Rice Gene Developments: A Review on Genomic Database
Received: 02-Jun-2022 / Manuscript No. rroa-22-67941 / Editor assigned: 04-Jun-2022 / PreQC No. rroa-22-67941 / Reviewed: 18-Jun-2022 / QC No. rroa-22-67941 / Revised: 24-Jun-2022 / Manuscript No. rroa-22-67941 / Published Date: 28-Jun-2022 DOI: 10.4172/2375-4338.1000309
Abstract
Rice (Oryza sativa L) is one of the most important crops worldwide. Its relatives, including phylogenetically related species of rice and paddy weeds with a similar ecological niche, can provide crucial genetic resources (such as resistance to biotic and abiotic stresses and high photosynthetic efficiency) for rice research. Although many rice genomic databases have been constructed, a database providing large-scale curated genomic data from rice relatives and offering specific gene resources is still lacking. Here, we present RiceRelativesGD, a user-friendly genomic database of rice relatives. RiceRelativesGD integrates large-scale genomic resources from 2 cultivated rice and 11 rice relatives, including 208 321 specific genes and 13 643 genes related to photosynthesis and responsive to external stimuli. Diverse bioinformatics tools are embedded in the database, which allow users to search, visualize and download the information of interest. To our knowledge, this is the first genomic database providing a centralized genetic resource of rice relatives. RiceRelativesGD will serve as a significant and comprehensive knowledgebase for the rice community.
Introduction
Crop breeding is crucial for guaranteeing food security and sustainable human population growth. Cultivated rice (Oryza sativa L.) is one of the most important crops worldwide and a model species for functional genomics of monocots. During the period of domestication, cultivated rice has lost many genes controlling important agronomic traits, such as resistance to abiotic and biotic stresses, which are potentially very useful for modern rice breeding. Many studies have revealed that genes regulating these traits are maintained in two closely related groups of the cultivated rice. One group includes species such as O. rufipogon that is phylogenetically closed to the cultivated rice [1]. The other group consists of paddy weeds that have a similar ecological niche as rice and are highly competitive and readily adapt to the agro ecosystem. Species of both groups are potential gene resources for modern rice molecular breeding programs aiming for improvement of agronomic traits. With the availability of sequencing technologies, genome-based molecular approaches can increase the efficiency of rice breeding to facilitate the accessibility of information from such enormous genomic data; several Oryza genome databases have been created to accommodate the genome data and various other types of data. The current online genomic resources of rice can be roughly divided into three categories depending on the main resources included. One is the de novo genome data [2].
Materials and Methods
Data Collection, Classification and Annotation
The current database included genomic datasets from 13 rice relatives that are publically available From the raw protein files of 13 genomes, the longest protein of each orthologous gene termed as ‘primary protein’ was extracted for gene family clustering analysis [3]. Based on the Markov Cluster algorithm, 34 570 gene families were identified using Orthofinder v2.2.7 with sequence search program ‘diamond’ . And according to the gene family clustering results, genes present in multiple species were defined as ‘multi-species family genes’, genes present only in a single species were defined as ‘species-specific family genes’, while genes that could not be clustered to any gene family were defined as ‘orphan genes’. Species-specific family genes, orphan genes and multiple-species family genes that could not be clustered with O [4]. sativa genes in the gene family analysis were further defined as ‘specific genes’ or, in other words, specific genes in RiceRelativesGD refer to genes without paralogs in O. sativa (japonica group) or O.sativa (indica group).
A Case Study for the Application of Rice Relatives GD
The database provides detailed information for each gene, including organism of origin, genomic location, family ID, gene structure, function annotations and sequences. EC_v6.g004014 belongs to the OG0001327 gene family, which has 34 genes from 7 species. In the interface of the gene family OG0001327, users can find function annotations, overview of the gene family and the phylogenetic tree of all genes of the family as well as download sequences. The phylogenetic tree of the gene family includes not only family members but also the Pfam domain information of each gene allowing researchers to easily observe the differences and similarities among the family members [5].
