Research Article Open Access
Isolation of Nucleotide Binding Site (NBS)-Leucine Rich Repeat (LRR) Resistant Gene Analogs (Rgas) In Arabica Coffee (Coffea Arabica L. Cv S.288)
Deepak Kumar* and H.L. SreenathPlant Biotechnology Division, Coffee Board, Unit of Central Coffee Research Institute (CCRI), Dr. S. Radhakrishnan Road, Manasagangothri, Mysore-570 006, Karnataka, India
- Corresponding Author:
- Deepak Kumar
Plant Biotechnology Division, Coffee Board
Unit of Central Coffee Research Institute (CCRI)
Dr. S. Radhakrishnan Road, Manasagangothri
Mysore-570 006, Karnataka, India
Tel: +91-8971703863
E-mail: deepakkumardeo@gmail.com
Received date: June 18, 2012; Accepted date: August 08, 2012; Published date: August 11, 2012
Citation: Deepak Kumar, Sreenath HL (2012) Isolation of Nucleotide Binding Site (NBS)-Leucine Rich Repeat (LRR) Resistant Gene Analogs (Rgas) In Arabica Coffee (Coffea Arabica L. Cv S.288). J Biotechnol Biomater 2:146. doi:10.4172/2155-952X.1000146
Copyright: © 2012 Deepak Kumar , et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
Cloning of resistance gene analogues against diverse pathogens from variety of plants in last decade has revealed that many of them share high level of conserved sequence motifs. The conserved backbone of amino acid motifs present in Nucleotide Binding Site (NBS) domain makes it possible to isolate resistance gene analogues by Polymerase chain reaction (PCR) with degenerate primers. Oligo-nucleotide primers combinations that target conserve motif of NBS domain as mentioned in earlier studies were used to amplify resistance gene analogues from Coffea arabica (S.288). PCR product amplified from genomic DNA as well as cDNA were cloned and sequenced. In the present study amplified resistance gene analogues from C.arabica genomic DNA and cDNA using Ploop-cof and GLPL-cof primers were cloned, sequenced using T7 / SP6 primers, analyzed at NCBI/SGN Nucleotide Data Bank. Analysis of these RGA leads to the understanding that difference in expression profile might be due to the difference present at sequence level of Resistant Gene Analogs (RGA) isolated from DNA and cDNA. Analysis also revealed presence of high level similarity at their sequences. Seven RGA isolated from genomic DNA of S.288 using non-degenerate primers, eleven more RGA were isolated from S.288 genomic DNA with degenerate oligo-nucleotide primers and thirty two RGA isolated from cDNA of S.288. Fifteen RGA clones isolated from cDNA prepared from rust race I infected leaf sample for 24 hours. BLASTN result showed these C.arabica RGA has a high level of similarity with C.canephora RGA. This confirm the integrity maintained among RGA even it was isolated from different coffee variety which has difference at there genome (C.arabica 2n= 44 and C.canephora 2n= 22). RGA isolated which has below 475 bp or above 530 bp in size along with both primer sequences in their end has either no match with any RGA or they match with microsatellite. There is one independent sequence from genomic DNA and four independent sequences from cDNA which has not given any BLASTN result, these sequences has primer sequence with them that indicates these may be belong to new type of RGA. The RGA reported in current study are mainly from the class A type of RGA.
Keywords
Nucleotide Binding Site (NBS); Leucine Rich Repeat (LRR); Resistant Gene Analogs (Rgas); Coffea arabica.
Introduction
Coffee is one of the important beverage crops for Global point of view. Coffee is a stimulating non-alcoholic beverage, which earns a substantial amount of foreign exchange to India. Coffee belongs to the genus Coffea, family Rubiaceae. The bean of the crop is used to prepare drinking coffee. Commercially arabica and robusta are the two important coffee species where arabica produces high quality beverage and originated in southwestern Ethiopia (Kaffa region -centre of diversity). Coffee is the second largest traded commodity in the world, next only to petroleum. About 3,40,306 ha of area is covered with coffee in India producing 4372000 bags and exported 3145000 bags in the year of 2008/09 earnings of US $395.04 million [1]. The disease affecting coffee are many, among them major disease of economical importance are leaf rust caused by Hemieleia vastatrix. Hemieleia vastatrix is a fungus belongs to the Phylum Basidiomycota, order Uredinales. Parasitic in nature to plant causes serious damage to the crop.
