Special Issue Article Open Access
Oxic and Anoxic Metabolism of Steroids by Bacteria
| Wael Ismail1 and Yin-Ru Chiang2* | |
| 1Biotechnology Program, College of Graduate Studies, Arabian Gulf University, Al-Manamah, Kingdom of Bahrain | |
| 2Microbiology Laboratory, Graduate Institute of Natural Products, College of Medicine, Chang-Gung University, Tao-Yuan, Taiwan | |
| Corresponding Author : | Dr. Yin-Ru Chiang Microbiology Laboratory Graduate Institute of Natural Products College of Medicine, Chang-Gung University 259 Wen-Hwa 1st Road Tao-Yuan, Taiwan Tel: 886-3-2118800 ext. 5372 Fax: 886-3- 2118421 E-mail: yinru915@mail.cgu.edu.tw |
| Received Spetember 05, 2011; Accepted November 03, 2011; Published November 11, 2011 | |
| Citation: Sahli E, Tekeli O (2012) Evaluation of Retinal Nerve Fiber Layer Thickness with Spectral Domain Oct in Primary Open Angle Glaucoma and Ocular Hypertension. J Clin Exp Ophthalmol 3:247. doi: 10.4172/2155-6199.S1-001 | |
| Copyright: © 2011 Ismail W, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Steroid compounds are produced by eukaryotes where they have a variety of chemical structures and play important physiological roles. Many bacteria are capable of transforming and completely degrading steroids under various growth conditions. The microbial metabolism of steroids has gained considerable interest due to its potential applications in industrial and environmental biotechnology. The oxic degradation pathways of steroids and some of the involved enzymes are well characterized. The key players in these pathways are oxygenases which depend on dioxygen as a co-substrate. On the contrary, much less is known about the mechanisms operating under anoxic conditions. Obviously, anoxic bacterial metabolism of steroids should proceed via oxygenase-independent reactions. So far, a few bacteria that can completely degrade steroids in the absence of oxygen were characterized. Surprisingly, all of them belong to denitrifying bacteria and utilize only nitrate as the alternative electron acceptor. Recent studies of anoxic metabolism of steroids using denitrifying bacteria revealed unique and interesting biochemical reactions and enzymes. Here we discuss the current understanding of the biochemistry and molecular biology of bacterial steroid metabolism under anoxic conditions. The aerobic metabolism of steroids is briefly presented for the sake of comparison. Future investigations on anoxic metabolism of steroids will unravel novel aspects of the regulation and evolution of catabolic pathways as well as unprecedented biocatalysts with useful applications in biotechnology.
| Keywords |
| Anaerobes; Biodegradation; Biotransformation; Cholesterol; Denitrifying bacteria; Steroids; Testosterone |
| Abbreviations |
| AcmA: Cholesterol Dehydrogenase/Isomerase; AcmB: cholest-4-en-3-one-Δ1-dehydrogenase; AcmC: Cholest-4-en-3- one Hydroxylase; AD: Androst-4-en-3,17-Dione; ADD: Androsta-1,4- Diene-3,17-Dione; APCI: atmospheric pressure chemical ionization; CoA: Coenzyme A; ESI: Electrospray Ionization; KSTD: 3-Ketosteroid- Δ1-Dehydrogenase; LC-APCI-MS: Liquid Chromatography- Atmospheric Pressure Chemical Ionization-Mass Spectrometry; SDR: Short-chain Dehydrogenase/Reductase superfamily; Sli: Sterolibacterium; Sdo: Steroidobacter. |
| Introduction |
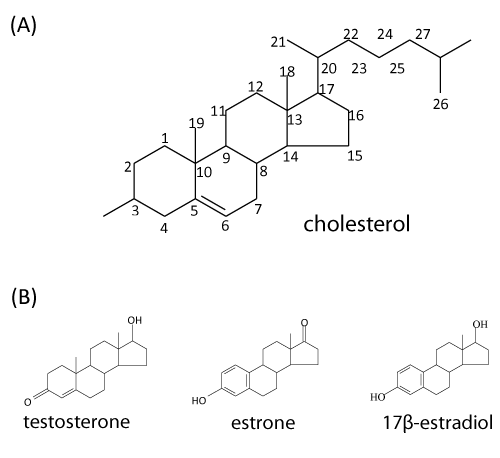
| Steroids constitute a wide variety of compounds that are abundant in nature and many derivatives have important physiological activities. In animals, all of the steroid hormones, also bile acids and vitamin D are derived from their common precursor, cholesterol. The adult human body contains around 200 to 300 g of cholesterol (Figure 1A). Since its first isolation from gallstones in 1784, cholesterol has been attracting much attention because of the relationships among diet, blood cholesterol levels, atherosclerosis, and heart disease [1]. Cholesterol and its related sterols (e.-g. phytosterols and ergosterol) are the important components of eukaryotic membrane, where it participates in modulation of membrane fluidity [2]. It is worthmentioning that many fungal species have the ability to transform steroid compounds by a variety of hydroxylations, dehydrogenation/reduction reactions, and sterol side chain cleavage [3]. However, to our knowledge, no fungal strain capable of completely degrading steroid compounds has been reported to date. Interestingly, steroids are absent from most prokaryotes. Only a few studies reported steroid biosynthesis in bacteria. Apparent exceptions are Methylococcus capsulatus and Nannocystis exedens [4,5]. These organisms contain sterols in amounts comparable to those present in eukaryotes. To replace the functions of steroids in membranes, most bacteria produce pentacyclic hopanoids [5,6]. |
| In addition to the role of sterols in the membranes of higher organisms, some steroids function as hormones for the regulation of biochemical activities in animals and plants [7]. In the human body, they control sexual development and fertility and hence, many steroids are used in medicine, for instance, for the treatment of cancer, arthritis, or allergies and in birth control [8]. The idea that steroidal hormones play an essential role in plant development and physiology is relatively recent. Brassinoide was firstly purified from Brassicus napus pollen [9] and so far approximately 60 related compounds have been identified [7]. Collectively, these compounds are referred to as brassinosteroids, which control a broad range of responses in plants, including seed germination, stem and root elongation, vascular differentiation, leaf expansion, and apical dominance [10]. |
| Steroid hormones are constantly released into the environment through man-made (e.-g., synthetic ethinyl estradiol) and natural sources (e.-g., human urine and livestock manure) [11]. The primary natural steroid hormones detected in the environment include estrone, 17β-estradiol, and testosterone [12,13] (Figure 1B). It was reported that pregnant women can excrete up to 259 μg of 17β-estradiol per day [14]. A variety of androgens and estrogens have been detected in effluents of American, Brazilian, Canadian, and German wastewater treatment plants and in surface water in American and Dutch rivers at concentrations in the ng l-1 range [12,15-18]. |
