Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
Google Scholar citation report
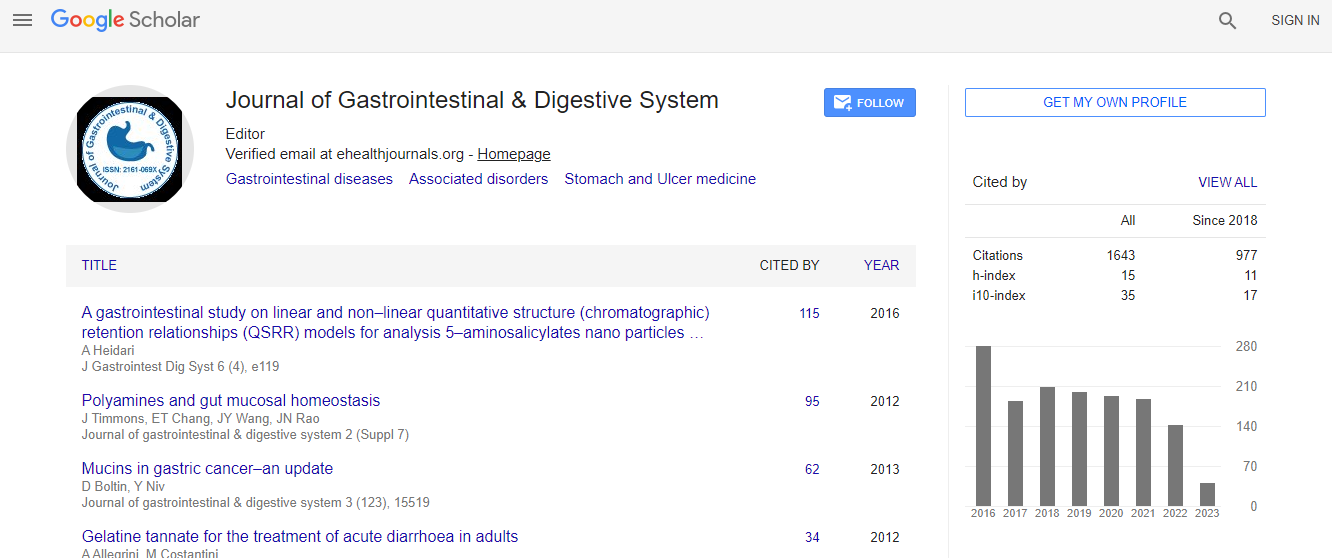
Citations : 2091
Journal of Gastrointestinal & Digestive System received 2091 citations as per Google Scholar report
Journal of Gastrointestinal & Digestive System peer review process verified at publons
Indexed In
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Open J Gate
- Genamics JournalSeek
- China National Knowledge Infrastructure (CNKI)
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Mahmud ElHefnawi
Biography
Dr. Mahmud ElHefnawi is the biomedical informatics and Chemoinformatics group leader at the Center of Excellence for Advanced Sciences (CEAS) and Associate Professor at the Informatics and Systems Department, National Research Center (NRC). He is also a part-time faculty/ Senior Research scientist at Nile University and was so at the Egyptian-Japanese University for Science and Technology (EJUST), and had affiliations/ part-time participations/ consultations at the Youssif-Jamil Science and Technology Research Center (YJ-STRC) and was a part-time adjunct faculty at the American University in Cairo teaching graduate level courses in Bioinformatics. He was the Information Technology Institute (ITI) bioinformatics track coordinator and co-founder. He has an H index of eight on scopus reflecting high citations of his works. He published more than thirty five Bioinformatics International publications on topics including rational drug design, docking, and optimizing gene therapy for Influenza, Hepatitis C Virus (HCV), and different cancers, structural bioinformatics and motif prediction applications in virology, and applications of phylogenetic analysis. These were in reputed journals like Bioinformatics (Oxford University Press), PLOS1 (Public library of science), Journal of Molecular Recognition (JMR), European Journal of Medicinal Chemistry,Virology (Elsevier), Genomics Proteomics Bioinformatics (Elsevier), Archives of Pharmaceutical Research and Virology Journal. He was the Principal Investigator for two STDF projects on structure-based drug design and docking of inhibitors for Hepatitis C virus, and optimizing gene therapy and siRNAs for Hepatitis C virus. He was also Copy for three projects at Youssif Jameel Science and Technology Research Centre (STRC), and American University in Cairo on bioinformatics, gene therapy, drug design, and nano-delivery. His interests span the computational as well as the medical and life sciences fields. From the informatics and engineering point: He worked on data mining and classification algorithms, reverse engineering of gene networks, HMM models, motif prediction algorithms, and other machine learning techniques. In the bioinformatics application side, he worked on: applications of the bioinformatics methods and techniques to study viral evolution, progression stages, RNA motifs and secondary structures, protein 3D structure and domain prediction, prognosis drug resistance mechanisms and modeling, modeling viral immune system, networks of interactions, rational drug design, small molecule inhibitors, and siRNA design as therapeutic agents for Hepatitis C virus, and Influenza. He has many national and international collaborations with research groups in Italy, Germany, AUC, TBRI, ITI, VacSera, Kasr el-Aini and NRC in interdisciplinary research involving IT, biomedical engineering, biomedical technology, computer-aided drug design and pharmaceuticals, and medicine, through use of advanced bioinformatics techniques and technologies. In supervisions, He had many PhD and MSc students from different faculties like engineering and computer science, biophysics, pharmaceutical sciences, biochemistry, microbiology, biotechnology, and medicine. His articles are highly cited ( H index=4, average impact factor = 3.5) and his publication rate and development is high (Please refer to the CV for details). Some sample publications are referenced here after the objective: I do strive to always Working in a high quality academic institute in research and teaching in my areas of expertise of bioinformatics, computer-aided drug design, computational biology, biotechnology, virology, cancer, molecular biology, and artificial intelligence/ machine-learning. ïâ· Focusing on drug discovery for Hepatitis C virus, influenza and different cancers. ïâ· Using machine learning approaches in drug discovery, bioinformatics and system biology. ïâ· Integration of the omics sciences for the understanding of and medicine, andconducting multi-desciplinary involving IT, biotechnology, nano delivery and nano technology and clinical trials. ïâ· Utilising the new areas of metagenomics and next-generation sequencing for understanding health and disease, and personalized medicine applications. ïâ· Mysara, M., Garibaldi, J.M. and Elhefnawi, M. MysiRNA-Designer (2011) "A Workflow for Efficient siRNA Design". PLoS One, 6, e25642.