Discussion
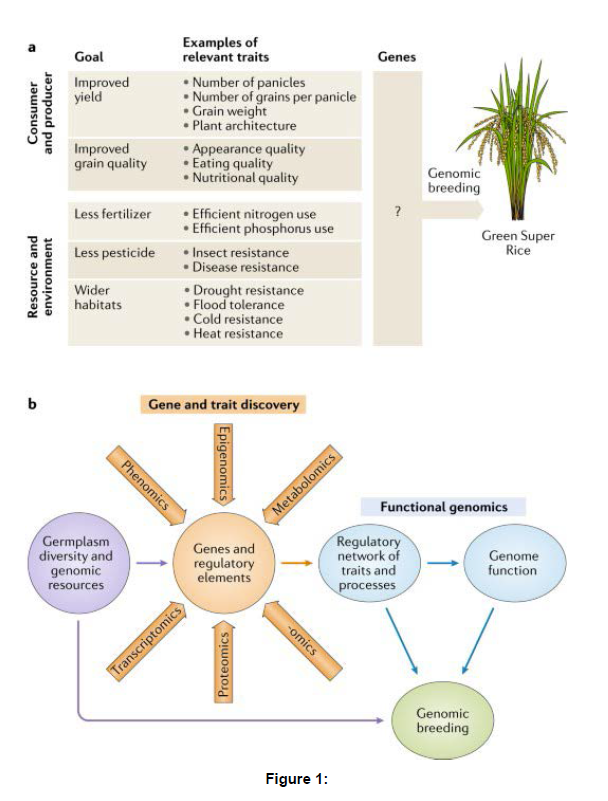
Rice relatives have become increasingly important for future improvement of rice varieties as they retain many competitive agronomic traits that have lost in rice during domestication and breeding with intensive artificial selection [6]. Re-introducing these genetic elements back into the rice genetic background would not only enhance the performance of rice but also alleviate the increased genetic load caused by domestication and breeding . Genomic data of rice relatives are essential and crucial sources for uncovering the genes lost in the cultivated rice currently, most rice genomic databases do not provide information on rice relatives [7]. Even though a few databases (e.g. Gramene, Ensemble Plants or PLAZA) integrate genomic resources of some rice relatives, they mainly focus on providing general information on the genomes, such as their orthologs and paralogs, gene gain/loss tree and genomic alignments, and pay no attention to a particular group of rice relatives or analysis of specific genes in rice relatives that could be valuable for modern rice breeding programs [8]. Rice Relatives GD fills the gap by providing not only more comprehensive genomic information of rice relatives for the rice community but also specific genes from rice relatives, including stress-related genes, photosynthesis genes and so on. RiceRelativesGD collected and organized published genomic data of rice and its relatives from relevant literatures [9]. Currently, a total of 208 321 specific genes from rice relatives are deposited in RiceRelativesGD. (Figure 1)
Conclusion
In conclusion, gene editing technologies, particularly the CRISPR/ Cas9 system, hold a greater significance in defining plant research in the recent times. It has truly emerged as the most effective tool for crop improvement owing to its ability to create mutations at desired targets in the genome with greater accuracy, efficiency, and simplicity. A major advantage of this process lies in the fact that the transgenes causing genetic modification can be easily eliminated from the genome through genetic segregation resulting in no differences between the gene-edited plants and those developed through conventional breeding. The development of CRISPR–Cpf1 system and base editing by far holds greater promise for editing rice genome with much more precision and efficiency. Furthermore, genome editing-based epigenetic regulation through the manipulation of DNA methylation and histone modification also holds greater promise in crop improvement as such modifications can be inherited into plant off springs without any change in the genomic sequence. A recent study involving CRISPR/ dCas9 fused with DNA methyl transferase 3a (DNMT3a) induced DNA methylation in the target regions of the mammalian cells.
References
- Lai Z, (1999) A shotgun optical map of the entire Plasmodium falciparum genome. Nat Genet 23: 309-313.
- Lin J, Qi R, Aston C, Jing J, Anantharaman TS,et al. (1999) Whole genome shotgun optical mapping of Deinococcus radiodurans. Science 285: 1558-1562.
- Mahairas GG, Wallace JC, Smith K, Swartzell S, HolzmanT,et al.(1999) Sequence-tagged connectors: A se-quence approach to mapping and scanning the human genome. Proc Natl Acad Sci 96: 9739-9744.
- Mao L, Wood TC, Yu Y, Budiman MA, Woo SS,et al. (2000)Rice transposable elements: A survey of 73,000 sequence-tagged-connectors (STCs). Genome Res 10: 982-990.
- Nagano H, Wu L, Kawasaki S, Kishima Y, Sano Y (1999) Genomic organization of the 260 kb surrounding the waxy locus in a japonica rice. Genome 42: 1121-1126.
- Talbot NJ(2003) On the trail of a cereal killer: Exploring the biology of (Magnaporthe grisea). Annu Rev Microbiol 57: 177-202.
- Dean RA, TalbotNJ, Ebbole DJ, Farman ML,Mitchell TK,et al.( 2005) The genome sequence of the rice blast fungus. Magnaporthe Grisea Nat 434-980.
- Wang ZY, Jenkinson JM,Holcombe LJ, Soanes DM,Veneault-Fourrey C,et al.( 2005) The molecular biology of appressorium turgor generation by the rice blast fungus (Magnaporthe grisea). Biochem Soc Trans 33: 384-388.
- Wilson RA, Talbot NJ(2009) Under pressure: Investigating the biology of plant infection by (Magnaporthe oryzae). Nat Rev Microbiol 7: 185.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Ibro G (2022) Rice Gene Developments: A Review on Genomic Database. J Rice Res 10: 309. DOI: 10.4172/2375-4338.1000309
Copyright: © 2022 Ibro G. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 2397
- [From(publication date): 0-2022 - Dec 15, 2025]
- Breakdown by view type
- HTML page views: 1922
- PDF downloads: 475