Research Article Open Access
Induction of Apoptosis by Benzamide in K562 Cells Was Triggered by Inhibition of Poly (ADP-Ribosyl) Action of Protein(S)
Utpal Ghosh1* and Nitai P. Bhattacharyya21Department of Biochemistry and Biophysics, University of Kalyani, Kalyani-741235, India
2Crystallography and Molecular Biology Division, Saha Institute of Nuclear Physics, 1/AF Bidhannagar, Calcutta 700 064, India
- Corresponding Author:
- Utpal Ghosh
Department of Biochemistry and Biophysics
University of Kalyani, Kalyani-741235, India
Fax: +091 033-2582-8282
E-mail: utpal8ghosh@gmail.com
Received date: November 10, 2012; Accepted date: January 22, 2013; Published date: January 27, 2013
Citation: Ghosh U, Bhattacharyya NP (2013) Induction of Apoptosis by Benzamide in K562 Cells Was Triggered by Inhibition of Poly(ADP-ribosyl)ation of Protein(S).J Biotechnol Biomater S13:008. doi:10.4172/2155-952X.S13-008
Copyright: © 2013 Ghosh U, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Keywords
PARP; Benzamide; siRNA; AIF; Apoptosis; Caspase-8; Caspase-9/caspase-6; Caspase-3; Poly (ADP-ribosyl)ation
Abbreviations
PAR: Poly (ADP-ribose); PARP: Poly (ADPribose) polymerase; AIF: Apoptosis inducing factor; NAD+: benzamide
Introduction
Poly (ADP-ribose) polymerase (PARP) family consists of 18 members, that Poly (ADP-ribosyl) late their target proteins and can thereby, change the function of the proteins [1]. Poly ADP-ribosylation is one kind of post-translational modification that can play crucial function in various cellular processes such as transcription, replication, cell cycle, telomere metabolism, DNA repair, and also apoptosis. Major amount of poly(ADP-ribosyl)ation reaction in cell is carried out by PARP-1, the well-characterized isoform of PARP. Investigations over the decade using chemical inhibitors of PARP, transgenic mice and cell lines reveal that PARP-1 participates in the recovery of damaged DNA. A large number of nuclear protein, including PARP-1 itself, are poly (ADP-rbosyl)ated, although the biological function of such modification is not very clear [2].
On the basis of observations that chemical inhibitors of PARP-1 increases the sensitivity of diverse DNA damaging agents, as detected by colony forming ability, PARP inhibitors have been considered as one of potential candidates for tumor treatment, in combination with radiation or alkylating agents like MMS [3-7]. Synergistic killing effect was observed in hepatocellular carcinoma, when PARP inhibitors applied with chromatin modifying enzymes [8]. PARP inhibitors are in clinical trials for breast cancer treatment with BRCA mutant population [9]. Poly (ADP-ribose) polymerase-1 (PARP-1) has been shown to involve in DNA damage induced apoptosis. But actual role of PARP- 1 or poly(ADP-ribosyl)ation reaction in apoptosis is very confusing. Chemical inhibitors of PARP decrease induction of nucleosomal ladders induced by various agents [10-12]. On the contrary, oxidant stress and temozolomide induced apoptosis are increased by such inhibition [13,14]. In view of the anti oxidant properties of the chemical inhibitors, it is difficult to contribute the role of PARP in this process [15]. The role of PARP-1 in the induced apoptosis may depend on the cell type, as well the inducing agents. PARP-1 is one of the major targets of activated caspase-3 [16,17], and PARP-1 is cleaved during the early stages of apoptosis. Nucleosomal ladder formation has been shown to be mediated by Ca++Mg++ dependent endonuclease DNase1L3, and is inactivated by poly (ADP-ribosyl)ation [18-20]. Poly (ADP-ribose) chain binds to caspase activated DNase (hCAD/DFF-40), at a specific site in vitro [21]. This implicated that inhibition of poly (ADP-ribosyl) ation may lead to the activation of DNases involved in apoptosis. PARP- 2, which is devoid of the DNA binding domain of the PARP-1, has been shown to be the substrate of the activated caspase-8 in nucleus [22]. DNA repair ability in PARP-1 knock out cells, as well as PARP-2 knock out cells is impaired. Even though the knock-out mice for PARP-1 and PARP-2 are viable, the double knock out for the two enzymes is lethal [23]. Nucleosomal ladder formation studied in cells from one of the three independent knockout mice showed increased nucleosomal ladders [24]. We have shown that benzamide, an inhibitor of PARP, treatment alone induced nuclear fragmentation, nucleosomal ladders, cytochrome-c release and caspase-3 activation in Chinese hamster V79 cells [25]. PARP inhibitors can induce apoptosis in myeloid leukemia cells, but detailed mechanism is not clear [26].
In the present communication using a human leukemic cell line K562, we have extended our earlier study [25], to decipher the detailed pathway involved in benzamide induced apoptosis.
Materials and Methods
Chemicals and antibodies
RNase A, benzamide, anti-AIF and anti-beta actin antibody, NBT/BCIP tablet were obtained from Sigma Chemicals (USA), and Proteinase K was from Life Technologies, USA. Nonidet P40 was obtained from Boeringer Mannheim, Germany. Anti-cytochrome-c were purchased from Santa Cruz Biotechnology (USA), caspase-3 and caspase-6/9 assay kit were from PharMingen (USA). Caspase-2 and caspase-8 assay kits, anti-PAR antibody were procured from ALEXIS Biochemicals, UK. The anti-mouse and anti-rabbit secondary antibodies, both alkaline phosphatase conjugate and horseradish peroxidase conjugate, were purchased from GeNei (India). Antibodies for Bid and the kit for fluorometric detection of caspase-8 were from Chemicon International, USA. Caspase-9/6 assay kit was procured from BD Biosciences (USA). Other molecular biology grade fine chemicals were procured locally.
Cell culture
Human leukemic cell line K562 was obtained from National Centre for Cell Sciences, Pune, India. K562 cells were routinely grown in culture flasks using RPMI medium (Life Technology, USA), supplemented with 10% bovine serum (complete medium) at 37°C in humidified atmosphere containing 5% CO2.
Detection of nuclear fragmentation and nucleosomal ladders
Methodology used for nuclear fragmentation and nucleosomal ladders were same as described earlier [25].
Transfection of K562 cells with the plasmid containing siRNA construct
We have used silenced PARP-1 in K562 using the same siRNA construct, as described earlier [27]. Here, the transfection was carried out by electroporation, as described in Basu and Banerjee [28]. The K562 cells transfected with pRNA-U6.1/Hygro vector with siRNA construct was designated as KsiI and the cells transfected with only the vector without siRNA construct was designated as K-vector. Almost 90% reduction of expression of PARP-1 was observed in KsiI cells, as detected by standard RT-PCR, where as K-vector shows same PARP-1 expression as that of untreated control K562 cells.
Western blot analysis for caspase-8, Bid, cytochrome c and AIF
Western blot analysis of cytochrome c and AIF was carried out using the methods described by Ghosh et al. [25]. In brief, K562 cells treated with benzamide for 24 h and untreated control cells were harvested, washed twice with PBS, and cytoplasmic extract was prepared using standard method [29]. Anti-caspase-8 (1:200 dilution), anti-Bid (1 μg/ml), anti-cytochrome c (1:2000), and anti-AIF (1:1000) were used to react overnight with separated proteins on the membrane in TBS containing 0.05% tween-20 at 4°C.