Disease resistance genes from various plant species have been cloned and sequenced. Some of them are distinguished by the presence of N-terminal nucleotide binding site (NBS) and C-terminal stretch of leucine-rich repeats (LRR). Although these gene products are structurally related, but DNA sequences are poorly conserved in the case of Arabidopsis [2]. Nucleotide binding sites (NBS) domains related to R-genes show a highly conserved backbone of amino-acid motifs, which makes it possible to isolates resistance gene analogues (RGAs) by PCR with low degeneracy primers. The amino-acid sequence of coffee RGAs were identified, that showed strong sequence similarity to almost all known non-TIR (Toll/Interleukin1 receptor)-type R-genes. The high degree of similarity between particular coffee RGAs and R-genes isolated from other angiosperm species, such as Arabidopsis, tomato and rice indicates an ancestral relationship and existence of common ancestors [3].
Part of nucleotide binding site has enough DNA identity to design primer for the polymerase chain reaction to amplify some of these regions which is related to the DNA sequences. Such primers are used to get certain resistance gene like (RGL) DNA fragments from Arabidopsis thaliana.
Most of these RGL DNA fragments were found in a clustered or dispersed multi-copy sequence organization. R-genes that have been molecularly characterized so far can be grouped into classes based on similarities in the function or amino acid sequence of the proteins they encode. Examples are: RPS2 [4,5], RPM1 [6], RPP5 [7], L6 gene from flax.
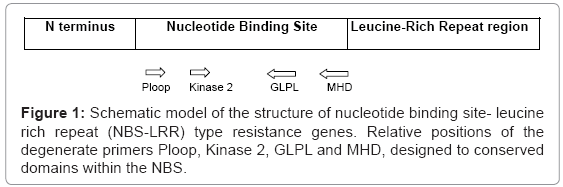
These R-genes are found in the diverse plant species and the conserved NBS-LRR features suggest a common function in the defence response against pathogen attack, probably as part of the signal transduction pathway. Knowledge about the genes involved in disease resistance in plants speed up the works carried out against plant diseases. The gene products of some of these can be distinguished by the presence of an N- terminal nucleotide-binding site and a C-terminal stretch of leucine rich repeats. The NBS sequences of R genes are characterized by the presence of up to seven conserved domains including the Ploop, Kinase2 and GLPL motifs [8] (Figure 1). The presence of these conserved domains has facilitated the cloning of resistance gene analogues from diverse species by PCR using degenerate oligo nucleotide primers. Thus the identification of RGAs represents a potentially powerful strategy for the generation of markers for map based cloning of resistance genes.
Nine distinct classes of RGAs of the NBS-like type, representing a highly diverse sample, were isolated from Coffea arabica and Coffea canephora species. Coffee RGA family suggests point mutation as the primary source of diversity. Coffee RGA family appeared to be closed related in sequence to cloned R-gene [3]. Coffee RGAs amino acid sequence showed strong sequence similarity to almost all known non-TIR (Toll/Interleukin I Receptor) type R-genes. High degree of similarity between coffee RGAs and R-genes isolated from other angiosperm species, such as arabidopsis, tomato and rice indicates an ancestral relationship. The data obtained from coffee species suggests that the evolution of NBS-encoding sequence involves the gradual accumulation of mutations and slow rates of divergence within distinct R-gene families.
The majority of the studies that utilize the PCR-based method use genomic DNA as the template. Although a large number of RGAs are amplified in such studies, many of the identified RGAs are probably pseudo genes [9] or non-functional genes. RGAs amplified from cDNA are, on average, more likely to be functional R genes than those amplified from genomic DNA, since the transcription of non-functional genes would be associated with an overall fitness cost to the plant. However, only very few reports of PCR-based amplification of RGAs from cDNA template were found [10,11].
