Mutational Analysis of FGFR3 and HRAS Genes in Bladder Cancer and Washing Cell Sediments of Moroccan Patients
Received: 22-May-2015 / Accepted Date: 25-Dec-2015 / Published Date: 30-Dec-2015 DOI: 10.4172/2161-1165.1000214
Abstract
Genetic alterations related to carcinogenesis, including punctual mutations, may have a potential as bladder cancer biomarker with high specificity and sensitivity. Specific acquired FGFR3 and HRAS mutations have been found to predominate in bladder cancer. The aim of this study is to determine the incidence of FGFR3 and HRAS mutations in bladder tumors in Morocco and to explore whether these molecular events can be detected in urine sediments of bladder cancer patients. In this order, DNA sequencing of both tissues and urine sediments was used to identify gene mutations. Among 42 DNA tumor specimens, FGFR3 mutations were found in 28.6%.On the sequenced exons 7 and 10 of FGFR3 gene, three different mutations were identified R248C, S249C and Y375C. Whereas, HRAS mutations were detected in 9.5% and two distinct HRAS mutations were found G12C and Q61L. FGFR3 mutations were mutually exclusive with HRAS mutations. FGFR3 mutations were predominant in low stage (p<0.001) and low grade (p=0.045) and this predominance was statistically significant. HRAS mutations were more frequent in low stage than in high stage but there is no significant association with the stage of tumors, whereas the predominance of HRAS mutations in high grade was statistically significant (p<0.001). Therefore, FGFR3 mutation may be considered as an early biomarker for bladder cancer and HRAS mutation may inform on the prognosis of this cancer. Gene mutations were also analyzed in urine and were detected in 56.25% for both FGFR3 and HRAS genes. Thus, mutation detection is feasible in urine sediments and a molecular approach will therefore increase the detection sensitivity.
Keywords: Bladder cancer; Mutations; Urine sediments; FGFR3; HRAS
164171Introduction
Worldwide, bladder cancer is the most common cancer, approximately accounting 386,300 new cases each year. The highest incidence rates are observed in the countries of Europe, North America, and Northern Africa. In this last area, the age standardized urinary bladder cancer incidence rates was evaluated at 14.5 in males and 2.4 in females [1]. In Morocco, according to the regional cancer registers, bladder cancer is the most common cancer with an incidence of 5.8 and 11.3 per 100,000 persons in Casablanca and Rabat, respectively [2,3].
The majority of malignant bladder tumors are of epithelial in origin, and more than 90% correspond to histologically defined “urothelial carcinomas” (UC), formely called “transitional cell carcinomas” (TCC) [4].
Approximately 75% to 85% of bladder neoplasms are diagnosed to be superficial bladder tumors [5]. Most of these patients with papillary superficial UC (70%) will recur frequently, without developing an invasive neoplasm. Only 2% to 5% of Ta [6] and 20% to 30% of T1 [7] bladder tumors will invade the bladder muscle. The remaining 15% to 25% carcinomas are invasive (T2, T3, T4) or metastatic lesions at the time of initial clinical presentation.
There is a need for improved clinical stratification methods that can distinguish patients with early-stage disease from those with high risk of recurrence. Conventional methods, including urine cytology, histopathology, or tumor-node-metastasis classification, have numerous limitations in early detection and risk assessment.
Contrarily a large scale of novel molecular methods, such as detection of activating mutation and methylation profiling have shown promising results and may have a potential as bladder cancer biomarker with high specificity and sensitivity [8]. Subsequent larger studies point out to the relevance of mutation of bladder cancer [9-11].
Mutations of FGFR3 were identified in a great portion of bladder carcinomas and at low frequency in cervical urothelial cell, and colorectal carcinomas. Many of these mutations occured at highly conserved sequences in the Ig-like domain III. In non-muscle invasive bladder tumors, mutated FGFR3 leads to constitutive activation of the RAS-MAPK-pathway and consequently to an augmented transduction of growth signals [12,13]. These mutations are found in approximately 70% of low-grade Ta and to a much lower extent of 10-20% in muscleinvasive bladder cancer [14,15].