*: this publication was in a highly-ranked journal , involved producing a workflow similar to what’s proposed in the project, and was very well received. ïâ· ElHefnawi, M., O. Alaidi, et al.: “ Identification of novel conserved functional motifs across most influenza a viral strains”, Virology J.A bioinformatics workflow for analysisof the virus was developed and applied resulting in many interesting findings. ïâ· ElHefnawi et. al.: “The design of optimal small interfering RNA molecules targeting diverse strains of influenza a virus”, Bioinformatics (Oxford University Press).This publication was in the leading bioinformatics journal and dealth with a new rational drug design approach for influenza a virus and its experimental validations. ïâ· Elhefnawi, M. M., A. A. Youssif, et al. (2010) "An integrated methodology for mining promiscuous proteins: a case study of an integrative bioinformatics approach for hepatitis C virus non-structural 5A protein". Adv Exp Med Biol 680: 299-305. A methodology for elucidation of structural, functional, and mechanistic knowledge on promiscuous proteins is proposed ïâ· ElHefnawi, M. M., S. Zada, et al. (2010) "Prediction of prognostic biomarkers for interferon-based therapy to hepatitis C virus patients: a meta-analysis of the NS5A protein in subtypes 1a, 1b, and 3a". Virol J 7: 130.*: here, datamining was used to find biomarkers for response prediction using class association rules and comparing the predictability of the different features. ïâ· Galal, S. A., A. S. Abdelsamie, et al. (2011) "Part I: Synthesis, cancer chemopreventive activity and molecular docking study of novel quinoxaline derivatives". Eur J Med Chem 46(1): 327-340. This work was on novel cancer therapeutics as is proposed in the project for liver cancer. ïâ· Zein, H. S., A. A. El-Sehemy, et al. (2011) "Generation, characterization, and docking studies of DNA-hydrolyzing recombinant F(ab) antibodies". JMR 24(5): 862-874.This publication was on drug design with molecular modeling of DNA-antibodies interactions; a very new topic. ïâ· Temerak A, Abdulla M and ElHefnawi M: Rational Drug Design for Identifying Novel multi-Kinase inhibitors as potential candidate therapies for Hepatocellular Carcinoma”, Accepted in Anti-cancer agents and medicinal chemistry, Benthalm Publishing, 2012. Here, a protocol for computer-aided drug design and optimization is applied to find new anti-cancer RTK inhibitors. ïâ· ElHefnawi M, Amin M, Omar M:” Multiple virtual screening approaches for finding new Hepatitis c virus RNA-dependent RNA polymerase inhibitors: Structure-based screens and molecular dynamics for the pursue of new polypharmacological inhibitors”, Accepted in BMC Bioinformatics InCoB Supplement issue, 2012. Here, different computer-aided drug design approaches are implemented to find new HCV inhibitors with good ADMET profiles. ïâ· Hamouda A , Saad M, ElHefnawi M: “ Structure-based activity prediction model for some benzimidazole inhibitors of hepatitis C virus NS5B polymerase and its application in the design of new analogs”, accepted in Medicinal Chemistry Research, August 2012. Here, a new lead optimization protocol is presented and applied to find new HCV Benzimedasole inhibitors with better physiochemical properties. ïâ· ElHefnawi M Et. Al.: ” Accurate prediction of response to Interferon-based therapy in Egyptian patients with Chronic Hepatitis C using different machine-learning techniques”, accepted in ASINUM2012 IEEE Explorer conference proceedings,and Springer Lecture notes, Turkey,September 2012. Yours sincerely Mahmoud ElHefnawi (Google scholar, Linked in, I am scientist, and facebook profile) 26,B, Gezira Wosta street, Zamalek Cairo, Egypt.
Research Interest
Influenza, Hepatitis C Virus (HCV), and different cancers, structural bioinformatics and motif prediction applications in virology, and applications of phylogenetic analysis.
Relevant Topics
Peer Reviewed Journals
Make the best use of Scientific Research and information from our 700 + peer reviewed, Open Access Journals

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi