Research Article Open Access
Predicting Therapeutic Results of Interferon Based Treatment by Modeling the First Week Viral Kinetics in Patients with Chronic Hepatitis C
Chun-Hsiang Wang*, Ruey-Chang Lin, Lein-Ray Mo, Kuo-Kuan Chang and Jen-Juan KuoDepartment of Hepatogastroenterology, Tainan Municipal Hospital, Tainan, Taiwan
- *Corresponding Author:
- Chun-Hsiang Wang, M.D
Department of Hepatogastroenterology
Tainan Municipal Hospital
Address: 670, Chongde Road, Tainan, Taiwan
Tel: 886-6-2609926
Fax: 886-6-2606351
E-mail: chunhsiangwang1@gmail.com
Received date: March 07, 2015; Accepted date: March 18, 2015; Published date: March 27, 2015
Citation: Wang CH, Lin RC, Mo LR, Chang KK, Kuo JJ (2015) Predicting Therapeutic Results of Interferon Based Treatment by Modeling the First Week Viral Kinetics in Patients with Chronic Hepatitis C. J Gastrointest Dig Syst 5:270. doi:10.4172/2161-069X.1000270
Copyright: © 2015 Wang CH, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Gastrointestinal & Digestive System
Abstract
Background: The present study aimed to develop an optimal model predictive of the treatment response one week after initiating antiviral therapy.
Patients and Methods: In all, 166 patients completed the treatment and were followed up for six months.
Results: A viral response was achieved in 14 (8.4%) genotype 1 (G1) and 32 (19.3%) non-genotype 1 (G0) patients at week one after initiation of therapy and 45 (27.1%) G1 and 90 (54.2%) G0 patients at week four. A sustained viral response (SVR) was achieved in 50 (30.1%) G1 and 83 (50.0%) G0 patients, in 13 (29.5%) G1 and 31 (70.5%) G0 patients whose antiviral response was achieved at one week, and in 37 G1 (32.2%) and 78 G0 (67.8%) patients whose antiviral response was achieved at four weeks. The HCV viral kinetic parameter π (i.e., 1: α, α was defined as the area under the line y [viral load]=– a[x{time}] between week 0 and week 1) was found to be the best performing test for predicting SVR (p<0.0001). Regression analysis identified π as the only independent predictor of SVR (p=0.043).
Conclusions: The use of bio-mathematical models of HCV kinetics can reliably predict SVR one week after starting therapy.
Abstract
Background: The present study aimed to develop an optimal model predictive of the treatment response one week after initiating antiviral therapy.
Patients and Methods: In all, 166 patients completed the treatment and were followed up for six months.
Results: A viral response was achieved in 14 (8.4%) genotype 1 (G1) and 32 (19.3%) non-genotype 1 (G0) patients at week one after initiation of therapy and 45 (27.1%) G1 and 90 (54.2%) G0 patients at week four. A sustained viral response (SVR) was achieved in 50 (30.1%) G1 and 83 (50.0%) G0 patients, in 13 (29.5%) G1 and 31 (70.5%) G0 patients whose antiviral response was achieved at one week, and in 37 G1 (32.2%) and 78 G0 (67.8%) patients whose antiviral response was achieved at four weeks. The HCV viral kinetic parameter π (i.e., 1: α, α was defined as the area under the line y [viral load]=– a[x{time}] between week 0 and week 1) was found to be the best performing test for predicting SVR (p<0.0001). Regression analysis identified π as the only independent predictor of SVR (p=0.043).
Conclusions: The use of bio-mathematical models of HCV kinetics can reliably predict SVR one week after starting therapy.
Keywords
Hepatitis C virus; Interferon; Ribavirin; Viral load; Polymerase chain reaction
Abbreviation
HCV: Hepatitis C Virus; HCC: Hepatocellular Carcinoma; EG-IFN: Pegylated interferon-α; RBV: Ribavirin; SVR: Sustained Virological Response; ROC: Receiver operating Curve; AUROC: Area Under the ROC Curve; BMI: Body Mass Index; G1: Genotype 1; G0: Non-Genotype 1; ALT: Alanine Aminotransferase
Background
The chronic hepatitis C virus (HCV) epidemic affects approximately 170 million people worldwide [1]. Infection may lead to end stage liver diseases, such as liver cirrhosis and hepatocellular carcinoma (HCC) [2], inevitably increasing the burden of HCV-related morbidity and mortality. Therefore, antiviral therapy is a matter of the utmost importance. The current standard-of-care therapy for chronic hepatitis C (i.e., once weekly pegylated interferon-α [PEG-IFN] plus daily oral ribavirin [RBV] for 24 to 48 weeks [3]) has improved the sustained virological response (SVR) rate. Although the therapeutic effectiveness of antiviral therapy against HCV infection has improved, the relapse rate and nonresponse rate still remain high. Furthermore, current antiviral treatment still has adverse effects. Thus, it is important to predict therapeutic efficacy earlier in the treatment phase. One study suggested that the decay of HCV RNA 48 hours after therapy is an independent predictor of SVR [4]. Accordingly, mathematical models have been developed to describe HCV kinetics during therapy [5-7]. Furthermore, several studies using mathematical models of HCV infection indicate that the final slope of HCV-RNA decay represents the rate of infected cell loss [7] and that the typical pattern of HCV RNA decline during treatment is biphasic [8]. Mathematical models of virus infection were initially used to study the dynamics of human immunodeficiency virus infection [9,10] and later extended to the study of both HCV [8,9,11] and hepatitis B virus infections [12]. The observations of these studies indicate that host factors as well as viral factors are associated with rates of response to antiviral therapy. The aim of the current study was to identify determinants from a range of the viral-kinetic parameters that could be used to accurately predict SVR after one week’s antiviral therapy. The HCV-RNA level in sera from naïve patients infected with HCV was monitored frequently to determine viral kinetics during therapy.
Patients and Methods
From July 2009 to December 2010, this study enrolled 181 patients to receive antiviral therapy for chronic hepatitis C. Patients were eligible for the study if they were 18–75 years old, chronically infected with HCV without a history of treatment for HCV infection, and serum HCV RNA positive. Ten patients were excluded from the study owing to (1) loss to follow-up; (2) diagnosis of HCC at study entry; (3) other liver diseases; (4) other forms of viral hepatitis; and (5) insufficient data. Five patients dropped out due to the adverse effects of the therapy. The remaining 166 patients (106 males; 60 females; mean age: 56.6 ± 10.0 years old; 68 genotype 1 [G1] and 98 non-genotype 1 [G0] patients) completed the scheduled treatment and scheduled follow up for six months. Clinical information including age, sex, body mass index (BMI), baseline aspartate transaminase (AST), baseline alanine aminotransferase (ALT), genotypes, and history of diabetes mellitus was collected from each patient. Serum biochemistry was measured before treatment and after 1, 4, 8, 12, 24, 36, 48, and 72 weeks of treatment.
Eligible patients were treated with PEG-IFN-α plus RBV therapy over a period of 24 or 48 weeks. HCV-RNA levels were measured using the Amplicor HCV test (Roche Molecular Systems, Branchburg, NJ) before treatment and after 1, 4, 12, 24, and 48 weeks of treatment and HCV genotypes were determined using a reverse hybridization assay (Innolipa HCV, Innogenetics, Ghent, Belgium). The primary outcome variable was SVR, defined as negative HCV RNA test 24 weeks after completion of antiviral therapy. Nonresponse was defined as a positive HCV RNA test 24 weeks after completion of antiviral therapy.
Informed consent was obtained from each patient and the study protocol conformed to the 1975 Declaration of Helsinki. During therapy, safety was evaluated by biochemical and hematologic tests at each visit. Patients were observed closely for signs of unusual side effects. The dose of PEG-IFN and/or ribavirin was reduced or discontinued if the side effects were severe or intolerable. The Institutional Review Board approved the study protocol before the investigation began.
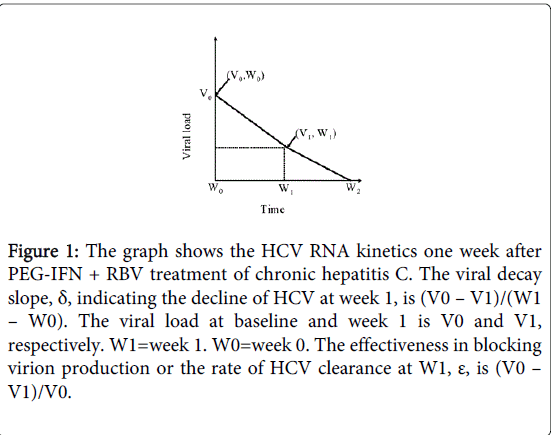
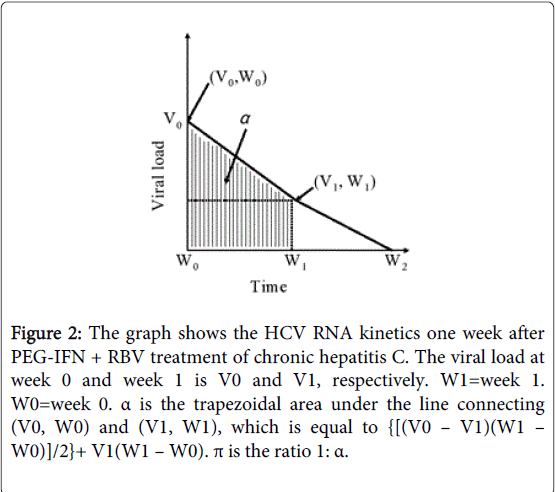
Five HCV kinetic parameters were measured including (a) the logarithm of the HCV RNA level at week 0, indicating baseline HCV viremia; (b) δ, the logarithm of the decline in HCV RNA level (Figure 1) between week 0 and week 1. (Antiviral treatment can interrupt viral infection. Thus, viral clearance causes serum virus level to fall. The degree of the decline in HCV RNA level is a reliable indicator of treatment effectiveness); (c) ε, the logarithm of the HCV RNA level at week 1 divided by the baseline level (Figure 1) and representing the rate of HCV clearance; (d) α, the trapezoidal area [the algebraic sum of areas] under the graph of the viral load between week 0 and week 1 (Figure 2). (In the current model, frequent viral load measurements must be made between week 0 and week 1. In our study, the trapezoidal area was an approximation to the integral of the function over the given interval. However, α was regarded as inversely related to SVR); (e) π, denoting the ratio 1: α and representing an SVR rate.
Figure 1: The graph shows the HCV RNA kinetics one week after PEG-IFN + RBV treatment of chronic hepatitis C. The viral decay slope, δ, indicating the decline of HCV at week 1, is (V0 – V1)/(W1 – W0). The viral load at baseline and week 1 is V0 and V1, respectively. W1=week 1. W0=week 0. The effectiveness in blocking virion production or the rate of HCV clearance at W1, ε, is (V0 – V1)/V0.
Figure 2: The graph shows the HCV RNA kinetics one week after PEG-IFN + RBV treatment of chronic hepatitis C. The viral load at week 0 and week 1 is V0 and V1, respectively. W1=week 1. W0=week 0. α is the trapezoidal area under the line connecting (V0, W0) and (V1, W1), which is equal to {[(V0 – V1)(W1 – W0)]/2}+ V1(W1 – W0). π is the ratio 1: α.