Somatic FGFR3 mutations in bladder tumors were localized in exon 7, 10 and 15 [16,17], and were shown to occur frequently in low stage and grade tumors [18] (Knowles MA, 2008). It has been shown that FGFR3 mutation analysis in urine samples from bladder cancer patients was possible to detect recurrent tumors [19,20] and the screening for the presence of reccurences using urine-based assays can potentially improve quality-of-life and reduce disease management costs [21,22]. Therefore, detection of FGFR3 mutations in urine could also be employed for general population screening aimed at early detection of primary tumors.
HRAS mutations predominate in bladder cancer and vary widely from 0% to 45% between studies [23,24]. In most cases including bladder cancer, the somatic missense Ras mutations found in cancer cells introduces amino acid substitutions at positions 12, 13, and 61. These changes impair the intrinsic GTPase activity and confer resistance to GAPs, thereby causing cancer-associated mutant Ras proteins to accumulate in the active, GTP bound conformation [25].
These mutations have been shown to occur in all stages and grades [26]. In addition, it has been reported that RAS and FGFR3 mutations were mutually exclusive [26]. Preliminary results show that a combined test for mutation of FGFR3 and HRAS could potentially detect 75% of primary tumours; including 88% of the pTa-T1G1-2 tumors but only 36% of the high-grade and –stage malignancies [15].
The relationship that exists between FGFR3 and HRAS guide us to conduct the present study which aims to determine the incidence of FGFR3 and HRAS mutations in bladder tumors in Morocco. To investigate the potential of these alterations as markers for noninvasive detection of bladder cancer, we explore whether these molecular events can be detected in urine sediments of bladder cancer patients. The presence of FGFR3 and HRAS mutations and their correlation with clinic-pathological parameters were also analyzed in this study.
Materials and Methods
Characteristics of the patients and tissue samples
DNA from Forty two patients with bladder disease was available from our laboratory DNA bank. The study design and population has been previously described [27]. Briefly bladder cancer patients underwent transurethral resection of bladder tumor (TURBT) from January 2010 to June 2011, at the Urology department of Military Hospital of Instruction Mohamed V (MHIMV) in Rabat, Morocco. The mean age of these patients was 64.8 ± 10.1, the median age was 67 years (range 42-84) and 38 of them were male (male:female 7.6:1). Paired tumor and urine samples were collected.
The corresponding hematoxylin-eosin-stained sections were examined at the Anatomopathology department at the same hospital; all the samples were confirmed to be histologically urothelial carcinoma (UC) of bladder. Tumors were graded according to the WHO (World Health Organization) criteria [28] as follows: 13 tumors were low grade and 29 tumors were high grade; and staged according to the TNM (Tumor Node Metastasis) guidelines [29] as follows: 9 pTa, 25 pT1, 7 pT2 and 1 pT4. Four DNA from urinary inflammatory lesions and Forty DNA blood samples from healthy volunteers were included as controls.
PCR amplification
Exons 7, 10 and 15 of the FGFR3, and exon 1 and 2 of the HRAS genes were amplified using the previously described specific primers [14,30] (Table 1). PCR mix was prepared in a final volume of 25 μl containing 1 μl of genomic DNA, 1X PCR buffer, 3 mM , 0.2 mM of each dNTP, 0.2 μM of each primer and 0.625 units AmpliTaq Gold DNA polymerase (Applied Biosystems, CA, USA).
| Exon | Sequence (5’ > 3’) | T (°C) | Fragment size (bp) | Reference | |
|---|---|---|---|---|---|
| FGFR3 | |||||
| 7 | F | AGTGGCGGTGGTGGTGAGGGAG | 63 | 116 | (Bakkar et al., [14]) |
| R | CAGCACCGCCGTCTGGTTGG | ||||
| 10 | F | CAACGCCCATGTCTTTGCAG | 59 | 165 | (Bakkar et al., [14]) |
| R | GAGCCCAGGCCTTTCTTGG | ||||
| 15 | F | AGGACAACGTGATGAAGATCG | 59 | 154 | (Bakkar et al., [14]) |
| R | GTGTGGGAAGGCGGTGTTG | ||||
| HRAS | |||||
| 1 | F | GGAGACCCTGTAGGAGGACC | 61 | 282 | (Quiros et al., [30]) |
| R | GAGGAAGCAGGAGACAGGG | ||||
| 2 | F | GAGAGGTACCAGGGAGAGGC | 65 | 358 | (Quiros et al., [30]) |
| R | ACATGCGCAGAGAGGACAG | ||||
Table 1: Primers and annealing temperature to amplify FGFR3 and HRAS genes. F: Forward; R: Reverse; bp: Base Pair.