We have taken up a study for isolation of RGAs in Coffea arabica Cv. S.288 with the objective of getting molecular markers linked to Coffee Leaf Rust resistance.
Materials and Methods
Plant materials
C. arabica S.288 plants were maintained in the nursery of Plant Biotechnology Centre, Mysore.
Bacterial strains
Cloning of the RGA fragments obtained from genomic DNA was carried out in E. coli strain DH 5α whereas the strain JM109 was used to clone RGA fragments amplified from cDNA.
Primers used for the study
We used 16 pairs of degenerate primers combinations and two pair of non degenerate primer on genomic DNA and cDNA sample (Table 1).
| Primer name | Conserved amino acid motif | Forward Reverse | Primer sequence |
|---|---|---|---|
| Non-degenerate primers designed on the basis of the first coffee RGAs isolated (Noir et al. 2001). | |||
| Ploop-Cof | GVGKTT | F | 5’-GGGGGTGGGGAAGACGACTC-3’ |
| GLPL-Cof | DGLPLAL | R | 5’-AGGGCGAGGGGGAGGCCATC-3’ |
| Degenerate primers first set designed on the basis (Noir et al. 2001). | |||
| Ploop1 | GGV/I/MGKTT | F | 5’-GGIGGIGTIGGIAARACNAC-3’ |
| Ploop2 | GGV/I/MGKTT | F | 5’-GGNGGNRTNGGNAAAACAAC-3’ |
| Ploop4 | GGV/I/MGKTT | F | 5’-GGNGGNRTNGGNAARACTAC-3’ |
| Ploop5 | GGV/I/MGKTT | F | 5’-GGNGGNRTNGGNAARACCAC-3’ |
| GLPL1 | GL/FPL/FAL/V | R | 5’-IARIGCIARIGGIARNCC-3’ |
| GLPL2 | GL/FPL/FAL/V | R | 5’-CAHHGCNAAHGGHAAHCC-3’ |
| GLPL3 | GL/FPL/FAL/V | R | 5’-CAANGCCAANGGCAANCC-3’ |
| GLPL4 | GL/FPL/FAL/V | R | 5’-CAGNGCNAGNGGNAGNCC-3’ |
| Degenerate primers second set on the basis (Chen et al. 2007) | |||
| F1 | Ploop (GGV/I/MGKTT) | F | 5’-GGHDYVGGKAARACWAC-3’ |
| F2 | Ploop (GMGGV/I/SGKTT) | F | 5’-GGDATGVSVGGHDYVGGKAARAC-3’ |
| F3 | Ploop (GGV/I/MGKTT) | F | 5’-GGNGGNRTHGGNAARACHAC-3’ |
| F4 | Ploop (GGV/I/MGKTT) | F | 5’-GGNGTNGGNAARACNAC-3’ |
| F5 | Ploop (GGV/I/MGKTT) | F | 5’-GGNGGNGTNGGNAANACNAC-3’ |
| F6 | Ploop (GGV/I/MGKTT) | F | 5’-GGNGGNRTNGGNAARACNAC-3’ |
| HDR1 | Hydrophobic domain (GL/FPL/FA/VL) | R | 5’-AKWGCYARRGGDARYCC-3’ |
| HDR2 | Hydrophobic domain (GL/FPL/FA/VL) | R | 5’-GCMRCCARAGGMARYCC-3’ |
| HDR3 | Hydrophobic domain (GL/FPL/FA/VL) | R | 5’-AGNGCHAGNGGYAANCC-3’ |
| HDR4 | Hydrophobic domain (GL/FPL/FA/VL) | R | 5’-AANGCHAGNGGYAANCC-3’ |
| HDR5 | Hydrophobic domain (GL/FPL/FA/VL) | R | 5’-AGIGCHAGNGGNAGNCC-3’ |
| HDR6 | Hydrophobic domain (GL/FPL/FA/VL) | R | 5’-ARNWYYTTVARDGCVARWGGVARWCC-3’ |
I= deoxy-inosine, R=A/G, H=A/C/T, N= A/T/G/C
Table 1: Primers details.