| Environmental concern about steroid hormones stems from their ability to alter the sexual behavior and endocrine systems of wildlife and aquatic species [13,19]. The removal of these compounds from the environment via microbial activities (biodegradation and bioremediation) has gained increasing interest. Moreover, the microbial biotransformation of steroid compounds has attracted great attention because of its potential impact on many biotechnological, pharmaceutical, and clinical applications [20,21]. Actually, the production of steroid drugs has been one of the best examples of successful application of microbial technology in industrial processes. The knowledge about oxic steroid metabolism by microorganisms has been used in biotechnological applications such as the preparation of expensive steroid hormones (such as androst-4-en-3, 17-dione) from the low-cost cholesterol and phytosterols [22]. |
| The ubiquity and abundance of steroids in the environment has made them common carbon sources for microorganisms. Obviously, soil bacteria can degrade steroids more efficiently in the presence of molecular oxygen. For example, Fan et al. [13] reported that under aerobic conditions 6% of 17β-estradiol and 63% of testosterone, respectively, could be mineralized to CO2 in agricultural soils. However, under anaerobic conditions, only 0.9% of 17β-estradiol and 46% of testosterone, respectively, was mineralized in agricultural soils. |
| Recent studies indicated that anoxic river-bed sediments and soil have the potential to be a reservoir for steroid compounds [23,24]. Thus, in order to improve the removal of steroids from the environment, it is necessary to understand the biochemical processes involved in anoxic mineralization of steroid hormones. Preliminary evidences for the anoxic degradation of steroids in the environment have been provided in the past decades. Studies on the degradation of the organic compounds in the lake sediments revealed that the total sterol content decreases faster than the total organic carbon pools, but more slowly than the pool of linear, long-chain aliphatic alcohols and fatty acids [25]. Denitrifying enrichments were established in which 2-5% of the introduced [C4-14C] cholesterol was recovered as 14CO2 [26]. Steroid hormones such as estrone and 17β-estradiol could be completely oxidized through microbial processes in the absence of molecular oxygen [13,27-29]. Over the last few years, a few bacterial strains able to mineralize steroids under denitrifying conditions were isolated and characterized [30-33]. However, in these studies, nothing is known about the degradation mechanisms operating in the absence of oxygen. Recently, Chiang and his colleagues adopted the denitrifying bacteria as the model organisms to investigate the intermediates, genes, and enzymes involved in anoxic steroid degradation [34-38]. A series of ongoing studies revealed unique and unprecedented biochemical principles that differentiate the anoxic from oxic steroids degradation by bacteria. |
| Bacterial steroid metabolism under oxic conditions |
| It was thought that oxic cholesterol degradation by bacteria is only an environmental issue of concern to the global carbon cycle [39]. Recently, an increasing number of studies revealed that oxic cholesterol catabolism is central to the unusual ability of Mycobacterium tuberculosis and its relatives to survive in host macrophages [21,40]. It was suggested that oxic degradation of cholesterol by M. tuberculosis might play important roles other than providing carbon and energy [40]. Although no other pathogens were reported to be capable of utilizing host cholesterol, intracellular parasites do interact with this compound for a variety of different purposes [41]. Those investigations provided insights into potential targets for novel therapeutics. |
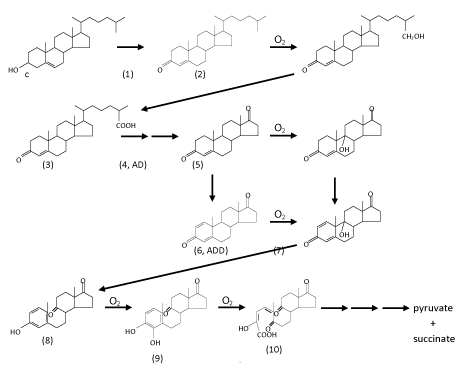
| In general, under oxic conditions, the bacterial degradation of cholesterol involves two processes: elimination of the C17-alkyl side chain and cleavage of the core ring system. The order of these two processes in vivo may differ among microorganisms [42]. For example, in the genus Mycobacterium, alkyl side chain elimination is followed by steroid ring opening [43]. Interestingly, bacteria appear to completely catabolize the A-ring to CO2 while incorporating side chain carbons into their lipid pools [40]. The complete aerobic mineralization of cholesterol by various genera of bacteria, such as Arthrobacter, Corynebacterium, Mycobacterium, Nocardia, Pseudomonas, and Rhodococcus was intensively studied in the 1960s to 1980s ( Figure 2) [44-52]. Those studies mainly focused on identifying of cholesterol degradation intermediates that were extracted from bacterial cultures, whereas genetic and biochemical aspects were neglected at that time. Based on those chemical studies, a general scheme of oxic bacterial cholesterol degradation was proposed by Kieslich [52], which proceeds as follows. |
| Oxidation of the A-ring of cholesterol |
| Generally, oxidation of the A-ring is thought to initiate cholesterol degradation [34,53,54]. The process includes three reactions: oxidation of the 3β-hydroxyl moiety, isomerization of Δ5 double bond into Δ4, and oxidation of C-1/C-2 of cholesterol. The first two reactions are catalyzed by either cholesterol oxidase or 3β-hydroxysteroid dehydrogenase [55] and result in the formation of cholest-4-en-3-one. The first enzyme (cholesterol oxidase) was purified and characterized from several bacterial strains [1,56-58]. FAD-dependent cholesterol oxidase is a bifunctional enzyme, catalyzing not only the oxidation of cholesterol to cholest-5-en-3-one, but also the further isomerization to form cholest- 4-en-3-one. It is therefore an alcohol dehydrogenase/oxidase belonging to the glucose-methanol-choline oxidoreductase family [58]. Oxidized flavin is the primary acceptor of the hydride abstracted from C-3. The reduced flavin then transfers the redox equivalents to molecular oxygen as the final acceptor. Cholesterol oxidase was proven to be suitable for the analysis of total cholesterol in serum, and has become the most widely used enzyme in clinical laboratories with the exception of glucose oxidase [5,59]. On the other hand, NAD (P)-dependent cholesterol dehydrogenase/isomerase that catalyzes the same reactions was also purified from different microorganisms [60-62], and has also been successfully used as a diagnostic enzyme for measuring cholesterol in blood [63]. Oxidation of C-1/C-2 of cholesterol and its derivatives involved in this oxic pathway is catalyzed by another FAD-dependent enzyme, 3-ketosteroid-Δ1-dehydrogenase (KSTD) [64-68]. For oxic steroid metabolism by microorganisms, the introduction of a double bond between C-1 and C-2 at an early stage is critical since it enables cleavage of the core ring [52-69]. |
| Degradation of the C-17 alkyl side chain of cholesterol |
| Oxic degradation of the acyl side chain of cholesterol was mainly studied at the phenomenological level [46-48,52]. After oxidation of the A-ring, the resulting cholest-4-en-3-one (compound 1 in (Figure 2)) is hydroxylated by a monooxygenase at its terminal methyl group (C-26), which is then oxidized to the corresponding sterol C-26-oic acid (compound 3). Theoretically, the aliphatic side chain is degraded via a series of β-oxidation reactions to form androst-4-en-3,17-dione (AD, compound 4) with the concomitant release of propionyl-CoA and acetyl-CoA. To our knowledge, no steroidal-CoA intermediate involved in this proposed pathway has so far been observed. Genome sequence data has revealed that actinomycetes posses a high number of genes encoding cytochrome P450s, compared with other prokaryotes [70,71]. However, the physiological functions of most of these enzymes in actinobacterial cells are still unclear [42]. Recently, the enzyme (cytochrome P450 125, also called steroid 26-monooxygenase) catalyzing the terminal hydroxylation of C-27sterol was purified and characterized from a variety of actinomycetes such as Rhodococcus jostii [42,43]. It is known that various cytochrome P450s have the ability to catalyze multistep oxidations [72-74]. For example, a cytochrome P450 from Pseudomonas putida is able to catalyze the sequential oxidations of linalool to form 8-hydroxylinalool and 8-oxolinalool. However, it is still unclear whether cytochrome P450 125 catalyzes the further oxidation of 26-hydroxy-4-cholesten-3-one to the corresponding carboxylic acid. To date, other enzymes involved in elimination of the alkyl side chain have not been fully identified. |
| Breakdown of the core ring system of cholesterol |
| After A-ring oxidation and alkyl side chain elimination, androsta- 1,4-diene-3,17-dione (ADD) is produced. The core ring system of ADD is cleaved via a series of oxygenase-catalyzing steps. The first step is the introduction of a hydroxyl group by a monooxygenase at C-9 in the B-ring (compounds 5 and 7 in (Figure 2)). However, 9α-hydroxy-androsta-1,4-dien-3,17-dione (compound 7) is very unstable and undergoes simultaneous aromatization of the A-ring and cleavage of the B-ring via a non-enzymatic reaction to form 9,10-secoandeosta1,3,5- trien-3-ol-9,17-dione (compound 8). Recently, the enzyme (3-ketosteroid 9α-hydroxylase, KshAB) that catalyzes the hydroxylation reaction was purified and characterized from M. tuberculosis and its environmental relatives [75-77]. KshAB is a twocomponent Rieske oxygenase. The oxygenase (KshA) component is a homotrimer containing a Rieske [2Fe-2S] cluster and mononuclear ferrous ion, whereas the reductase (KshB) component is a monomer containing a plant type [2Fe-2S] cluster and a FAD. Of the two potential substrates, KshAB has twice the specificity for 1,4-androstadiene-3,17- dione as for 4-androstene-3,17-dione. In addition, transformation of both substrates is well coupled to the consumption of O2 [76]. It is also worthmentioning that o-phenanthroline, a highly specific Fe2+ chelator, and a series of divalent metal ions, especially Zn2+, are able to significantly inhibit the hydroxylation activity of KshAB [77]. The next step is hydroxylation at C-4 in the A-ring (compound 9). After that, the A-ring is split via the well-known meta-cleavage by a dioxygenase to form compound 10. The resulting compound is then metabolized in a series of reactions to pyruvate and succinic acid. Overall, at least four transformations that require molecular oxygen as a co-substrate occur during the aerobic mineralization of cholesterol. However, except for KshAB, the other oxygenases involved in core ring cleavage still remain to be purified and characterized. |
| Organization of genes responsible for oxic cholesterol degradation |
| Many genes hypothetically responsible for oxic cholesterol degradation were found in the genome of Rhodococcus sp. strain RHA1 [21]. A microarray analysis revealed 572 genes that were up-regulated by at least 2-fold during growth on cholesterol compared to growth on pyruvate. Many of the up-regulated genes are scattered throughout the 9.7-Mbp genome of Rhodococcus sp. However, six clusters of upregulated genes were clearly discerned. The most striking of these was a cluster of 51 genes that occur within a 235-kbp stretch. Many genes in this cluster encode proteins with significant sequence identities with enzymes involved in the catabolism of steroid rings A and B by Comamonas testosteroni TA441 [78,79]. A mycobacterial gene cluster, mce4, which encodes a cholesterol import system was recently identified [40]. They showed that the cholesterol import system is not required for establishing infection in mice or for growth in resting macrophages. However, this function is essential for persistence in the lungs of chronically infected animals; however, it is worthmentioning that the functions of many genes within these clusters still remain to be experimentally proven, especially genes proposed to be involved in the degradation of the alkyl side chain of cholesterol. |