DNA amplification was performed on Gen Amp PCR system 9700 appartus (Applied Biosystems) under the following conditions: 9 min at 95°C fo initial denaturation, 35 cycles of 30s at 94°C for, 30s at specific annealing temperature of each primer set (Table 1) and 30s 72°C, followed by a final elongation step of 7 min at 72 °C. Water was used as negative control. Ten μl of PCR products were loaded into 2% agarose gel and sperated by electrophoresis in TBE buffer. The gels were then stained with ethidium bromide and visualized under UV illumination.
DNA sequencing
PCR products were purified by Exosap-IT clean up system (USB, USA) and the sequencing was performed with BigDye®Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster city, CA, USA), according to manufacturer’s protocol, on an ABI 3130XL DNA analyzer (Applied Biosystems, Foster city, CA, USA).
The sequences obtained were then matched with gene reference sequence collected from the GeneBank database. The sequence aligenment was performed using Clustal W program in BioEdit Software. According to the position of targeted mutation, we used either forward or reverse primer in sequencing. If a mutation was identified, matched DNA urine sediments were also sequenced. Results were confirmed by sequencing both strands.
Statistical analysis
The association between mutations and stage/grade was evaluated by calculating p values according to χ2 test using MedCalc version 9. P<0.05 was considered significant.
Results
Oncogenic mutations of FGFR3 and HRAS in bladder cancer cases
Exon 7, 10 and 15 of the FGFR3, and exon 1 and 2 of the HRAS genes were screened for mutations in DNA from 42 bladder cancer cases and there paired urine sediments. The various FGFR3 and HRAS mutations identified are listed in Table 2.
| Exon | Nucleotide position and base change | Amino acid change | Number of tumors | |
|---|---|---|---|---|
| FGFR3 gene | 7 | 742 C>T | Arg/Cys (R248C) | 1 |
| 7 | 746 C>G | Ser/Cys (S249C) | 9 | |
| 10 | 1124 A>G | Tyr/Cys (Y375C) | 2 | |
| HRAS gene | 1 | 34G>T | Gly/Cys (G12C) | 2 |
| 2 | 182 A>T | Gln/Leu (Q61L) | 2 |
Table 2: Frequencies of individual mutations of GFR3 and HRAS genes in primary tumors of 42 patients.
Overall, 28.6% (12 of 42) of the tumors contained an FGFR3 mutation. The FGFR3 mutation in exon 15 was not detected in any of the 42 urinary bladder cancer cases. On the sequenced exons 7 and 10 of FGFR3 gene, three different mutations were identified R248C, S249C and Y375C (Table 2), which are known mutation hotspots in bladder cancer. In exon 7, The S249C mutation consisting in the substitution of the nucleotide C by the nucleotide G at position 746 occurs more frequently and was detected in 21,4% (9/42) of analyzed cases.. Whereas, the R248C (substitution of C by T at position 742 in exon 7) and Y375C (substitution of A by G at position 1124 in exon 10) were detected only in 2,3% (1/42) and 4,7% (2/42) respectively.
The HRAS mutations were screened for mutations in 42 bladder cancer cases and were detected in 9.5% (4 of 42) of the tumors. Two distinct HRAS mutations were found G12C (substitution of G by T at position 34 in exon 1) and Q61L (substitution of A by T at position 182 in exon 2) and were distributed equally among the tumors (Table 2).
Codon and mutated nucleotide numbered according to the coding DNA Sequence of FGFR3 (Accession number for genomic reference sequence: NM_000142.4) and HRAS gene (Accession number for genomic reference sequence: NM_005343.2).