Statistical analysis
Continuous variables are expressed as means ± SD. Between-group differences in continuous variables were analyzed with the use of the Student’s t-test (with 95% confidence intervals) or the Mann-Whitney U test. A p-value of less than 0.05 was considered to be statistically significant. Stepwise logistic regression was used to identify predictors of SVR that might be confounders. Receiver operating curve (ROC) statistics were used for various HCV kinetic predictive model comparisons. The ROC curve (sensitivity plotted against 1–specificity) was constructed. The area under the ROC curve (AUROC) was considered a measure of how well a parameter distinguishes between two diagnostic groups and how well a diagnostic variable performs. The Youden index J (defined as max [sensitivity + specificity-1]) represents the optimal cut-off point on the ROC curve and indicates a variable's best performance [13]. Graphically, J is the maximum vertical distance between the ROC curve and the diagonal line. Ideally, the AUROC would have a value of 1. Univariate and multivariate logistic regression statistical models were implemented for the analysis of clinical factors (age, sex, BMI, baseline ALT, genotypes, and history of diabetes) and HCV kinetic factors predictive of SVR. SPSS version 14.0 software (SPSS Inc. USA) was used to analyze the data.
Results
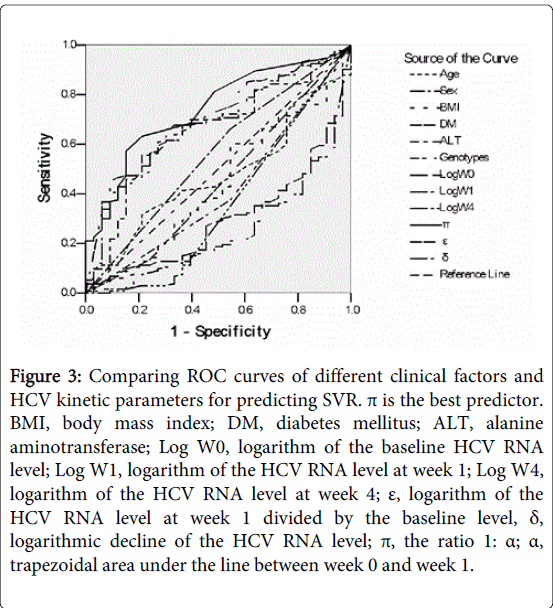
Baseline characteristics of patients are shown in Table 1. An SVR was achieved in 133 patients (80.1%; G1: 50 [30.1%], G0: 83 [50.0%]). Among 46 patients (14 G1 and 32 G0) with a antiviral response at the end of the first week, 44 (13 [29.5%] G1 and 31 [70.5%] G0) but not 2 (1 G1 [50%] and 1 G0 [50%]) patients achieved an SVR. Among 135 patients (45 G1 and 90 G0) with a viral response at the end of the fourth week, 115 (37 G1 [32.2%] and 78 G0 [67.8%]) but not 20 (8 G1 [40%] and 12 G0 [60%]) patients achieved an SVR. Univariate analysis identified baseline ALT level (p=0.024), ε (p<0.001), α (p<0.001), π (p<0.001), δ (p=0.022), and HCV RNA level at week 0 (p=0.001), week 1 (p<0.001), and week 4 (p<0.001) as significant predictors of SVR. Comparing different clinical parameters and HCV kinetic variables, ROC analysis identified π as the most powerful predictor of SVR (p<0.0001, AUROC=0.74) (Table 2 and Figure 3).
| Characteristics | |
| Mean age, y/o | 56.5 ± 10.1 |
| Sex | |
| M, n (%) | 106 (63.9) |
| F, n(%) | 60 (36.1) |
| Diabetes mellitus, n (%) | 16 (16.) |
| Mean hemoglobin, g/dL | 14.3 ± 1.7 |
| Mean neutrophils, ×103/μL | 5682.5 ± 1457.2 |
| Mean platelets, ×103/μL | 16 ± 5.4 |
| Prothrombin time, sec | 11.2 ± 0.73 |
| BMI | 25.1 ± 3.3 |
| AST, IU/L | 105.5 ± 59.2 |
| ALT, IU/L | 158.3 ± 90.8 |
| Genotypes | |
| G1, n (%) | 68 (41) |
| G0, n (%) | 98 (59) |
| Log ofbaseline HCV RNA,106 IU/mL | 5.4 ± 0.9 |
| AST: Aspartase Transaminase; ALT, Alanine Aminotransferase; BMI, Body Mass Index; G1, Genotype 1; G0, Non-Genotype 1 | |
Table 1: Baseline demographic characteristics, clinical characteristics, and Laboratory Data.
