al. (1991) Trichloroethylene oxidation by the membrane-associated methane monooxygenase in type I, type II and type X methanotrophs. Biodegradation 2: 151-164.
- Osei K, Rhinesmith S, Gaillard T, Schuster D (2004) Impaired Insulin Sensitivity, Insulin Secretion, and Glucose Effectiveness Predict Future Development of Impaired Glucose Tolerance and Type 2 Diabetes in Pre-Diabetic African Americans Implications for primary diabetes prevention. Diabetes Care 27: 1439-1446.
- Lontoh S, Semrau JD (1998) Methane and Trichloroethylene Degradation by Methylosinus trichosporium OB3b Expressing Particulate Methane Monooxygenase. Applied and Environmental Microbiology 64: 1106-1114.
- Bælum J, Nicolaisen MH, Holben WE, Strobel BW, Sørensen J, et al. (2008) Direct analysis of tfdA gene expression by indigenous bacteria in phenoxy acid amended agricultural soil. The ISME Journal 2: 677-687.
- Forrester SB, Han JI, Dybas MJ, Semrau JD, Lastoskie CM (2005) Characterization of a mixed methanotrophic culture capable of chloroethylene degradation. Environmental Engineering Science 22: 177-186.
- Lee SW, Keeney DR, Lim DH, Dispirito AA, Semrau JD (2006) Mixed pollutant degradation by Methylosinus trichosporium OB3b expressing either soluble or particulate methane monooxygenase: can the tortoise beat the hare? Applied and Environmental Microbiology 72: 7503-7509.
- Yoon S and Semrau JD (2008) Measurement and modeling of multiple substrate oxidation by methanotrophs at 20 C. FEMS Microbiology Letters 287: 156-162.
- Dalton H (2005) The Leeuwenhoek Lecture 2000 the natural and unnatural history of methane-oxidizing bacteria. Philosophical Transactions of the Royal Society B: Biological Sciences 360: 1207-1222.
- Scott D, Brannan J, Higgins I (1981) The effect of growth conditions on intracytoplasmic membranes and methane mono-oxygenase activities in Methylosinus trichosporium OB3b. Microbiology 125: 63-72.
- Prior SD, Leak DJ, Stanley SH (1984) Regulation and control of methane monooxygenase. Microbial growth on C1 compounds. American Society for Microbiology, Washington, USA, pp: 75-82.
- Morton JD, Hayes KF, Semrau JD (2000) Bioavailability of chelated and soil-adsorbed copper to Methylosinus trichosporium OB3b. Environmental Science & Technology 34: 4917-4922.
- Hakemian AS, Rosenzweig AC (2007) The biochemistry of methane oxidation. Annu Rev Biochem 76: 223-241.
- Kim HJ, Graham DW, DiSpirito AA, Alterman MA, Galeva N, et al. (2004) Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science 305: 612-1615.
- Behling LA, Hartsel SC, Lewis DE, DiSpirito AA, Choi DW, et al. (2008) NMR, mass spectrometry and chemical evidence reveal a different chemical structure for methanobactin that contains oxazolone rings. Journal of the American Chemical Society 130: 12604-12605.
- Choi DW, Zea CJ, Do YS, Semrau JD, Antholine WE, et al. (2006) Spectral, kinetic, and thermodynamic properties of Cu (I) and Cu (II) binding by methanobactin from Methylosinus trichosporium OB3b. Biochemistry 45: 1442-1453.
- De Visscher A, Boeckx P, Van Cleemput O (2007) 12 Artificial Methane Sinks. Greenhouse gas sinks, p: 184.
- Boeckx P, Van Cleemput O, Villaralvo I (1996) Methane emission from a landfill and the methane oxidising capacity of its covering soil. Soil Biology and Biochemistry 28: 1397-1405.
- Barlaz MA, Green RB, Chanton JP, Goldsmith CD, Hater GR (2004) Evaluation of a biologically active cover for mitigation of landfill gas emissions. Environmental Science & Technology 38: 4891-4899.
- Huber-Humer M, Gebert J, Hilger H (2008) Biotic systems to mitigate landfill methane emissions. Waste Management & Research 26: 33-46.
- Scheutz C, Kjeldsen P, Bogner JE, De Visscher A, Gebert J, et al. (2009) Microbial methane oxidation processes and technologies for mitigation of landfill gas emissions. Waste Management & Research 27: 409-455.
- Knief C, Dunfield PF (2005) Response and adaptation of different methanotrophic bacteria to low methane mixing ratios. Environmental Microbiology 7: 1307-1317.
- Baani M, Liesack W (2008) Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proceedings of the National Academy of Sciences 105: 10203-10208.
- Bothe H, Jensen KM, Mergel A, Larsen J, Jørgensen C, et al. (2002) Heterotrophic bacteria growing in association with Methylococcus capsulatus (Bath) in a single cell protein production process. Applied microbiology and biotechnology 59: 33-39.
- Gursel I, Hasirci V (1995) Properties and drug release behaviour of poly (3-hydroxybutyric acid) and various poly (3-hydroxybutyrate-hydroxyvalerate) copolymer microcapsules. Journal of Microencapsulation 12: 185-194.
- Khmelenina VN, Kalyuzhnaya MG, Starostina NG, Suzina NE, Trotsenko YA, et al. (1997) Isolation and characterization of halotolerant alkaliphilic methanotrophic bacteria from Tuva soda lakes. Current Microbiology 35: 257-261.
- Kaluzhnaya M, Khmelenina V, Eshinimaev B, Suzina N, Nikitin D, et al. (2001) Taxonomic characterization of new alkaliphilic and alkalitolerant methanotrophs from soda lakes of the Southeastern Transbaikal region and description of Methylomicrobium buryatense sp. nov. Systematic and Applied Microbiology 24: 166-176.
- Conrado RJ, Gonzalez R (2014) Envisioning the bioconversion of methane to liquid fuels. Science 343: 621-623.
- Semrau JD, DiSpirito AA, Vuilleumier S (2011) Facultative methanotrophy: false leads, true results, and suggestions for future research. FEMS Microbiology Letters 323: 1-12.
- Zischka H, Lichtmannegger J, Schmitt S, Jägemann N, Schulz S, et al. (2011) Liver mitochondrial membrane crosslinking and destruction in a rat model of Wilson disease. The Journal of Clinical Investigation 121: 1508-1518.
Citation: Aimen H, Khan AS, Kanwal N (2018) Methanotrophs: The Natural Way to Tackle Greenhouse Effect. J Bioremediat Biodegrad 9: 432. DOI: 10.4172/2155-6199.1000432
Copyright: © 2018 Aimen H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 12396
- [From(publication date): 0-2018 - Nov 19, 2025]
- Breakdown by view type
- HTML page views: 11080
- PDF downloads: 1316