In total, 38.1% (16/42) of the analyzed tumors had mutation of either HRAS gene or FGFR3 gene. All the mutated cases harbored no more than one mutation and the FGFR3 mutations were mutually exclusive with HRAS mutations. .
No mutation was detected in the four urinary inflammatory lesions and control cases for both FGFR3 and HRAS genes.
Association between mutations and clinicopathological parameters
In our studied cohort, thirty-four of 42 studied bladder tumors (80.95%) were non muscle invasive tumors (9 pTa and 25 pT1), whereas 8 tumors (19.05%) were muscle invasive tumors (7 pT2 and 1 pT4). This distribution of tumors in our study was consistent with previous epidemiological studies on the stage, supporting the validity of our cohort of patients.
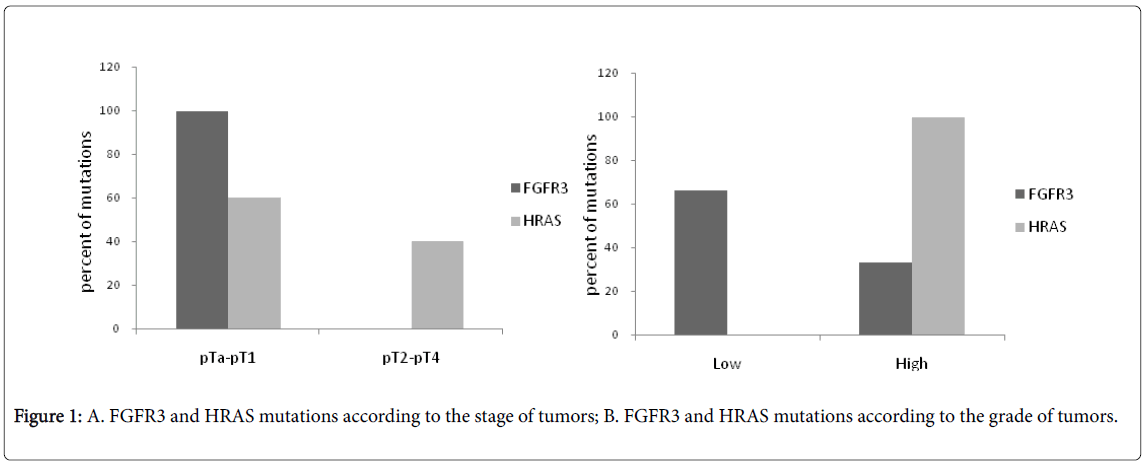
The distribution of FGFR3 and HRAS mutations according to stage and grade was represented in Figure 1A and 1B; respectively. FGFR3 mutations were predominant in low stage (2/12 pTa and 10/12 pT1) (p<0.001) and low grade (8/12) (p=0.045) and this predominance was statistically significant. HRAS mutations were more frequent in low stage (3/4 pT1) than in high stage but there is no significant association with the stage of tumors, whereas the predominance of HRAS mutations in high grade (4/4) was statistically significant (p<0.001).
Mutations in urine samples from patients with bladder cancer
To assess the value of the individual markers for noninvasive detection of bladder cancer, once a mutation was detected in the tumor DNA, we analyzed DNA isolated from paired urine sediments. Overall, mutations were detected in 56.25% (9 of 16) of mutated cases for both FGFR3 and HRAS genes. In FGFR3 gene, mutations in urine sediments were identified in 58.33% (7 of 12) of mutated cases in paired tumors, whereas HRAS mutations were found in 50% (2 of 4) of mutated cases.