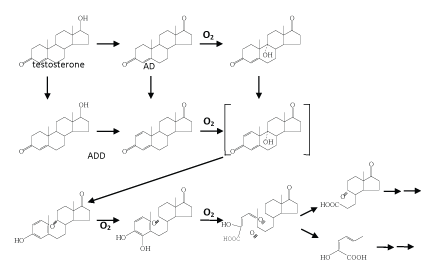
| Oxic testosterone degradation demonstrated in C. testosteroni |
| The best studied case in the topic of oxic androgen degradation by bacteria is testosterone [78,79,80-87]. Interestingly, the proposed oxic testosterone pathway demonstrated in C. testosterone ( Figure 3) is highly similar to that of oxic cholesterol degradation (Figure 2). A variety of oxygenases required in this pathway were isolated and characterized [82,88]. It is worthmentioning that detailed studies on the regulation of genes involved in testosterone degradation were also performed [78,79,89-91]. In C. testosteroni ATCC11996, the teiR gene was proven to encode a positive regular of testosterone degradation [79]. TeiR is a membrane-bound protein with a steroid-binding domain. In addition, TeiR is able to bind to promoters for transcription of genes involved in testosterone degradation. In addition, proteomics study was also carried out to search for testosterone-induced enzymes [92]. To date, however, it is still unclear whether testosterone alone is able to induce the production of all enzymes involved in the complete catabolic pathway. |
| Oxic estrogen degradation by bacteria |
| Several bacterial strains, such as Narcadia sp. strain E110 [93], Novoshingobium tardaugens sp. nov. strain ARI-1 [94], Sphingomonas sp. strain D12 [95], Sphingomonas sp. strain ED8 [96], Rhodococcus zopfii [97], Rhodococcus equi [97], and Nitrosomonas europaea [98], were isolated and identified as estrogen-degrading microorganisms. However, compared to the extensive studies on oxic cholesterol and testosterone degradation, much less is known about oxic estrone degradation by bacteria. So far, the pathways of oxic estrogen degradation by bacteria are not well described. This is partially due to the presence of benzene (the A-ring) in the core ring system of C18 estrogens, which is extremely difficult to attack by bacteria. Combe et al. [93] reported that exposure of estrone to Narcadia sp. strain E110 resulted in the formation of three possible degradative products. However, in their experiments, 13C- or 14C-labeled substrate was not utilized, therefore, the possibility that these compounds are not directly derived from estrone could not completely be excluded. In addition, Kurisu and colleagues [96] utilized Sphingomonas sp. as a model organism to study the degradation of estradiol in the presence of molecular oxygen. By incubating whole cells with estradiol and 3-chlorocatechol (a metacleavage inhibitor), two steroid intermediates, 4-hydroxyesterone and 4-hydroxyestradiol, were produced. To our knowledge, no other estrogen-derived intermediates involved in this oxic catabolic pathway are reported. |
| Oxic degradation of bile salts by bacteria |
| Bile salts are surface-active steroid compounds derived from cholesterol. Their main physiological function is aiding the digestion of lipids in the intestinal tract of vertebrates. It was reported that bile salts are toxic to many bacteria at a concentration of ≥10 mM [99]. Various groups of bacteria including actinobacteria and proteobacteria are capable of completely degrading bile salts in an oxic environment [99- 103]. Based on the structures of degradative intermediates, a proposed oxic pathway for bacterial bile salt degradation was established by Hayakawa [103]. That process is not the focus of this review. For a recent review see Philipp [104]. In brief, the four-ringed core of bile salts is cleaved in a manner highly similar to that for oxic testosterone degradation. Interestingly, it seems that in the oxic degradation of steroid compounds (except C18 estrogens) by bacteria, androst-4-en- 3,17-dione (AD) can be considered a central intermediate. Further degradation of AD proceeds via the common 9,10-seco-pathway. |
| Anoxic steroid transformation by bacteria |
| To date, very little is known about the mechanisms that operate under anaerobic conditions. It is obvious that anaerobic catabolism of steroids involves novel, oxygen-independent steps. The beststudied anaerobic reactions involve the incomplete transformation of cholesterol and bile salts by bacteria. |
| Anoxic transformation of cholesterol by intestinal bacteria |
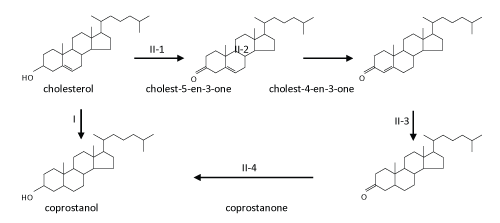
| The typical transformation of cholesterol at aerobic-anaerobic boundaries and in anaerobic environments involves reduction of the carbon-carbon double bond at C-5 of cholesterol to form coprostanol by intestinal bacteria [105-108], either directly by cholesterol reductase or via an alternative indirect, complicated pathway [108] ( Figure 4). Many strictly anaerobic cholesterol-reducing bacteria were isolated and characterized from the gut contents of mammals and were found to belong to the genus Eubacterium such as E. coprostanoligenes [107,109-111]. The enzyme responsible for the direct reduction is called cholesterol reductase [112]. Coprostanol, unlike cholesterol, is poorly absorbed by the human intestines. Therefore, coprostanol frequently constitutes more than 50% of the total fecal sterols in humans [113]. |
| Anoxic transformation of bile salts by intestinal bacteria |
| Anoxic microbial transformation of bile salts by anaerobes was recently reviewed [114,115]. In the human body, cholesterol is transformed to primary bile acids such as cholic acid and chenodeoxycholic acid in the liver, and these are further metabolized by the liver via conjugation (N-acyl amidation) to glycine or taurine. Bile salts are actively absorbed from the intestines. However, bile salt absorption is incomplete, and many of them are lost into the colon [114]. It was reported that intestinal bacteria can transform primary bile salts into 15~20 different bile acid metabolites [100]. Well-studied biotransformations include deconjugation of primary bile acids at the C-24 position; oxidation of α-hydroxyl groups at the C-3, C-7, and C-12 positions; generation of various oxo bile acids; and reduction of oxo bile acids to corresponding β-hydroxyl bile acids. It was hypothesized that deconjugation may be a mechanism for the detoxification of bile salts, in addition to obtaining carbon, nitrogen, and energy from deconjugated amino acids. On the other hand, dehydrogenation reactions may function in relation to energy generation and attempts to maintain low concentration of more-hydrophobic (also more-toxic) bile acids in the environment [115]. In all cases studied, cyclic carbon rings of bile salts remain intact [104,114,116]. |