The blot was washed 4 times with 5.0 ml TBS containing 0.05% Tween-20 (TBS-T), followed by treatment with horseradish peroxidase-conjugate anti-mouse (for caspase-8), and anti-rabbit (for Bid and AIF) immunoglobin G (1:2000 dilution in TBS-T) for 2½ h at room temperature. Alkaline phosphatase (ALP) conjugate anti-mouse antibody was used (1:2000) for cytochrome C for 2 h incubation at room temperature. The membrane was then washed 4 times with TBS-T and developed using either NBT/BCIP, the substrate for ALP or ECL western blotting detection reagents (Amarsham Biosciences).
Fluorometric determination of caspse-2, caspase-8, caspase-9/6 and caspase-3
Caspase-2, caspase-8, -6/9 and -3 activity assay were performed according to the protocols given by the manufacturers. In short, after treatment with 5 mM benzamide for various time interval or different doses of benzamide for a fixed time of incubation (24 h for caspase-3 and 20 h for caspase-8 and 9, since the caspase-9 and 8 are in the upstream of caspase-3 in apoptosis pathway, we had chosen time points), K562 cells were processed according to the protocol provided by the manufacturers and our earlier study [30]. To check the enzyme-substrate specificity, the inhibitors of the caspases (supplied by the manufacturer of the kits) were used together with their respective substrates during the assay and found inhibited by the inhibitors, showing the specificity of the assays (data not shown).
Detection of cellular NAD+
Estimation of cellular NAD+ was carried out spectrofluorimetrically, following the method originally developed for in vitro assay of NAD+, described by Putt and Hergenrother [31], with slight modification as we did earlier [30,32]. Exponentially growing cells were treated with 5 mM benzamide for different times of interval (0-48 h) and estimated NAD+.
Immunoblotting of poly (ADP-ribosyl)ated proteins
K562 cells were treated with various doses (0-10 mM) of benzamide for 4 h and Poly (ADP-ribosyl)ation of proteins were detected using ant-PAR antibody as our earlier method [32].
Comet assay
DNA damage (single stand break) was measured using the alkaline single cell electrophoresis assay, following the method as described by Singh et al. [33], with slight modification. Comet was observed under a Nikon fluorescent microscope and subjected to image analysis using comet assay software (CASP). DNA damage was quantitated by tail moment measurement, calculated by multiplying the total intensity of the comet tail by the tail length, measured from the centre of the comet head.
Statistical calculations
Each experiment was repeated 3-4 times and the mean of apoptotic parameter, say for example, caspase-3 activity at each dose of benzamide was compared with the mean caspase-3 activity of untreated control in a particular cell type and designated as ‘*’ (p ≤ 0.05), ‘**’ (0.05 ≤ p ≤ 0.01) and ‘***’ (p ≤ 0.001) in graphs or tables. Again, the differences of caspase-3 activity, for example, in K-vector and KsiI cells at each dose were also calculated and the p-values were designated as ‘#’ (p ≤ 0.05). The significance level for any such apoptotic parameter was calculated using ANOVA with Dunnett’s test (SPSS software).
Results
Nuclear fragmentation and nucleosomal ladder induced by benzamide in K562 cells
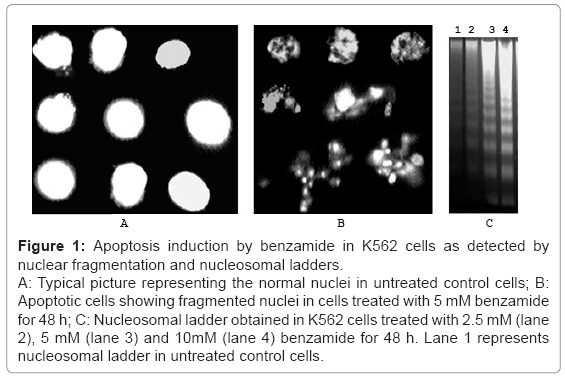
Benzamide treatment for 48 h increased the nuclear fragmentation in a dose (2.5-10 mM) dependent manner in K562 cells, as detected by the fragmented nucleus under microscope after staining with Hoechst dye. Typical normal nuclei and apoptotic nuclei are shown in the figures 1a and 1b, respectively. Summary of the result is shown in the table 1. Significant increase (p=0.0078) in the nuclear fragmentation was observed in 2.5 mM benzamide treatment for 48 h. At higher doses, the nuclear fragmentation was higher. Time dependent increase of nuclease fragmentation was also observed with 5 mM benzamide treatment, and significant increase was observed at and higher than 24 h (p=0.008), as shown in table 2.
Figure 1: Apoptosis induction by benzamide in K562 cells as detected by nuclear fragmentation and nucleosomal ladders.
A: Typical picture representing the normal nuclei in untreated control cells; B: Apoptotic cells showing fragmented nuclei in cells treated with 5 mM benzamide for 48 h; C: Nucleosomal ladder obtained in K562 cells treated with 2.5 mM (lane 2), 5 mM (lane 3) and 10mM (lane 4) benzamide for 48 h. Lane 1 represents nucleosomal ladder in untreated control cells.
| Concentrations of benzamide (mM) | % apoptotic cells ± SD (p value) |
|---|---|
| 0 | 5.6 ± 0.85 |
| 2.5 | 13.85 ± 2.76 (0.0078) |
| 5.0 | 38.09 ± 6.94 (0.0013) |
| 10.0 | 50.95 ± 7.29 (0.0005) |
P-values were calculated at each dose with respect to untreated control and shown within bracket.
Table 1: Induction of apoptosis by different concentrations of benzamide in K562 cells when treated for 48 h.
| Time of treatment (h) | % apoptotic cells ± SD (p value) |
|---|---|
| 0 | 8.1 ± 2.2 |
| 4 | 9.6 ± 1.27 (0.364) |
| 12 | 13.4 ± 3.83 (0.1062) |
| 16 | 16.12 ± 5.29 (0.0724) |
| 24 | 25.29±5.7 (0.008) |
| 36 | 33.75 ± 5.30 (0.0015) |
| 48 | 44.5 ± 6.5 (0.0008) |
P-values were calculated at each dose with respect to untreated control and shown within bracket.
Table 2: Induction of apoptosis by 5 mM benzamide in K562 cells at different time of treatment.
Nucleosomal ladders were increased in a dose dependent manner. A typical experiment for DNA ladder formation in K562 cells by the treatment with various doses of benzamide for 48 h is shown in figure 1C. As shown (lane 2), 2.5 mM benzamide treatment for 48 h increased the nucleosomal ladders appreciably, and at higher doses, the nucleosomal ladders increased further. This result showed that benzamide induced nucleosomal ladders in K562 cells in a dose dependent manner.