Discussion
In urothelial tumors somatic mutations in the FGFR3, HRAS, NRAS, KRAS and PIK3CA genes may be of use for early detection of primary and recurrent tumors in urine-based assays, for prognosis prediction, and as a companion diagnostic for targeted therapies [15]. In view of these findings, we investigated the incidence of FGFR3 and HRAS genes mutations in bladder cancer from Moroccan patients and to our knowledge, this is the first study investigating mutations in both FGFR3 and HRAS genes in bladder cancer in North Africa. We found that FGFR3 gene mutations are a frequent event occurring in bladder carcinomas and are similar in terms of frequency with previous studies [31]. Indeed, we found tree somatic point mutations of FGFR3 gene (R248C, S249C and Y375C) in 28.6% of bladder tumors (12 of 42). The three mutations detected in our patients group which induce strong constitutive activation of the receptor, were also identified previously in different series of bladder cancer patients [15]. Among the mutated cases, S249C mutation was predominant (75%) which is consistent with other reports [15,26,31]. The mutations were significantly associated with low-grade (p=0.045) and low-stage (p<0.001) and distribution of the mutations among different domains of FGFR3 was in accordance with the most of previously published data [32]. Since the FGFR3 activation is a key event in the development of noninvasive bladder tumors [32] and that FGFR3 mutations in bladder cancer are related to low stage and grade, they make FGFR3 the first marker for non-aggressive disease.
Regarding the frequency of HRAS mutations, our data were comparable with other studies [33,34] and two distinct HRAS mutations in exon 1 and 2 were found in 9.5% (4 of 42) of the analyzed tumors. HRAS mutations were more frequent in low stage than in high stage but there is no significant association with the stage of tumors, whereas the predominance of HRAS mutations in high grade was statistically significant (p<0.001). In previous study, Activating HRAS mutations were detected with an estimated overall frequency of 10-15% without a clear association with tumor grade or stage [15,26].
In bladder cancer, the common HRAS gene mutations occur in the hotspot codons 12, 13 (exon1), or 61 (exon2). These mutations cause specific substitutions to amino acid and result in the loss of GTPase activity [35]. This enzyme enables hydrolysis of RAS protein from active conformation (RAS-GTP) to inactive form (RAS-GDP). In the lack of GTPase activity, the active form is permanently maintained, thereby inducing activation of downstream kinase cascades that results in continuous mitogenic signaling leading to uncontrolled cell division and the formation of a tumor [36].
As for RAS, upstream FGFR3 is also able to activate the MAPK signaling pathway. Superficial papillary tumors are characterized by a high frequency of FGFR3 mutations [24,34] leading to constitutive activation of the RAS-MAPK pathway [12,13]. In our study, as well as in other, FGFR3 and HRAS mutations were mutually exclusive in bladder cancer, probably due to their redundancy in the same signaling pathway [26].
Cytology is the non-invasive test widely used for patient follow-up, whereas this method had several limits in detection of non-muscle invasive bladder cancer [37]. Numerous studies have shown that it is possible to detect bladder oncogene mutations in DNA isolated from urine sediments [33,38]. In our study, the sensitivity of mutation identification is the fraction of mutations detected in urine divided by mutations found in tumor biopsies. The FGFR3 mutation is detected in 58.3% of urine samples. This frequency is comparable to other studies [33]. For the HRAS gene mutation, the sensitivity in the present study (50%) was higher than previous reports [38,39]. In some cases, mutations were detected in tumors but not in the corresponding urine. DNA isolated from urine sediments may not be sufficient for a molecular analysis due to the limited numbers of bladder tumor cells exfoliated in urine [33].Thus, tumor cell population should be enriched for a better outcome in detection of molecular events. This may be achieved either by improving the procedure for urine sampling or by using filter devices or antibody-based assays to selectively capture the tumor cells [40,41].
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. (2011) Global cancer statistics. CA Cancer J Clin 61: 69-90.
- Benider A, Bennani-Othmani M, Harif M, et al. (2004) Registre des cancer de la region du grand Casablanca.
- Dinney CP, McConkey DJ, Millikan RE, Wu X, Bar-Eli M, et al. (2004) Focus on bladder cancer. Cancer Cell 6: 111-116.
- Haukaas S, Daehlin L, Maartmann-Moe H, Ulvik NM (1999) The long-term outcome in patients with superficial transitional cell carcinoma of the bladder: a single-institutional experience. BJU Int 83: 957-963.
- Bryan RT, Wallace DM (2002) 'Superficial' bladder cancer - time to uncouple pT1 tumours from pTa tumours.BJU Int 90: 846-852.
- Serizawa RR, Ralfikiaer U, Steven K, Lam GW, Schmiedel S, et al. (2001). Integrated genetic and epigenetic analysis of bladder cancer reveals an additive diagnostic value of FGFR3 mutations and hypermethylation events. International Journal of Cancer 129:78-87.
- Black PC, Agarwal PK, Dinney CP (2007) Targeted therapies in bladder cancer--an update. Urol Oncol 25: 433-438.
- Junker K, van Oers JM, Zwarthoff EC, Kania I, Schubert J, et al. (2008) Fibroblast growth factor receptor 3 mutations in bladder tumors correlate with low frequency of chromosome alterations. Neoplasia 10: 1-7.
- López-Knowles E, Hernández S, Malats N, Kogevinas M, Lloreta J, et al. (2006) PIK3CA mutations are an early genetic alteration associated with FGFR3 mutations in superficial papillary bladder tumors.Cancer Res 66: 7401-7404.
- Agazie YM, Movilla N, Ischenko I, Hayman MJ (2003) The phosphotyrosine phosphatase SHP2 is a critical mediator of transformation induced by the oncogenic fibroblast growth factor receptor 3.Oncogene 22: 6909-6918.
- di Martino E, L’Hote CG, Kennedy W, Totnlinson DC,Knowles MA (2009). Mutant ?broblast growth factor receptor 3 induces intracellular signaling and cellular transformation in a cell type- and mutation-speci?c manner. Oncogene28 : 4306-4316.
- Bakkar AA, Wallerand H, Radvanyi F, Lahaye JB, Pissard S, et al. (2003) FGFR3 and TP53 gene mutations define two distinct pathways in urothelial cell carcinoma of the bladder.Cancer Res 63: 8108-8112.
- Kompier LC, Lurkin I, van der Aa MN, van Rhijn BW, van der Kwast TH, et al. (2010) FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy.PLoS One 5: e13821.
- Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, et al. (1999) Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas.Nat Genet 23: 18-20.
- Sibley K, Cuthbert-Heavens D, Knowles MA (2001) Loss of heterozygosity at 4p16.3 and mutation of FGFR3 in transitional cell carcinoma.Oncogene 20: 686-691.
- Knowles MA (2008) Molecular pathogenesis of bladder cancer.Int J Clin Oncol 13: 287-297.
- van Oers JM, Lurkin I, van Exsel AJ, Nijsen Y, van Rhijn BW, et al.(2005). A simple and fast method for the simultaneous detection of nine fibroblast growth factor receptor 3 mutations in bladder cancer and voided urine. Clinical cancer research: an official journal of the AACR 11:7743-7748.
- Zuiverloon TC, van der MN, van der Kwast TH, Steyerberg EW, et al.(2010) Fibroblast growth factor receptor 3 mutation analysis on voided urine for surveillance of patients with low-grade non-muscle-invasive bladder cancer. Clinical cancer research: an official journal of the AACR 16:3011-3018.
- van der Aa MN, Steyerberg EW, Sen EF, Zwarthoff EC, Kirkels WJ, et al. (2008) Patients' perceived burden of cystoscopic and urinary surveillance of bladder cancer: a randomized comparison.BJU Int 101: 1106-1110.
- Müezzinoglu T, Ceylan Y, Temeltaş G, Lekili M, Büyüksu C (2005) Evaluation of pain caused by urethrocystoscopy in patients with superficial bladder cancer: a perspective of quality of life.Onkologie 28: 260-264.
- Bos JL (1989) ras oncogenes in human cancer: a review.Cancer Res 49: 4682-4689.
- Olderøy G, Daehlin L, Ogreid D (1998) Low-frequency mutation of Ha-ras and Ki-ras oncogenes in transitional cell carcinoma of the bladder.Anticancer Res 18: 2675-2678.
- Trahey M, McCormick F (1987) A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants.Science 238: 542-545.
- Jebar AH, Hurst CD, Tomlinson DC, Johnston C, Taylor CF, et al. (2005) FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma.Oncogene 24: 5218-5225.