| Anoxic steroid Mineralization by Denitrifying Bacteria |
| Bacteria that can completely degrade steroid compounds in the absence of oxygen |
| Some nitrate-reducing β-proteobacteria were isolated on oxygen containing or unsaturated monoterpenes as the sole carbon source and electron donor, including Thauera spp. [117] and Alcaligenes spp. [118]. The metabolism of monoterpenes and isoprenoids in anaerobic ecosystems was reviewed [114]. In all cases studied, denitrifyers are able to completely oxidize their isoprenoid substrates to CO2. Due to their facultative metabolism, denitrifying bacteria are more versatile than other groups of microorganisms and are potential sources of novel biotransformations of natural compounds and xenobiotics. |
| In the last few years, two bacterial strains were isolated and reported to have the ability to mineralize cholesterol under denitrifying conditions [30,31]. Both strains, 72Chol DSMZ 12783 and Sterolibacterium (Sli.) denitrificans Chol-1ST DSMZ 13999 are strictly respiratory with only nitrate or oxygen as the electron acceptor (Table 1). It was reported that Sli. denitrificans Chol-1ST is unable to grow on cholesterol with sulfate, thiosulfate, fumarate, or Fe(III) as the electron acceptor [31] (Table 1). Analysis of the 16S rDNA sequence of the two bacterial strains showed that both of them are β-proteobacteria, and are closely related to the genera Thauera and Azoarcus [30,31]. According to their 16S rRNA gene sequences, Sli. denitrificans Chol-1ST shows the highest sequence similarity with strain 72Chol. Physiological characterization showed that Sli. denitrificans Chol-1ST and strain 72Chol used only a limited number of non-polar substrates, including sterols and saturated longchain fatty acids [31]. It is worthmentioning that Sli. denitrificans Chol- 1ST, but no 72Chol, can also grow anaerobically on the C-19 steroid hormone, 4-androstene-3,17-dione (Table 1). When Sli. denitrificans Chol-1ST was grown on cholesterol with nitrate, nitrite accumulation was detected. Nitrite is further reduced to dinitrogen after the nitrate in the bacterial culture is completely consumed [34]. |
| It was also reported that another β-proteobacterium, Denitratisoma oestradiolicum DSMZ 16959 can anaerobically mineralize 17β-estradiol or estrone with nitrate as the terminal electron acceptor [32] ( Table 1). This strain represents the first known bacterium to grow on estrogens under anaerobic conditions. Not surprisingly, phylogenetic analysis of its 16S rRNA gene sequence revealed that its closest relatives are the cholesterol-degrading Sli. denitrificans Chol-1ST and strain 72Chol, with ca. 94% sequence identity [32]. It seems that D. oestradiolicum has a broader substrate spectrum than its cholesterol-degrading relatives, because this strain is able to use some polar compounds such as pyruvate, fumarate, and succinate as the sole carbon and energy source (Table 1). It can also grow anaerobically on some short-chain fatty acids (e.g., acetate, propionate, valerate, and caproate). Interestingly, D. oestradiolicum is unable to use cholesterol or long-chain fatty acids as a carbon source [32] (Table 1). Recently, Fahrbach et al. [33] isolated and characterized a γ-proteobacterium, Steroidobacter (Sdo.) denitrificans which can use 17β-estradiol or testosterone as the sole carbon and energy source under denitrifying conditions (Table 1). Phylogenetic analysis of its 16S rRNA gene sequence revealed that this strain has no close relatives and represents a distinct genus within the γ-proteobacteria [33]. When Sdo. denitrificans DSMZ18526 was grown on steroid substrates with nitrate, nitrite accumulation was not detected. It was shown that nitrate was first reduced to dinitrogen monoxide, which was further reduced to dinitrogen after the nitrate in the bacterial culture was completely consumed [33]. |
| Overall, these steroid-degrading bacteria share some common physiological and phylogenetic characteristics (Table 1). They are all rod-shaped proteobacteria, and have relatively narrow substrate spectra. These Gram-negative bacterium are mesophilic (optimally at 28-32˚C), neutrophilic (optimally at around pH 7.0), non-sporeforming, and strictly respiratory. When growing anaerobically on steroid compounds, these bacteria can only use nitrate as the terminal electron acceptor. Under denitrifying conditions, all of them can completely reduce nitrate to dinitrogen. These bacteria show no growth on complex media under either aerobic or anaerobic conditions [30-33]. |
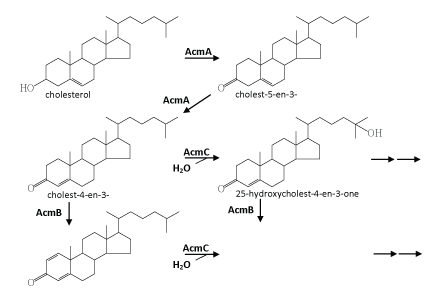
| Chiang et al. [34] published the first study on the complete anaerobic catabolism of a steroid compound. In this and subsequent studies, chemical, molecular biological, and biochemical approaches were applied to investigate the initial metabolic intermediates, genes, and enzymes involved in anaerobic cholesterol metabolism by Sli. denitrificans Chol-1ST [34-36]. An outline for the initial reactions in the anaerobic cholesterol-degrading pathway was proposed (Figure 5). This anaerobic pathway includes some common but also some fundamentally different intermediates compared to the aerobic cholesterol-degrading pathway (Figure 2). These initial anaerobic reactions involve the oxidation of ring A and the aliphatic side chain of cholesterol. However, so far it is still unclear how the core ring system of cholesterol is cleaved by Sli. denitrificans Chol-1ST. Obviously oxygenases cannot function I this case and therefore must be substituted by unprecedented oxygen-independent enzymes. |