Benzamide induced cleavage of procaspase-8 and activation of caspase-8 in K562 cells
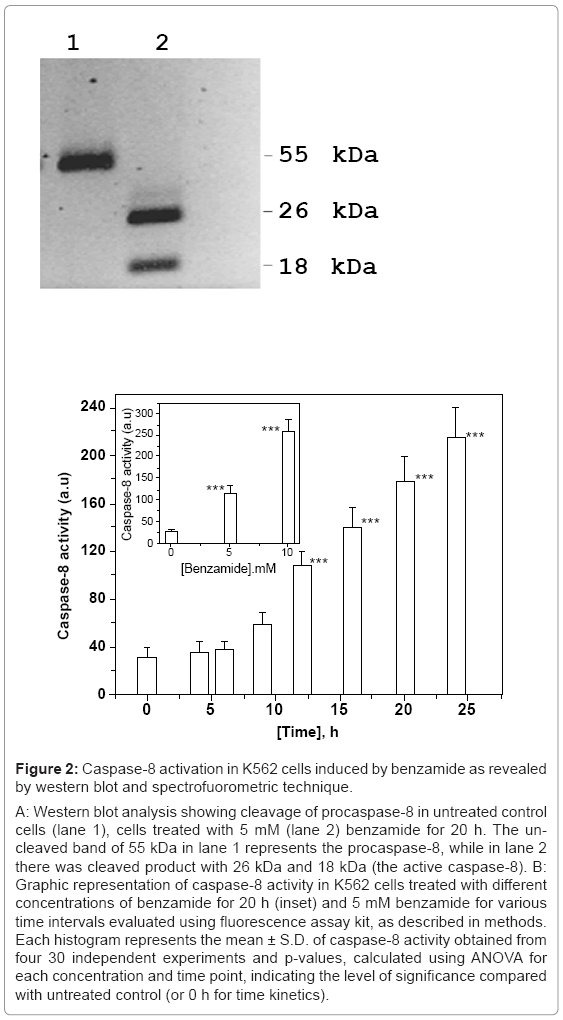
Caspase-8 in its inactive form remains as procaspase-8 of 55 KDa in cytosol, and after two-step proteolysis, active form of caspase-8 is produced. In the first step, 55 KDa procaspase-8, is cleaved into 43 KDa and 12 KDa fragments. The 12 KDa fragment is subsequently cleaved into 10 KDa fragment. The 43 KDa fragment is converted into 26 KDa and the active caspase-8 of 18 KDa fragment. The procaspase-8 cleavage after 20 h of treatment with benzamide is shown in figure 2a. The 26 KDa and 18 KDa (active form of caspase-8) fragments were clearly observed (lane 2) after 10 mM benzamide treatment, where as uncut was in lane 1(untreated control). We also confirmed the activation of caspase-8 by benzamide treatment spectrofluorometricaly, using commercially available kit. Benzamide induced caspase-8 activation in a dose (Figure 2b) and time dependent manner (Figure 2b). The time kinetics of caspase-8 activation revealed that significant activation started after 9 h (p=0.0152) treatment with 5 mM benzamide, as shown in figure 2b. Treatment with 5 mM and 10 mM benzamide for 20 h increased ~ 4 and ~ 10 fold activity of caspase-8, in comparison to untreated control cells. This increase was statistically significant, indicating that the caspase-8 activity was increased in a dose and time dependent manner by benzamide treatment.
Figure 2: Caspase-8 activation in K562 cells induced by benzamide as revealed by western blot and spectrofuorometric technique.
A: Western blot analysis showing cleavage of procaspase-8 in untreated control cells (lane 1), cells treated with 5 mM (lane 2) benzamide for 20 h. The uncleaved band of 55 kDa in lane 1 represents the procaspase-8, while in lane 2 there was cleaved product with 26 kDa and 18 kDa (the active caspase-8). B: Graphic representation of caspase-8 activity in K562 cells treated with different concentrations of benzamide for 20 h (inset) and 5 mM benzamide for various time intervals evaluated using fluorescence assay kit, as described in methods. Each histogram represents the mean ± S.D. of caspase-8 activity obtained from four 30 independent experiments and p-values, calculated using ANOVA for each concentration and time point, indicating the level of significance compared with untreated control (or 0 h for time kinetics).
Detection of activation of caspase-2, caspase-9/6 and caspase-3 in K562 cells treated with benzamide
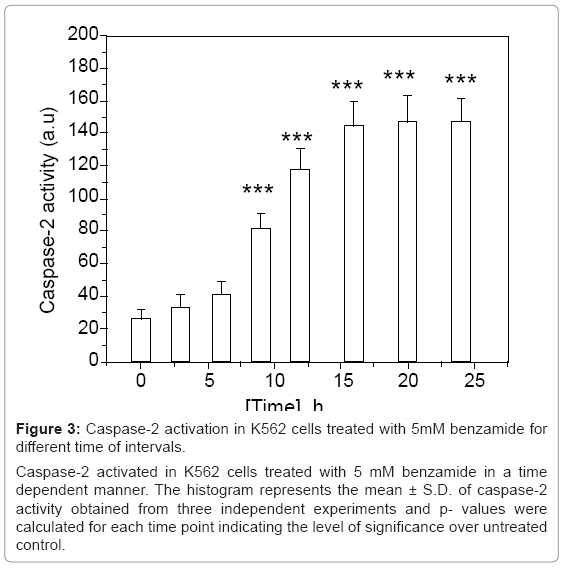
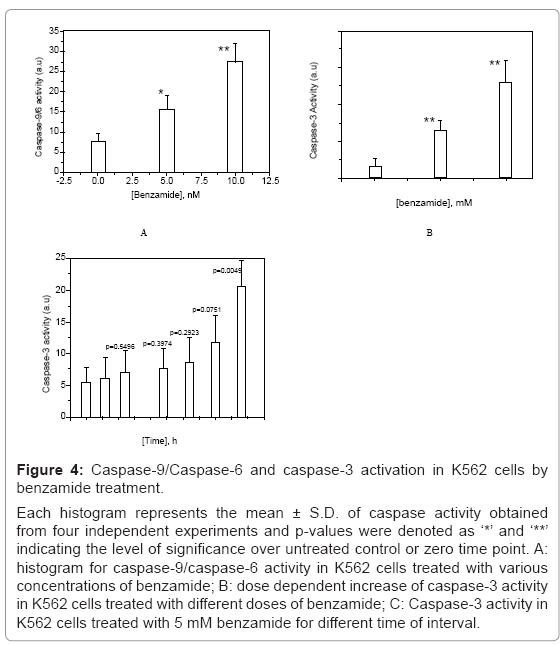
We observed caspase-2 activation in K562 cells treated with 5 mM benzamide for various time of intervals as detected by spectrofluorometric measurement, as described earlier. The intensity of activation of caspase-2 increased with the increase of incubation time and significant activation was observed after 6 h treatment with 5 mM benzamide (p=0.0468). The result is shown in figure 3. Caspase- 9/6 activity was increased about 4 fold by 10 mM benzamide treatment for 20 h (p=0.0034), as shown in figure 4a. We had also detected dose dependent increase of caspase-3 activity in K562 cells after 24 h treatment, as shown in figure 4b. Although, there was time dependent increase in caspase-3 activity in K562 cells treated with 5 mM benzamide as shown in figure 4c, the significant increase (p=0.0049) was found after 24 h treatment.
Figure 3: Caspase-2 activation in K562 cells treated with 5mM benzamide for different time of intervals.
Caspase-2 activated in K562 cells treated with 5 mM benzamide in a time dependent manner. The histogram represents the mean ± S.D. of caspase-2 activity obtained from three independent experiments and p- values were calculated for each time point indicating the level of significance over untreated control.
Figure 4: Caspase-9/Caspase-6 and caspase-3 activation in K562 cells by benzamide treatment.
Each histogram represents the mean ± S.D. of caspase activity obtained from four independent experiments and p-values were denoted as ‘*’ and ‘**’ indicating the level of significance over untreated control or zero time point. A: histogram for caspase-9/caspase-6 activity in K562 cells treated with various concentrations of benzamide; B: dose dependent increase of caspase-3 activity in K562 cells treated with different doses of benzamide; C: Caspase-3 activity in K562 cells treated with 5 mM benzamide for different time of interval.
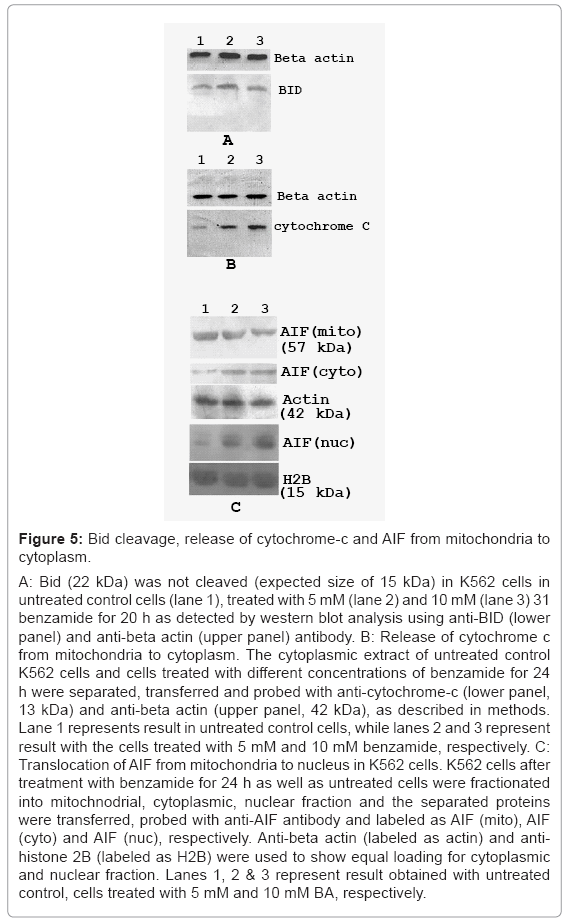
Bid cleavage, cytochrome-C release and translocation of apoptosis inducing factor (AIF) from the mitochondria to the nucleus in K562 cells treated with benzamide
It is known that activated caspase-8 may directly activate the executor caspases-3. It may also cleave the Bid (22 kDa), a pro-apoptotic Bcl-2 family member, and induces cytochrome-c release from mitochondria into cytoplasm, after its truncation by activated caspase-8. To find out the pathway involved in our experiment, we have carried out western blot using anti-Bid antibody. The truncated product, tBid (15 kDa) was not detected in K562 cells treated with benzamide for 20 h, as shown in figure 5a (lower panel). Thus, activated caspase-8 did not cleave the Bid in our experimental conditions.
We investigated the release of cytochrome c into cytoplasm, after 24 h treatment with benzamide. Under the experimental conditions, a small quantity of cytochrome-c was detected in the cytoplasmic fraction in untreated control cells, but there was dose dependent increase of cytochrome-c release in the cytoplasm by benzamide treatment in K562 cells, as shown in figure 5b (lower panel). The intensities of the bands as calculated using the software ImageMaster indicated that the release of cytochrome-c induced by 10 mM benzamide increased about 7 fold, compared with untreated control cells. This result showed that benzamide treatment also increased the cytochrome-c release to the cytoplasm.
Treatment with 5 mM and 10 mM benzamide decreased AIF in mitochondrial fraction, while increased in the cytoplasmic fraction, as well as nuclear fraction in a dose dependent manner in K562 cells. The result of this experiment is shown in figure 5c. Densitometric analysis of the bands revealed that about 32% and 56% of AIF (considering AIF in mitochondria in untreated cells as 100%) were released from mitochondria (labeled as AIF (mito)), after treatment with 5 mM and 10 mM benzamide, respectively. The concentration dependent increase of AIF in cytoplasmic as well as in nuclear fraction after treatment with benzamide were observed as shown in figure 5c. The blot was stripped and re-probed with anti-beta actin (for cytoplasmic fraction) and anti-histone 2B (for nuclear fraction) antibody, showing equal loading of protein in each lane. This result indicated that benzamide treatment induced translocation of AIF from mitochondria to nucleus. We obtained similar result with HeLa cells treated with benzamide (data not shown).
Induction of nuclear fragmentation and caspase-3 activation in siPARP-1 K562 cells and parental K562 cells after treatment with benzamide
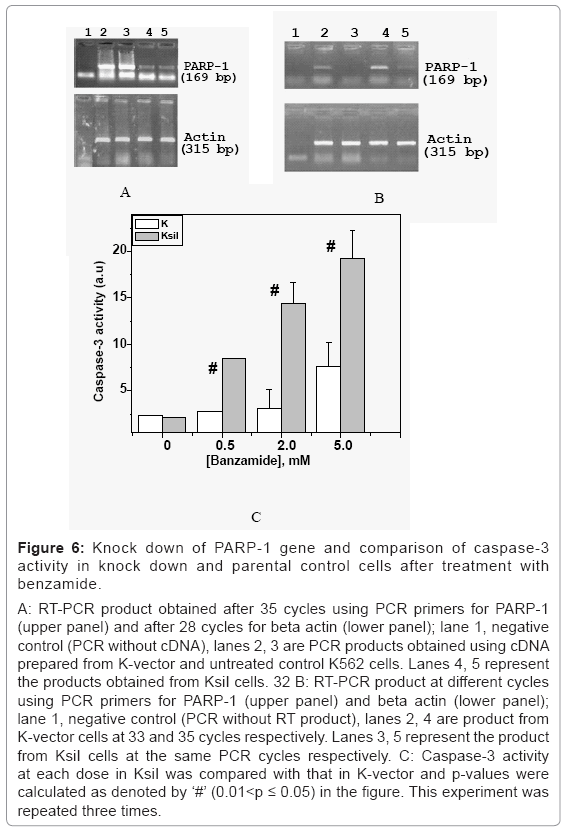
K562 cells were transfected with plasmid containing siRNA construct and selected for the hygromycin resistance, as described in the methods. Resistant clones were pooled and continued to grow in the presence of the hygromycin (100 μg/ml). The cells transfected with vector, with or without siRNA construct, was named as KsiI and K-veector, respectively. The PARP-1 expression in control parental K562 was same as that of K-vector, as shown in lane 2 and 3 in figure 6a (upper panel), but significant reduction of PARP-1 was observed in KsiI cells, as shown in lane 4 and 5 in upper panel of the same figure 6A, as detected by RT-PCR. There was no appreciable band for PARP-1 in KsiI cells after different cycles of PCR, as shown in lane 3 and 5, while cycle-dependent increase of fairly intense band of PARP-1 in K-vector cells was visible in lane 2 & 4 in the same respective cycles, as shown in figure 6b.
Figure 6: Knock down of PARP-1 gene and comparison of caspase-3 activity in knock down and parental control cells after treatment with benzamide.
A: RT-PCR product obtained after 35 cycles using PCR primers for PARP-1 (upper panel) and after 28 cycles for beta actin (lower panel); lane 1, negative control (PCR without cDNA), lanes 2, 3 are PCR products obtained using cDNA prepared from K-vector and untreated control K562 cells. Lanes 4, 5 represent the products obtained from KsiI cells. 32 B: RT-PCR product at different cycles using PCR primers for PARP-1 (upper panel) and beta actin (lower panel); lane 1, negative control (PCR without RT product), lanes 2, 4 are product from K-vector cells at 33 and 35 cycles respectively. Lanes 3, 5 represent the product from KsiI cells at the same PCR cycles respectively. C: Caspase-3 activity at each dose in KsiI was compared with that in K-vector and p-values were calculated as denoted by ‘#’ (0.01<p ≤ 0.05) in the figure. This experiment was repeated three times.
Induction of caspase-3 in KsiI cells at different doses of benzamide (0.5, 2, 5 mM) showed significantly higher than that in K-vector as shown in figure 6c. Here, ‘#’ in the figure represented p-values (less than 0.05), calculated with respect to K-vector cells at corresponding doses. For example, caspase-3 activity in KsiI at 0.5 mM (p=0.026) and 2 mM (p=0.000) was significantly higher, compared with K-vector cells treated with the same doses of benzamide, respectively. But, there is no significant caspase 3 activity in K-vector or parental K562 cells treated with 2 mM. Similarly, nuclear fragmentation observed in KsiI cells at each dose was significantly higher than that in K-vector as shown in table 3. The nuclear fragmentation in KsiI at 2.0 mM was significantly (as denoted by ‘**’(0.001<p ≤ 0.01)), higher than untreated control KsiI, but the increase of nuclear fragmentation in K-vector cells at the same dose 2.0 mM is not significant. Again, nuclear fragmentation in KsiI was found significantly higher than K-vector, at both 2.0 mM and 5.0 mM, as denoted by ‘#’ (0.01<p ≤ 0.05). We have earlier showed that PARP-1 knock down HeLa cells were significantly sensitive to benzamide treatment, as detected by nuclear fragmentation and caspase-3 activation [29]. Therefore, benzamide induced apoptosis was due to inhibition of PARP-1 and sensitivity of siPARP-1 cells to PARP inhibitor like benzamide, as detected by apoptosis is not cell-type specific event.
| Concentrations of BA (mM) | % nuclear fragmentation ± SD in K-vector cells | % nuclear fragmentation ± SD (p value) in KsiI cells |
|---|---|---|
| 0 | 6.8 ± 1.9 | 5.3 ± 2.9 |
| 0.5 | 7.1 ± 1.8 | 12.2 ± 3.9 |
| 2.0 | 14.3 ± 3.9 | 29.3 ± 5.8 # ** |
| 5.0 | 28.8 ± 7.3 ** | 55.2 ± 6. 9 # *** |
p-values at each dose in KsiI and K-vector was calculated with respect to untreated control of respective cell type (‘**’ denotes 0.001<p ≤ 0.01, ‘***” denotes p ≤ 0.001). Significant difference of nuclear fragmentation at each dose between two cell type were also calculated and denoted as ‘#’ (0.01<p ≤ 0.05). p-values greater than 0.05 left blank
Table 3: Induction of apoptosis by different concentrations of BA in K- vector and KsiI cells treated for 24 h.
Inhibition of poly (ADP-ribosy)lation of proteins in the physiological conditions by the benzamide treatment in K562 cells
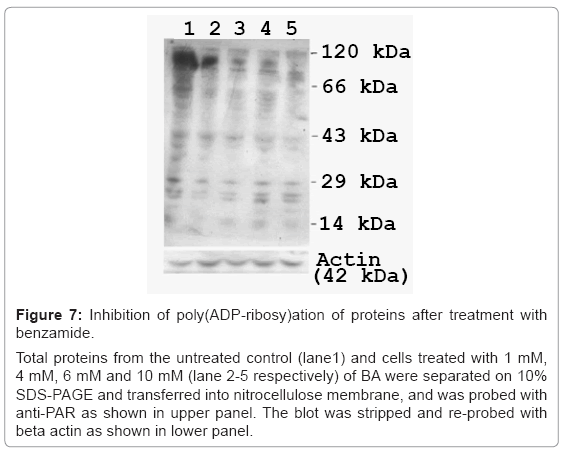
It is well known that PARP poly (ADP-ribosy)lates its target proteins, using cellular NAD+ as the substrate. Using the antibody specific for PAR, we observed that a number of proteins were poly (ADP-ribosyl)ated in the physiological condition of growth, and various doses of benzamide treatment (4 h) inhibited such poly (ADP-ribosyl)ation, in a dose dependent manner, as shown in figure 7 (upper panel). The same blot was stripped and re-probed with anti-beta actin (lower panel) showing equal loading in each lane.
Figure 7: Inhibition of poly(ADP-ribosy)ation of proteins after treatment with benzamide.
Total proteins from the untreated control (lane1) and cells treated with 1 mM, 4 mM, 6 mM and 10 mM (lane 2-5 respectively) of BA were separated on 10% SDS-PAGE and transferred into nitrocellulose membrane, and was probed with anti-PAR as shown in upper panel. The blot was stripped and re-probed with beta actin as shown in lower panel.
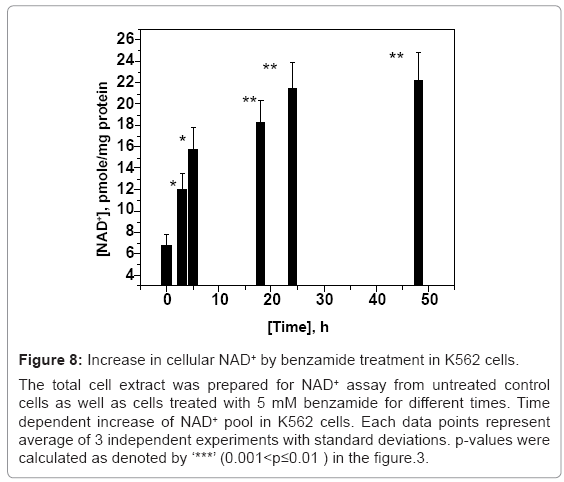
Cellular NAD+ level was measured using the in vitro method, originally described by Putt and Hergenrother [31], and adapted for detection of NAD+ level in total cell extract. We have earlier showed increase of NAD+ level and decrease of poly(ADP-ribosyl)ation in K562 cells treated with benzamide [32]. It is known that N’-methyl-N’-nitro-N-nitrosoguanidine (MNNG) treatment reduce the NAD+ pool drastically [34,35].
We observed similar decrease in the cellular NAD+ pool, and such decrease was inhibited by pre-treatment with benzamide, before MNNG treatment. This observation validated that the similar method can be used for cellular NAD+ level detection, and hence, PARP activity. We have determined NAD+ pool in K562 cells treated with 5 mM benzamide for different time of interval. We observed that benzamide treatment increased NAD+ level in a time-dependent manner. The lowest time point where there was significant (p=0.02) increase in the NAD+ level with 5 mM benzamide treatment was 5 h, as shown in figure 8. The estimated value of the intracellular NAD+ concentration in untreated K562 cells (6.7 pmole/mg protein) is similar to what has been published earlier [36]. This result (Figures 7 and 8) showed that benzamide treatment increase the cellular NAD+ level, and thus, inhibited PARP in a dose and time dependent manner.
Figure 8: Increase in cellular NAD+ by benzamide treatment in K562 cells.
The total cell extract was prepared for NAD+ assay from untreated control cells as well as cells treated with 5 mM benzamide for different times. Time dependent increase of NAD+ pool in K562 cells. Each data points represent average of 3 independent experiments with standard deviations. p-values were calculated as denoted by ‘***’ (0.001<p≤0.01 ) in the figure.3.
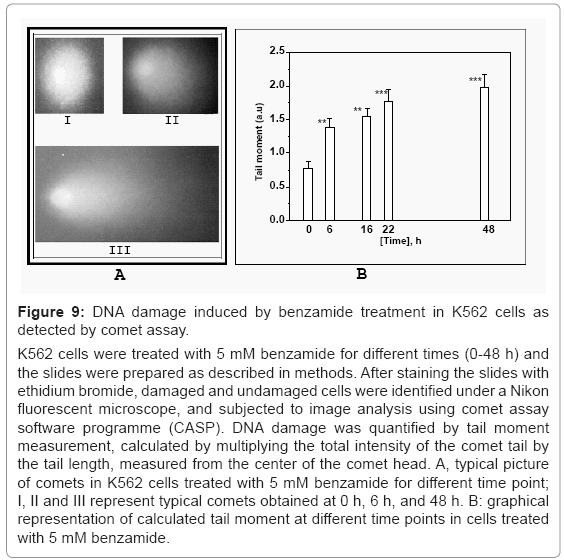
Benzamide induced DNA damage in K562 cells as detected by comet assay
To test whether the apoptosis inducing ability of benzamide was through its ability to induce DNA damage, we determined the DNA damage in K562 cells treated with 5 mM benzamide for various times (0-48 h), by comet assay. Typical picture of comet at different time points 0 h, 6 h and 48 h were shown in I, II and III, respectively in figure 9a. The tail moment was increased in a time dependent manner (Figure 9b). This tail moment represents the index of DNA breaks. Benzamide was capable of forming appreciable DNA breaks, leading the cell to start to polarize into head and tail of a comet when electric field was applied. Significant tail moment was observed at and above 6 h treatment of benzamide.
Figure 9: DNA damage induced by benzamide treatment in K562 cells as detected by comet assay.
K562 cells were treated with 5 mM benzamide for different times (0-48 h) and the slides were prepared as described in methods. After staining the slides with ethidium bromide, damaged and undamaged cells were identified under a Nikon fluorescent microscope, and subjected to image analysis using comet assay software programme (CASP). DNA damage was quantified by tail moment measurement, calculated by multiplying the total intensity of the comet tail by the tail length, measured from the center of the comet head. A, typical picture of comets in K562 cells treated with 5 mM benzamide for different time point; I, II and III represent typical comets obtained at 0 h, 6 h, and 48 h. B: graphical representation of calculated tail moment at different time points in cells treated with 5 mM benzamide.
Discussion
In the present communication, we showed that benzamide treatment alone increased nuclear fragmentation, nucleosomal ladders, caspase-2, caspase-8, caspase-9/6 and caspase-3 activity in K562 cells in a dose and time dependent manner. Further, such treatment increased the cleavage of pro-caspase-8, release of cytochrome-c in cytoplasm, and translocation of AIF from the mitochondria to nucleus. There was no cleavage of Bid. This result showed that benzamide triggered the “intrinsic” (mitochondria mediated), as well as “extrinsic” (caspase-8 mediated) pathway of apoptosis in K562 cells. Time dependent increase of the comet tail, and hence, tail moment by 5 mM benzamide indicating that benzamide directly or indirectly induced DNA damage. Out of these whole apoptotic parameters, the significant earliest event was inhibition of poly (ADP-ribosyl)ation of cellular protein, as detected by NAD+ assay (5 h) and western blot using anti-PAR antibody (4 h). Furthermore, benzamide treatment increased nuclear fragmentation and caspase-3 activation significantly higher in KsiI cells, in comparison with the parental K-vector cells.
We reported earlier that benzamide treatment increased apoptosis in Chinese Hamster V79 cell [25]. We also observed that nuclear fragmentation and caspase-3 activation in PARP-1 knock down HeLa cells was significantly higher than parental HeLa cells [30]. Thus, the observed induction of apoptosis by benzamide was not cell-type specific. Here, comparisons of all the events in time frame showed inhibition of poly(ADP-ribosyl)ation (4 h), significant increase in NAD+ level (5 h), tail moment in comet assay (6 h), activation of caspase-2 (6 h), caspage-8 (9 h), caspase-3 (24 h), and induction of nuclear fragmentation (Table 2, 24 h) indicated that benzamide treatment decreased poly (ADP-ribosyl)ation of unidentified proteins that led to DNA damage, activation of caspases, release of cytochrome c and translocation of AIF to nucleus, and nuclear fragmentation in our experimental conditions. So, the whole apoptotic cascades were originated from inhibition of PARP in cells.
The classical theory of ‘intrinsic apoptosis’ tells that intracellular stress induces to activate pro-apoptotic genes of Bcl-2 family proteins, making the mitochondrial membrane porus, which leads to release of cytochrome c from mitochondria. This cytochrome c can bind to Apaf-1 to form apoptosome, which activates caspase-9 by the cleavage of pro-caspasse-9, and subsequently, caspase-3 gets activated, followed by activation of DNAses. Here also, we found release of cytochrome c, activation of caspase-9, caspase-3, but how inhibition of ADP-ribosylation triggers this cascade was not known. We do not know the protein(s) whose inhibition of poly (ADP-ribosyl)ation triggers apoptosis. But, there are some DNases such as Ca2+/Mg2+-dependent endonuclease (e.g DNAS1L3), which can be activated upon inhibition of poly(ADP-ribosyl)ation [18-21]. Possibly, such kind of DNases gets activated after treatment with benzamide that resulted DNA breaks, as observed in our cases as detected by comet assay (6 h). This DNA breaks might be leading to activation of caspase-2 and cytochrome c release, because it has been shown earlier that cytotoxic stress causes activation of caspase-2, followed by mitochondrial membrane permeabilization, to release cytochrome-c [37].
The ‘extrinsic pathway’ of apoptosis means extracellular signal comes into cells through death receptor proteins that lead to activate caspase-8, which eventually activates downstream caspases like caspase-3. Here also, we observed activation of caspase-8 after treatment with benzamide. How pro-caspase-8 was turned into active caspase-8 by benzamide treatment is unclear. Whether, caspase-8 was poly (ADP-ribosy)lated in the physiological condition of growth and remains inactivated, is to be tested. It is known that PARP-1 is necessary for the release of AIF from the mitochondria to the cytoplasm, and subsequently to the nucleus, and its activation to cleave the chromosome into nuclear fragmentation [38]. However, in our experimental conditions, where basal level PARP was inhibited by benzamide; translocation of AIF into nucleus was facilitated. It has been observed that release of AIF from mitochondria is interconnected with the activation of caspase-3 in FAS/APO-1/CDC and ceramide induced apoptosis. On the basis of this observation, it has been proposed that caspase-8 may in some experimental conditions affect the release of AIF [39,40]. Bid was not cleaved in our experimental condition. In this scenario, how cytochrome-c release was facilitated is unclear. As discussed above, activation of caspase-2 might lead to the release of cytochrome-c. It has been known that the release of AIF may influence the release of cytochrome-c, and thus, upstream to the cytochrome-c release [40]. Alternatively, as discussed below, benzamide treatment typically induce the apoptosis mediated through DNA damage response [41].
The concentrations of benzamide that inhibit PARP activity in vitro by 50% (IC50) varies from 1.3 μM to 22 μM [42,43], but we have used about thousand fold higher than that (2.5 mM to 10 mM). Intracellular concentration of benzamide in our experimental condition is not known. However, it has been shown in human fibroblast cells that an extra cellular benzamide concentration of 1 mM gave intra nuclear concentrations of 4-8 μM [44]. Thus, even though we have used mM concentrations of benzamide in growth medium, the intracellular concentrations of benzamide might be in the μM ranges.
It is not possible to indicate the specific member of the PARP family that had been inhibited by benzamide inducing the effects. Given the known role of PARP-1 and PARP-2 in the recovery of damaged DNA and alteration of large number of genes in PARP-1(-/-) cells [45,46], it is likely that the observed effect could be due to PARP-1inhibition by benzamide, since in our experiment with K562 cells, where the expression of PARP-1 was specifically reduced by siRNA, the increase in the nuclear fragmentation as well as caspase-3 activity induced by benzamide treatment were higher in comparison to the parental K562 cells. Actually the role of PARP in apoptosis is very complex, and it can act like a double-edged sword. Hyper-activation of PARP-1, generally due to huge DNA breaks, severely depletes ATP pool, resulting necrotic death of cells. Moderate activation of PARP-1 generally helps DNA repair in cells. That is why PARP inhibitors could be used as chemo-sensitizer or radio-sensitizer. The PARP inhibitor like iniparib, oliparib are capable of inducing apoptosis, and now-a-days such PARP inhibitors are used in clinical trial for treatment with cancer having BRCA1/2 mutants [47,48]. Hence, PARP inhibitors are lethal to the BRCA mutant breast cancer as single agent or as chemo-sensitizer [49].
Conclusion
Benzamide treatment inhibited poly(ADP-ribosyl)ation of unknown proteins that triggered DNA breaks in the cells, and subsequently, induced caspase-2 and cytochrome c release in cytoplasm. Either caspase-2 activation or DNA induced cytochrome c release which facilitated AIF translocation into nucleus and caspase-independent apoptosis. Release of cytochrome c induced apoptosis by classical way through activation of caspase-9/6, followed by caspase-3 and nuclear fragmentation. Caspase-8 activation also induced caspase-3 and subsequently, down-stream cascades in our experimental condition, although mechanism of activation of caspase-8 is not clear. The proposed pathway of apoptosis induced by benzamide was shown schematically in figure 10.
Acknowledgements
Partial financial support (Nitai P Bhattacharyya) from Department of Science and Technology (DST), New Delhi, Govt. of India is duly acknowledged.
References
- Amé JC, Spenlehauer C, de Murcia G (2004) The PARP superfamily. Bioessays 26: 882-893.
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG (1999) Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342 : 249-268.
- Virág L, Szabó C (2002) The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev 54: 375-429.
- Tentori L, Portarena I, Graziani G (2002) Potential clinical applications of poly(ADP-ribose) polymerase (PARP) inhibitors. Pharmacol Res 45: 73-85.
- Chalmers AJ (2004) Poly(ADP-ribose) polymerase-1 and ionizing radiation: sensor, signaller and therapeutic target. Clin Oncol (R Coll Radiol) 16: 29-39.
- Cleaver JE, Morgan WF (1991) Poly(ADP-ribose)polymerase: a perplexing participant in cellular responses to DNA breakage. Mutat Res 257: 1-18.
- Kedar PS, Stefanick DF, Horton JK, Wilson SH (2012) Increased PARP-1 association with DNA in alkylation damaged, PARP-inhibited mouse fibroblasts. Mol Cancer Res 10: 360-368.
- Zhang JX, Li DQ, He AR, Motwani M, Vasiliou V, et al. (2012) Synergistic inhibition of hepatocellular carcinoma growth by cotargeting chromatin modifying enzymes and poly (ADP-ribose) polymerases. Hepatology 55: 1840-1851.
- Maxwell KN, Domchek SM (2012) Cancer treatment according to BRCA1 and BRCA2 mutations. Nat Rev Clin Oncol 9: 520-528.
- Kuo ML, Shen SC, Yang CH, Chuang SE, Cheng AL, et al. (1998) Bcl-2 prevents topoisomerase II inhibitor GL331-induced apoptosis is mediated by down-regulation of poly(ADP-ribose)polymerase activity. Oncogene 17: 2225-2234.
- Pacini A, Quattrone A, Denegri M, Fiorillo C, Nediani C, et al. (1999) Transcriptional down-regulation of poly(ADP-ribose) polymerase gene expression by E1A binding to pRb proteins protects murine keratinocytes from radiation-induced apoptosis. J Biol Chem 274: 35107-35112.
- Richardson DS, Allen PD, Kelsey SM, Newland AC (1999) Effects of PARP inhibition on drug and Fas-induced apoptosis in leukaemic cells. Adv Exp Med Biol 457: 267-279.
- Walisser JA, Thies RL (1999) Poly(ADP-ribose) polymerase inhibition in oxidant-stressed endothelial cells prevents oncosis and permits caspase activation and apoptosis. Exp Cell Res 251: 401-413.
- Tentori L, Orlando L, Lacal PM, Benincasa E, Faraoni I, et al. (1997) Inhibition of O6-alkylguanine DNA-alkyltransferase or poly(ADP-ribose) polymerase increases susceptibility of leukemic cells to apoptosis induced by temozolomide. Mol Pharmacol 52: 249-258.
- Czapski GA, Cakala M, Kopczuk D, Strosznajder JB (2004) Effect of poly(ADP-ribose) polymerase inhibitors on oxidative stress evoked hydroxyl radical level and macromolecules oxidation in cell free system of rat brain cortex. Neurosci Lett 356: 45-48.
- Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, et al. (1995) Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature 376: 37-43.
- Kim JW, Won J, Sohn S, Joe CO (2000) DNA-binding activity of the N-terminal cleavage product of poly(ADP-ribose) polymerase is required for UV mediated apoptosis. J Cell Sci 113 : 955-961.