- Berrada N. Amzazi S. Ameziane El Hassani R, et al. (2010) Epigenetic alterations of adenomatous polyposis coli (APC). Retinoic acid receptor beta (RARbeta) and survivin genes in tumor tissues and voided urine of bladder cancer patients. Cell Mol Biol (Noisy-le-grand) Suppl 58:OL1744-1751.
- Epstein JI, Amin MB, Reuter VR, Mosto FK (1998). The World Health Organization/intemational Society of Urological Pathology consensus classi?cation of urotltelial (transitional cell) neoplasms of the urinary bladder. Bladder Consensus Conference Committee. The American joumal of surgical pathology 22:1435-1448.
- Sobin LH, Gospodariwicz M, Wittekmd C (2009). TNM Classi?cation of Malignant Ttnnors.UICC International Union Against Cancer. Oxford: Wiley-Blackwell. 262-265.
- Quiros RM, Ding HG, Gattuso P, Prinz RA, Xu X (2005). Evidence that one subset of anaplastic thyroid carcinomas are derived from papillary carcinomas due to BRAF and p53 mutations. Cancer 103:2261-2268.
- Billerey C, Chopin D, Aubriot-Lorton MH, Ricol D, Gil Diez de Medina S, et al. (2001) Frequent FGFR3 mutations in papillary non-invasive bladder (pTa) tumors.Am J Pathol 158: 1955-1959.
- van Rhijn BW, Lurkin I, Radvanyi F, Kirkels WJ, van der Kwast TH, et al. (2001) The fibroblast growth factor receptor 3 (FGFR3) mutation is a strong indicator of superficial bladder cancer with low recurrence rate.Cancer Res 61: 1265-1268.
- Serizawa RR, Nørgaard N, Horn T, Vibits H (2011) Hemangioma of the prostate--an unusual cause of lower urinary tract symptoms: case report.BMC Urol 11: 4.
- Platt FM, Hurst CD, Taylor CF, Gregory WM, Harnden P, et al. (2009) Spectrum of phosphatidylinositol 3-kinase pathway gene alterations in bladder cancer.Clin Cancer Res 15: 6008-6017.
- Shinohara N, Koyanagi T (2002) Ras signal transduction in carcinogenesis and progression of bladder cancer: molecular target for treatment?Urol Res 30: 273-281.
- Capon DJ, Chen EY, Levinson AD, Seeburg PH, Goeddel DV (1983) Complete nucleotide sequences of the T24 human bladder carcinoma oncogene and its normal homologue. Nature302:33-37.
- Konety BR (2006) Molecular markers in bladder cancer: a critical appraisal.Urol Oncol 24: 326-337.
- Buyru N, Tigli H, Ozcan F, Dalay N (2003) Ras oncogene mutations in urine sediments of patients with bladder cancer.J Biochem Mol Biol 36: 399-402.
- Fitzgerald JM, Ramchurren N, Rieger K, Levesque P, Silverman M, Libertino JA (1995) Identification of H-ras mutations in urine sediments complements cytology in the detection of bladder tumors. Journal of the National Cancer Institute 87:129-133.
- Rothacker J, Ramsay RG, Ciznadija D, et al. (2007) A novel magnetic bead-based assay with high sensitivity and selectivity for analysis of telomerase in exfoliated cells front patients with bladder and colon cancer. Electrophoresis 28:4435-4446.
- Zlieng S, Lin H, Lin J-Q, Balic M, et al. (2007) Membrane microfilter device for selective capture. electrolysis and genomic analysis of human circulating tumor cells. Journal of Chromatography 1162: 154-161.
Citation: Berrada N, Amzazi S, Abbar M, Ameur A, Khyatti M, et al. (2015) Mutational Analysis of FGFR3 and HRAS Genes in Bladder Cancer and Washing Cell Sediments of Moroccan Patients. Epidemiology (sunnyvale) 5:214. DOI: 10.4172/2161-1165.1000214
Copyright: © 2015 Berrada N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 11837
- [From(publication date): 12-2015 - Jul 01, 2025]
- Breakdown by view type
- HTML page views: 10802
- PDF downloads: 1035