| Some common intermediates involved in cholesterol catabolism |
| The initial steps are concerned with the oxidation and activation of the A-ring (Figure 5). First, the hydroxyl group at C-3 of cholesterol is oxidized to a carbonyl group leading to cholest-5-en-3-one. Subsequent isomerization at C-5 yields cholest-4-en-3-one. The same intermediates also occur in oxic cholesterol degradation [52] (Figure 2). Furthermore, as in the oxic pathway, these two reactions (oxidation and isomerization) are catalyzed by a bi-functional enzyme (AcmA) of Sli. denitrificans, with the difference that FAD-dependent cholesterol oxidases from the aerobic pathway require molecular oxygen as a cosubstrate [58]. Moreover, in the indirect anaerobic cholesterol reduction pathway (Figure 3), cholesterol is also transformed to coprostanol via these two intermediates (cholest-5-en-3-one and cholest-4-en- 3-one) [105,119]. Thus, the transformation of cholesterol to cholest- 4-en-3-one appears to be a common strategy to activate the A-ring of cholesterol. The fact that Sli. denitrificans can grow with cholest- 5-en-3-one or cholest-4-en-3-one as the sole carbon source under anaerobic conditions corroborates the conclusion that the two steroids are true intermediates of the anaerobic cholesterol catabolic pathway (unpublished data). The acmA gene was cloned and overexpressed in Escherichia coli, and the recombinant protein was purified, and shown to catalyze the expected reactions under both aerobic and anaerobic conditions [36]. The AcmA protein belongs to the short-chain dehydrogenase/reductase (SDR) superfamily [120] that comprises a growing number of NAD(P)-dependent non-metallo-oxidoreductases that are ca. 250~350 amino acids in length and bind NAD(P) with a Rossman fold motif [121]. However, the catalytic properties, substrate specificity, and potential inhibitors of AcmA are still not well studied. |
| The second step involves a dehydrogenation reaction at C-1 and C-2 that yields cholesta-1,4-dien-3-one which has a characteristic conjugated double bond system at the A-ring. This dehydrogenation reaction is catalyzed by a flavoprotein (AcmB) [35]. The acmB gene of Sli. denitrificans was cloned, and heterologously overexpressed in E. coli, and the recombinant AcmBhis protein was purified and characterized [35]. AcmB contains one molecule of FAD per monomer, and is able to use dichlorophenol indophenol and methylene blue as artificial electron acceptors. However, nothing is known about its natural electron acceptors. It seems that AcmBhis forms large molecular aggregates (> 600 kDa) in the absence of detergents, whereas typical KSTDs are monomeric [64,122]. In general, all steroids, which could serve as substrates for the enzyme, have in common a carbonyl group at the C-3 position. Interestingly, progesterone is an excellent substrate, even much better than cholest-4-en-3-one, which may be attributed to better solubility and/or better accessibility to the active site, because it lacks the aliphatic side chain [35]. In addition, the recombinant AcmBhis was competitively inhibited by corticosterone and estrone. AcmB was also shown to catalyze the predicted reaction both in the presence and absence of molecular oxygen. |
| Cholesta-1,4-dien-3-one is a key intermediate in oxic cholesterol degradation, because the introduction of a double bond between C-1 and C-2 to form a phenolic structure is required for the cleavage of the A-ring (via a classic meta-cleavage) [52] (Figure 2). However, this reaction requires a dioxygenase and molecular oxygen and thus cannot work under anaerobic conditions. So far it is unclear what the role of producing the 1,4-dien-3-one structure is in anaerobic cholesterol degradation. |
| Unprecedented intermediates involved in anaerobic cholesterol metabolism |
| The subsequent reactions (the formation of 25-hydroxycholest-4- en-3-one and 25-hydroxycholesta-1,4-diene-3-one) are proposed to fundamentally differ from those described for the aerobic cholesteroldegrading pathway. Further metabolism may proceed via cleavage of the ring A or the degradation of the aliphatic side chain. It is known that monooxygenases (also called mixed function oxidases) catalyze many hydroxylation reactions in steroid hormone metabolism in vertebrates, and usually use NADPH as the electron donor [1]. However, hydroxylation leading to 25-hydroxycholest-4-en-3- one and 25-hydroxycholesta-1,4-diene-3-one is not catalyzed by a monooxygenase, because an electron acceptor rather than an electron donor is required [34]. It seems that the resulting tertiary alcohol at the C-25 cannot be further oxidized. However, a rearrangement of the tertiary hydroxyl group at the C-25 to the C-26 or C-27 (a primary hydroxyl group) of cholesterol cannot be ruled out, which migh facilitate subsequent β-oxidation of the aliphatic side chain of cholesterol. Rearrangement of a tertiary hydroxyl group to a primary one (from linalool to geraniol) was reported in anaerobic linalool degradation by the denitrifying bacterium, Thauera linalootentis [114]. |
| Enzymes catalyzing such anaerobic hydroxylation normally belong to the molybdenum-containing hydroxylase enzyme family, and they use water as a source of the oxygen atom incorporated into their substrates (see for instance ethylbenzene dehydrogenase purified from the denitrifying Azoarcus strain EbN1; [123,124]). Ethylbenzene dehydrogenase is a molybdenum/iron-sulfur/heme protein of 155 kDa, which consists of three subunits (96, 43, and 23 kDa) in an αβγ structure. Ethylbenzene dehydrogenase is a periplasmic enzyme which catalyzes the anaerobic hydroxylation of ethylbenzene to form (S)-1- phenylethanol. The reported 43-kbp DNA fragment of S. denitrificans DSMZ 13999 genome does not encode proteins similar to ethylbenzene dehydrogenase [36]. Nonetheless, it was proposed that a complex molybdoenzyme might catalyze the anaerobic hydroxylation of the C-25 of the cholesterol side chain by S. denitrificans. |