- Yakovlev AG, Wang G, Stoica BA, Boulares HA, Spoonde AY, et al. (2000) A role of the Ca2+/Mg2+-dependent endonuclease in apoptosis and its inhibition by Poly(ADP-ribose) polymerase. J Biol Chem 275: 21302-21308.
- Yakovlev AG, Wang G, Stoica BA, Simbulan-Rosenthal CM, Yoshihara K, et al. (1999) Role of DNAS1L3 in Ca2+- and Mg2+-dependent cleavage of DNA into oligonucleosomal and high molecular mass fragments. Nucleic Acids Res 27: 1999-2005.
- Boulares AH, Zoltoski AJ, Contreras FJ, Yakovlev AG, Yoshihara K, et al. (2002) Regulation of DNAS1L3 endonuclease activity by poly(ADP-ribosyl)ation during etoposide-induced apoptosis. Role of poly(ADP-ribose) polymerase-1 cleavage in endonuclease activation. J Biol Chem 277: 372-378.
- Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR (2000) Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem 275: 40974-40980.
- Benchoua A, Couriaud C, Guégan C, Tartier L, Couvert P, et al. (2002) Active caspase-8 translocates into the nucleus of apoptotic cells to inactivate poly(ADP-ribose) polymerase-2. J Biol Chem 277: 34217-34222.
- Ménissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, et al. (2003) Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. Embo J 22: 2255-2263.
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, et al. (1997) Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A 94: 7303-7307.
- Ghosh U, Pandit B, Dutta J, Bhattacharyya NP (2004) Induction of apoptosis by benzamide and its inhibition by aurin tricarboxylic acid (ATA) in Chinese hamster V79 cells. Mutat Res 554: 121-129.
- Gaymes TJ, Shall S, MacPherson LJ, Twine NA, Lea NC, et al. (2009) Inhibitors of poly ADP-ribose polymerase (PARP) induce apoptosis of myeloid leukemic cells: potential for therapy of myeloid leukemia and myelodysplastic syndromes. Haematologica 94: 638-646.
- Ghosh U, Das N, Bhattacharyya NP (2007) Inhibition of telomerase activity by reduction of poly(ADP-ribosyl)ation of TERT and TEP1/TP1 expression in HeLa cells with knocked down poly(ADP-ribose) polymerase-1 (PARP-1) gene. Mutat Res 615: 66-74.
- Basu U, Banerjee S (2004) An engineered EBV vector expressing human factor VIII and von Willebrand factor in cultured B-cells. J Gene Med 6: 760-768.
- Martin SJ, Newmeyer DD, Mathias S, Farschon DM, Wang HG, et al. (1995) Cell-free reconstitution of Fas-, UV radiation- and ceramide-induced apoptosis. Embo J 14: 5191-5200.
- Ghosh U, Bhattacharyya NP (2009) Induction of apoptosis by the inhibitors of poly(ADP-ribose)polymerase in HeLa cells. Mol Cell Biochem 320: 15-23.
- Putt KS, Hergenrother PJ (2004) An enzymatic assay for poly(ADP-ribose) polymerase-1 (PARP-1) via the chemical quantitation of NAD(+): application to the high-throughput screening of small molecules as potential inhibitors. Anal Biochem 326: 78-86.
- Ghosh U, Bhattacharyya NP (2005) Benzamide and 4-amino 1,8 naphthalimide treatment inhibit telomerase activity by down-regulating the expression of telomerase associated protein and inhibiting the poly(ADP-ribosyl)ation of telomerase reverse transcriptase in cultured cells. Febs J 272: 4237-4248.
- Singh NP, McCoy MT, Tice RR, Schneider EL (1988) A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res 175: 184-191.
- Ying W, Garnier P, Swanson RA (2003) NAD+ repletion prevents PARP-1-induced glycolytic blockade and cell death in cultured mouse astrocytes. Biochem Biophys Res Commun 308: 809-813.
- Radons J, Heller B, Bürkle A, Hartmann B, Rodriguez ML, et al. (1994) Nitric oxide toxicity in islet cells involves poly(ADP-ribose) polymerase activation and concomitant NAD+ depletion. Biochem Biophys Res Commun 199: 1270-1277.
- Hageman GJ, Stierum RH (2001) Niacin, poly(ADP-ribose) polymerase-1 and genomic stability. Mutat Res 475: 45-56.
- Lassus P, Opitz-Araya X, Lazebnik Y (2002) Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science 297: 1352-1354.
- Chen M, Zsengellér Z, Xiao CY, Szabó C (2004) Mitochondrial-to-nuclear translocation of apoptosis-inducing factor in cardiac myocytes during oxidant stress: potential role of poly(ADP-ribose) polymerase-1. Cardiovasc Res 63: 682-688.
- Susin SA, Zamzami N, Castedo M, Daugas E, Wang HG, et al. (1997) The central executioner of apoptosis: multiple connections between protease activation and mitochondria in Fas/APO-1/CD95- and ceramide-induced apoptosis. J Exp Med 186: 25-37.
- Cregan SP, Fortin A, MacLaurin JG, Callaghan SM, Cecconi F, et al. (2002) Apoptosis-inducing factor is involved in the regulation of caspase-independent neuronal cell death. J Cell Biol 158: 507-517.
- Norbury CJ, Zhivotovsky B (2004) DNA damage-induced apoptosis. Oncogene 23: 2797-2808.
- Cheung A, Zhang J (2000) A scintillation proximity assay for poly(ADP-ribose) polymerase. Anal Biochem 282: 24-28.
- Banasik M, Komura H, Shimoyama M, Ueda K (1992) Specific inhibitors of poly(ADP-ribose) synthetase and mono(ADP-ribosyl)transferase. J Biol Chem 267: 1569-1575.
- Kun E, Kirsten E, Milo GE, Kurian P, Kumari HL (1983) Cell cycle-dependent intervention by benzamide of carcinogen-induced neoplastic transformation and in vitro poly(ADP-ribosyl)ation of nuclear proteins in human fibroblasts. Proc Natl Acad Sci U S A 80: 7219-7223.
- Cardoso RS, Espanhol AR, Passos GA, Sakamoto-Hojo ET (2002) Differential gene expression in gamma-irradiated BALB/3T3 fibroblasts under the influence of 3-aminobenzamide, an inhibitior of parp enzyme. Mutat Res 508: 33-40.
- Simbulan-Rosenthal CM, Ly DH, Rosenthal DS, Konopka G, Luo R, et al. (2000) Misregulation of gene expression in primary fibroblasts lacking poly(ADP-ribose) polymerase. Proc Natl Acad Sci U S A 97: 11274-11279.
- von Minckwitz G, Martin M (2012) Neoadjuvant treatments for triple-negative breast cancer (TNBC). Ann Oncol 23: vi35-vi39.
- Patel AG, De Lorenzo SB, Flatten KS, Poirier GG, Kaufmann SH (2012) Failure of iniparib to inhibit poly(ADP-Ribose) polymerase in vitro. Clin Cancer Res 18: 1655-1662.
- Plummer R (2011) Poly(ADP-ribose) polymerase inhibition: a new direction for BRCA and triple-negative breast cancer? Breast Cancer Res 13: 218.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 14607
- [From(publication date):
specialissue-2013 - Dec 20, 2025] - Breakdown by view type
- HTML page views : 9929
- PDF downloads : 4678