| Variables | Area | Standard error | p value | 95% Confidence interval | |
| Lower limit | Upper limit | ||||
| Age | 0.507 | 0.602 | 0.93 | 0.374 | 0.575 |
| Sex | 0.615 | 0.079 | 0.149 | 0.447 | 0.669 |
| BMI | 0.412 | 0.082 | 0.823 | 0.324 | 0.548 |
| Diabetes mellitus | 0.486 | 0.08 | 0.865 | 0.332 | 0.561 |
| ALT | 0.62 | 0.077 | 0.129 | 0.556 | 0.757 |
| Genotypes | 0.428 | 0.08 | 0.371 | 0.306 | 0.525 |
| Log W0 | 0.198 | 0.045 | 0 | 0.206 | 0.387 |
| Log W1 | 0.199 | 0.059 | 0 | 0.161 | 0.339 |
| Log W4 | 0.334 | 0.085 | 0.037 | 0.237 | 0.473 |
| ε | 0.728 | 0.064 | 0.004 | 0.612 | 0.794 |
| δ | 0.682 | 0.7 | 0.022 | 0.582 | 0.772 |
| π | 0.774 | 0.06 | 0.001 | 0.652 | 0.828 |
| BMI: Body Mass Index; ALT: Alanine Aminotransferase; Log W0: Logarithm of Baseline: HCV RNA level; Log W1: Logarithm of HCV RNA level at week 1; Log W4: Logarithm of HCV RNA level at week 4; ε: Logarithm of HCV RNA level at week 1 divided by baseline level; δ: Logarithm of the decline in HCV RNA level (viral decay slope); π: The ratio of 1: α (α=trapezoidal area under the line between week 0 and week 1). | |||||
Table 2: Comparison of different clinical parameters and HCV kinetic variables for Predicting SVR based on area under the ROC Curve.
Figure 3: Comparing ROC curves of different clinical factors and HCV kinetic parameters for predicting SVR. π is the best predictor. BMI, body mass index; DM, diabetes mellitus; ALT, alanine aminotransferase; Log W0, logarithm of the baseline HCV RNA level; Log W1, logarithm of the HCV RNA level at week 1; Log W4, logarithm of the HCV RNA level at week 4; ε, logarithm of the HCV RNA level at week 1 divided by the baseline level, δ, logarithmic decline of the HCV RNA level; π, the ratio 1: α; α, trapezoidal area under the line between week 0 and week 1.
The optimal cut-off point (the Youden index J) of the ROC curve of π was calculated as ≧0.2 (sensitivity=71.6%, specificity=68.8%, PPV=92%). Multivariate logistic regression analysis of the HCV kinetic factor (π) and clinical factors (age, sex, BMI, genotypes, baseline ALT level) identified π as the only independent predictor of SVR (p=0.043).
Discussion
The present study shows that HCV kinetic variables derived from bio-mathematical models can be used as predictors of SVR one week after starting therapy. The viral factors affecting the response to IFN-based therapy include genotype and viral load. Patients with a low pretreatment HCV RNA level are more likely to respond to therapy than those with a high pretreatment level. Furthermore, a larger proportion of patients with a rapid viral response (RVR) and negative viremia at week 4 (i.e., 60-72%) achieve SVR than those without an RVR [14]. Neumann and colleagues [6] showed that HCV RNA levels generally begin declining 7-10 hours after initiation of IFN therapy, which may mainly represent the time needed for IFN signal transduction and IFN stimulated gene expression. The above studies suggest that hepatitis C virus during antiviral treatment is highly dynamic, particularly in the initial phase of treatment.
Earlier monitoring of HCV RNA level after starting antiviral treatment to detect RVR (a potent predictor of SVR after four weeks of treatment) is an important guide to therapeutic strategies [15,16]. However, in our study, 20 patients (14.8%) with RVR did not achieve SVR. Therefore, HCV RNA level might be not the only factor that determines a therapeutic response. Furthermore, HCV viremia neither correlates with the severity of liver disease nor predicts the therapeutic outcome of HCV infection. Therefore, viral kinetics after initiating antiviral treatment might be a predictor of the real world therapeutic response in most situations. Based on the extrapolation of kinetic data demonstrating HCV decline, we were able not only to measure HCV RNA level earlier (i.e., one week after initiating antiviral treatment) but also to test five determinants of HCV RNA decay right after initiating treatment. Theoretically, models of the dynamic response to HCV therapeutic strategies can be used to individualize and optimize therapy by predicting SVR. Our study is the first to specifically examine change in viral kinetics at week 1 after initiation of antiviral treatment. Based on this pioneer design, five HCV viral kinetics models were developed from the results of the first week’s treatment. Mathematical parameters in our study were estimated from HCV RNA levels. Among the five parameters mentioned above, π was defined as the ratio 1: α. In previous studies, integral calculus was used to calculate α [17,18] (the area between the line and the x-axis). Another method would be to divide the distance between week 0 and week 1 into a number of equal segments. The current study used the trapezoidal area (the sum of the areas of each segment of the trapezoid) to approximate the area under the curve. Multivariate logistic analysis identified π as the only independent predictor of SVR. The predictive capability of π was validated by ROC curve analysis and compared to that of other baseline parameters (such as viral load before treatment, gender, and age), previously studied for their effect on treatment response [19,20]. The optimal cut-off point of π was calculated as ≧0.2, which is located at the upper left corner of the ROC curve. Therefore when the ratio of the area under the straight line between the logarithm of the HCV RNA level at week 0 and that at week 1 is 0.2 or higher, SVR has a positive predictive value of 92%. The change in HCV RNA level is used herein as a predictor of therapeutic response because viral kinetics during antiviral therapy may reflect interactions between virus and host and thereby account for treatment outcome. Treatment failure or resistance to antiviral therapy can be due to viral or cellular factors. Neither HCV alone nor host factors can accurately predict SVR. Therefore viral kinetics during therapy may take into account both viral and host factors. Little data is available indicating whether viral kinetic modeling has an important role in the analysis of HCV-RNA decay after the initiation of antiviral therapy. A key strength of this study is its use of viral kinetics to estimate the initial viral decline during the first week’s treatment, to estimate viral elimination and the achievement of SVR, and to guide the adoption of different anti-HCV treatment strategies to optimize therapy.
Understanding and careful studies of HCV kinetics during antiviral treatment may help individualize and optimize combination therapy. In the current study, mathematical models provided detailed information on HCV RNA kinetics during the time of antiviral treatment. Our findings have important implications for predicting treatment outcomes and managing response-guided combination antiviral therapies. We speculated that the area under the line between week 0 and week 1 is inversely related to the SVR rate. In other words, the smaller the area, the higher the SVR rate. Therefore, the HCV RNA titer after one week of antiviral treatment may be a useful tool for optimizing therapeutic strategies. However, a large population study is needed to overcome the limitation of small sample size and validate the model.
Acknowledgements
The research of this article did not receive any financial support. The authors want to thank Dr. Lein-Ray Mo for his original idea. We also thank experts and medical writers from MEDCOM ASIA for their preparing manuscript.
References
- (2000) World Health Organization Hepatitis C. Fact sheet No. 164. Revised. Oct.
- Sharma P, Lok A (2006) Viral hepatitis and liver transplantation.Semin Liver Dis 26: 285-297.
- Foster GR (2004) Past, present, and future hepatitis C treatments.Semin Liver Dis 24 Suppl 2: 97-104.
- Nomura H, Miyagi Y, Tanimoto H, Higashi M, Ishibashi H (2009) Effective prediction of outcome of combination therapy with pegylated interferon alpha 2b plus ribavirin in Japanese patients with genotype-1 chronic hepatitis C using early viral kinetics and new indices.J Gastroenterol 44: 338-345.
- Dixit NM, Layden-Almer JE, Layden TJ, Perelson AS (2004) Modelling how ribavirin improves interferon response rates in hepatitis C virus infection.Nature 432: 922-924.
- Neumann AU, Lam NP, Dahari H (1998) Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282: 103-107.
- Herrmann E, Lee JH, Marinos G, Modi M, Zeuzem S (2003) Effect of ribavirin on hepatitis C viral kinetics in patients treated with pegylated interferon.Hepatology 37: 1351-1358.
- Dahari H, Ribeiro RM, Perelson AS (2007) Triphasic decline of hepatitis C virus RNA during antiviral therapy.Hepatology 46: 16-21.
- Perelson AS, Herrmann E, Micol F, Zeuzem S (2005) New kinetic models for the hepatitis C virus.Hepatology 42: 749-754.
- Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD (1996) HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time.Science 271: 1582-1586.
- Dahari H, Shudo E, Ribeiro RM, Perelson AS (2009) Mathematical modeling of HCV infection and treatment.Methods Mol Biol 510: 439-453.
- Nowak MA, Bonhoeffer S, Hill AM, Boehme R, Thomas HC, et al. (1996) Viral dynamics in hepatitis B virus infection.Proc Natl Acad Sci U S A 93: 4398-4402.
- YOUDEN WJ (1950) Index for rating diagnostic tests.Cancer 3: 32-35.
- Ferenci P, Fried MW, Shiffman ML, Smith CI, Marinos G, et al. (2005) Predicting sustained virological responses in chronic hepatitis C patients treated with peginterferon alfa-2a (40 KD)/ribavirin.J Hepatol 43: 425-433.
- Yu JW, Wang GO, Li KG, Li AC (2007) Predictive value of rapid virological response and early virological response on sustained virological response in HCV patients treated with pegylated interferon -2a and ribavarin. J Gastroenterol Hepatol 22:832-836.
- McGovern BH, Nagami EH, Birch CE, Bowen MJ, Reyor LL, et al. (2009) Rate of sustained virologic response in relation to baseline hepatitis C virus (HCV) RNA level and rapid virologic clearance in persons with acute HCV infection.J Infect Dis 200: 877-881.
- Anton H (1999) Calculus: A New Horizon. 6th ed. New York: John Wiley and Sons.
- Ayres F Jr, Mendelson E (1990) Shaumas Outline of Theory and Problems of Differential and Integral Calculus. 3rd ed. New York: McGraw Hill.
- Layden TJ, Layden JE, Reddy KR, Levy-Drummer RS, Poulakos J, et al. (2002) Induction therapy with consensus interferon (CIFN) does not improve sustained virologic response in chronic hepatitis C.J Viral Hepat 9: 334-339.
- Layden-Almer JE, Ribeiro RM, Wiley T, Perelson AS, Layden TJ (2003) Viral dynamics and response differences in HCV-infected African American and white patients treated with IFN and ribavirin.Hepatology 37: 1343-1350.
Relevant Topics
- Constipation
- Digestive Enzymes
- Endoscopy
- Epigastric Pain
- Gall Bladder
- Gastric Cancer
- Gastrointestinal Bleeding
- Gastrointestinal Hormones
- Gastrointestinal Infections
- Gastrointestinal Inflammation
- Gastrointestinal Pathology
- Gastrointestinal Pharmacology
- Gastrointestinal Radiology
- Gastrointestinal Surgery
- Gastrointestinal Tuberculosis
- GIST Sarcoma
- Intestinal Blockage
- Pancreas
- Salivary Glands
- Stomach Bloating
- Stomach Cramps
- Stomach Disorders
- Stomach Ulcer
Recommended Journals
Article Tools
Article Usage
- Total views: 13857
- [From(publication date):
April-2015 - Aug 29, 2025] - Breakdown by view type
- HTML page views : 9296
- PDF downloads : 4561