| A novel aerobic pathway for cholesterol metabolism in Sli. denitrificans ? |
| Sli. denitrificans can utilize cholesterol as the sole carbon and energy source with nitrate or molecular oxygen as the terminal electron acceptor [31]. Chiang and colleagues provided some lines of evidence to support their suggestion that the anaerobic and aerobic cholesterol metabolism by Sli. denitrificans might follow the same route, at least during initial reactions [36]. The involvement of a common strategy to metabolize a single substrate under aerobic and anaerobic growth conditions was already shown in some cases. Denitrifying bacteria are subjected to fluctuations in oxygen availability inherent in their natural environment. Consequently, these organisms have to readily switch their metabolic machinery between aerobic and anaerobic modes of metabolism. Therefore, initiating metabolism of a substrate via the same enzymes regardless of the prevailing conditions and then channeling the last common intermediate into separate pathways depending on the prevailing conditions would increase their metabolic competence [36]. A similar case was reported for aerobic and anaerobic metabolism of benzoate in denitrifying bacteria. Regardless of oxygen availability, benzoate is converted to benzoyl-CoA by the same enzyme, benzoate- CoA-ligase of Thauera aromatica [125]. In this case, benzoyl-CoA then induces a separate aerobic or anaerobic pathway depending on the availability of molecular oxygen [125, 126]. |
| Genes involved in the anaerobic metabolism of cholesterol |
| So far, the entire genome of Sli. denitrificans Chol-1ST is still unknown. However, genes that participate in the initial steps of the anaerobic cholesterol catabolic pathway were identified by Chiang and colleagues using a reverse genetics approach [36]. Using a 857- bp DNA probe derived from cholest-4-en-3-one-Δ1-dehydrogenase encoding gene to screen a cosmid gene library of genomic DNA of Sli. denitrificans, they obtained a 43-kbp fragment of chromosomal DNA that harbors some genes proposed to be involved in anaerobic cholesterol metabolism. Among them, the metabolic functions of acmA (which codes for cholesterol dehydrogenase/isomerase) and acmB (which codes for cholest-4-en-3-one-Δ1- dehydrogenase) were unravelled [35,36]. It seems that these genes are not well organized into gene clusters [36]. However, most of the genes involved in anaerobic cholesterol metabolism are yet to be discovered. Thus, it cannot be excluded that the missing genes of anaerobic cholesterol metabolism might be clustered. Sequencing of the Sli. denitrificans genome should resolve this issue. A similar phenomenon was reported for genes involved in aerobic cholesterol metabolism by Rhodococcus sp. RHA1. Many of them are also scattered throughout the 9.7-Mbp genome of RHA1 [21]. In addition, it is also reported that upstream and downstream of the gene encoding 3-ketosteroid-Δ1-dehydrogenase of Rhodococcus rhodochrous are genes which are apparently unrelated to aerobic steroid metabolism [66]. |
| A novel testosterone degradation pathway demonstrated in Sdo. denitrificans DSMZ 18526 |
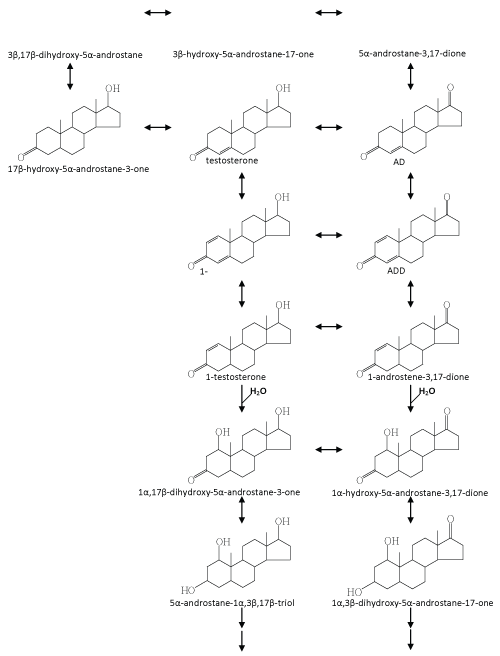
| Fahrbach et al. [33] isolated and characterized a γ-proteobacterium, Steroidobacter (Sdo.) denitrificans DSMZ 18526, which is able to completely degrade testosterone under denitrifying conditions. In a recent investigation they adopted cell suspensions of Sdo. denitrificans DSMZ 18526 to transform [4-14C] testosterone. They applied liquid chromatography-atmospheric pressure chemical ionization-mass spectrometry (LC-APCI-MS) to search for testosterone-derived intermediates [127]. In total, nine intermediates were identified, and their data suggest that several dehydrogenation/hydrogenation processes take place concurrently in the A- and D-rings of testosterone. In their experiments, hydroxylated steroid intermediates were not detected probably because they heavily depended on APCI for the MS analysis. In general, the signal intensity of steroid compounds in APCImass spectra is much better compared to that in electrospray ionization (ESI)-mass spectra. However, the use of APCI results in the formation of a range of dehydrated fragment ions in addition to the protonated molecular ion ([M+H]+). Unfortunately, in many cases, the protonated molecular ions are tiny and can easily be overlooked. According to our experience, both ionization techniques are required in order to obtain believable molecular ions of steroid intermediates. |
| Recently, Chiang et al. [37] and Leu et al. [38] carried out a series of in vitro assays to search for testosterone-derived intermediates. In the latter study, they found some hydroxylated steroid intermediates with a –OH group in the C-1α position, which were never reported to be involved in microbial steroid catabolism. In addition, in a fedbatch culture, Sdo. denitrificans DSMZ 18526 was grown on [4- 14C] testosterone with nitrate. During denitrifying growth, 14CO2 accumulation was detected. They thus presented direct evidence to show that testosterone was mineralized to CO2 by Sdo. denitrificans [38]. According to current investigations, an anoxic testosterone catabolic pathway demonstrated in Sdo. denitrificans DSMZ 18526 is proposed (Figure 6). Testosterone is first oxidized to ADD via two dehydrogenation reactions at C-1/C-2 and the hydroxyl group at C-17 of testosterone. In the next step, ADD is reduced to androst-1-en-3,17- dione, and as NADH seems to be the electron donor. Considering that a series of oxidation reactions is involved in the oxic pathway (Figure 4), it is still unclear why such a reduction reaction is involved in the anoxic testosterone catabolic pathway. From the perspective of organic chemistry, an alkene undergoes an electrophilic addition reaction. A tertiary carbon (C-5) is present in ADD, which can much more easily be attacked by a molecule of water (a nucleophile), compared to the secondary carbons (C-1 and C-2) in the A-ring. Considering that the next step is a hydration reaction occurring at C-1/C-2 of 1-testosterone (Figure 6), therefore, in order to hydroxylate C-1 of ADD, the reduction of C-4/C-5 may be a prerequisite. The authors provided several lines of evidence supporting the suggestion that the hydroxyl group is introduced via a novel hydration reaction, which implies a novel type of hydratase acting on C-1 and C-2 of steroid compounds is present in S. denitrificans DSMZ 18526. Taking into account that a variety of anaerobes utilize enoyl-CoA hydratase and fumarase, which are respectively involved in the β-oxidation pathway and citric acid cycle to oxidize different organic compounds, it is not surprising to see that anaerobes apply hydration reactions to oxidize and activate steroid substrates in the absence of oxygen. Further transformation processes seem to be very efficient; therefore, to date, formation of subsequent intermediates was not detected. According to current data, we expect that in the absence of molecular oxygen, cleavage of the core ring system of testosterone might start at the-A ring for the following reasons: (i) the monooxygenase responsible for the addition of a hydroxyl group at C-9 of ADD cannot work under anoxic conditions; (ii) the biochemical reactions we found so far occur mainly on the A-ring of steroid substrates [37,38,127]; and (iii) the A-ring structure of intermediates involved in the anoxic testosterone catabolic pathway are very similar to intermediates involved in the anoxic cyclohexanol catabolic pathway established by Dangel et al. [128]. In subsequent steps of the anoxic pathway, the two hydroxyl groups at C-1 and C-3 of WPS5α/β and WPS6 might be oxidized to carbonyl groups. A hydrolytic reaction might then occur to cleave the C-C bond between C-1/C-2 or C-2/C-3 of the steroid A-ring. |
| Interestingly, this bacterial strain is also able to utilize testosterone in the presence of oxygen. So far, it is unclear whether Sdo. denitrificans DSMZ 18526 uses the established oxic catabolic pathway (as demonstrated in C. testosteroni) to degrade testosterone. From an energetic perspective, it is more economic for a species to degrade a substrate using a common pathway in both the absence and presence of molecular oxygen. This is especially true for denitrifying bacteria, which are periodically subjected to fluctuations in oxygen availability inherent in their natural environment. Consequently, these organisms have to switch their metabolic machinery between oxic and anoxic modes of metabolism. Therefore, initiating the metabolism of a substrate via the same enzymes regardless of the prevailing conditions and then channeling the last common intermediate into separate pathways depending on the prevailing conditions would increase their metabolic competence. To date, nothing is known about the genes and enzymes involved in anoxic testosterone degradation. |
| Perspectives |
| The study of the microbial metabolism of organic compounds has contributed interesting answers to many intriguing questions concerning microbial physiology, biochemistry, and genetics. The anoxic metabolism of organic compounds is of particular interest because it has to deal with relatively inert and complex substrates in the absence of oxygen, an indispensable co-substrate for the aerobic metabolism of organic compounds. Over the past two decades, study of the anoxic metabolism of organic compounds has revealed many novel biochemical principles and aspects that paved the way for the development of useful biocatalysts [129]. Moreover, anaerobic metabolism is a vital process for environmental restoration since bioremediation in the subsurface is mediated by anaerobic bacteria. |
| Despite our better understanding of oxic steroid metabolism, the study of the anoxic metabolism of these compounds is still in its infancy. Further studies on the anoxic metabolism of steroids by bacteria should address the following issues: (i) whether cleavage of the core ring system of steroid compounds begins from the A-ring, (ii) whether elimination of the C17-alkyl side chain precedes cleavage of the core ring system in the anoxic cholesterol catabolic pathway, (iii) if a common pathway for steroid metabolism by denitrifying bacteria is potentially involved in both oxic and anoxic conditions, (iv) if steroidcatabolic genes cluster, and (v) purification and characterization of novel steroid-transforming enzymes. |
| Facultative anaerobes like Sli. denitrificans or Sdo. denitrificans are interesting model organisms due to their ability to switch between oxic and anoxic modes of metabolism depending on the availability of oxygen. Studies with such organisms will provide insights into mechanisms of adaptation to fluctuations in oxygen tensions inherent in their niches. Sequencing of the bacterial genome together with in-depth biochemical analysis should help unravel the underlying mechanisms. In addition, as a post-genomics tool, metabolomics is an emerging and vibrant field in its exponential growth phase. It can be expected that the use of metabolomics tools such as isotope distribution analysis and molecular connectivity analysis will significantly provide a better understanding of the anoxic steroid metabolism. Definitely, ongoing research and future studies on the anoxic metabolism of steroids will allow us to decipher novel aspects of the evolution and regulation of catabolic pathways. Furthermore, interesting biocatalysts which can be used in biotechnology will be discovered. |
| Acknowledgements |
| This work was funded by the National Science Council (NSC 100-2311-B-182- 005-MY3) of Taiwan. |
References
|
Tables and Figures at a glance
| Table 1 |
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 |
 |
 |
 |
| Figure 4 | Figure 5 | Figure 6 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 18119
- [From(publication date):
specialissue-2012 - Oct 04, 2025] - Breakdown by view type
- HTML page views : 13318
- PDF downloads : 4801