Genomic DNA Isolation
Leaf sample of C. arabica S.288 was washed thoroughly with tap water and, rinsed with sterile distilled water. The leaf was wiped dry with tissue paper and ground to fine powder with a mortar and pestle using liquid nitrogen. Around 100 mg of powdered leaf was used to isolate genomic DNA using CTAB extraction buffer. RNA contamination was removed by RNAase treatment.
For RGA isolation from Ploop 4 and GLPL 4, genomic DNA was isolated by DNeasy Plant Mini Kit (Qiagen). DNA isolation protocol was followed as per the manufacturer instruction. RNAase A (Pancreatic RNAase A Bangalore Genei), treatment was also performed according to the instruction provided.
DNA Quantification
DNA was quantified using spectrophotometer (Beckman DU 640 B).
RNA Isolation and Purification of mRNA
For RNA isolation young green leaves of S.288 plant, was detached from the plant immediately frozen in liquid nitrogen and ground to fine powder using pre-chilled mortar and pestle (DEPC treated and baked). RNA extraction was performed using Qiagen RNeasy kit according to manufacturer’s protocol with modification required for Coffee leaf tissue.
RNA Quantification
RNA was quantified by spectrophotometer (Beckman DU 640B). Poly-A mRNA was isolated using mRNA purification kit (Bangalore Genei) according to users manual.
First Strand cDNA Synthesis
Complementary DNA was prepared by using Bangalore Genei cDNA synthesis kit as per the manufacturer’s protocol. The reaction was stopped by incubation at 92oC for 2 minutes. The cDNA was snapped chilled on ice and used as template for PCR reaction.
PCR Amplification of RGAs
Oligonucleotide primers which were used for the present study designed based on the Ploop and GLPL motifs of existing RGA’s. The non degenerate primers and degenerate primers were used for isolation of RGA fragments from genomic DNA and cDNA of C. arabica S.288. PCR amplification was performed in a 50 μl reaction volume on a PTC 200 thermal cycler (MJ research). Either 100 ng of first strand leaf cDNA or 60 ng of genomic DNA was used as template for the PCR amplification in a reaction mixture composing Taq Polymerase assay buffer 1X final concentration (Bangalore Genei), 1 μM of each primer, 400 μM dNTP (10 mM), 1.5 mM MgCl2 and 3 Units Taq DNA polymerase (Bangalore Genei). The PCR program parameters include initial denaturation at 94°C for 5 minutes followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 58°C for 45 seconds, and elongation at 72°C for 1 minute 30 seconds and finally extension at 72°C for 10 minutes. The PCR amplified products were size fractionated on 1% of agarose gel in 1X TBE buffer under constant voltage of 80V for 45 minutes.
Cloning of RGA Fragments and Screening of Clones
The DNA bands were cut from the gel using razor and extracted by Gel Extraction Kit (Qiaquick) as instructed by the manufacturer instruction. The TA based cloning system (pGEM-T Easy TA cloning kit, Promega) was used to clone the RGA fragments according to the user’s guidelines in 1:2 ratio. Ligation was performed in 20 μl reaction volume overnight at 16°C using 3 units of T4 DNA ligase and 1X of buffer provided with the kit.
Blue white colonies appear which was counted for the efficiency of transformation. High transformation efficiency 8 x 107 indicates the successive ligation reaction and transformation. Single/independent round white colonies were picked up from the transformation petriplate using a sterile loop/tooth pick and re inoculated on selection plate containing Ampicilline, IPTG and X-gal. Those clones retain the white colour were used to inoculate 3 ml of LB medium supplemented with antibiotic. Overnight grown cultures were pelleted down by centrifuging at maximum speed for 1 minute. Plasmid DNA was isolated from the cells using the Qiagen Mini-Prep Kit according to users manual. PCR was performed with RGA/M13 primers to amplify them from DNA/cDNA were employed to screen positive clones. The PCR was conducted in similar program conditions as described before. The amplified product from the selected clone plasmid were analysed on 1% agarose gel. Those clones which retain the desired fragment used for cloning were picked and sent for sequencing. These clones were properly numbered. Glycerol stock prepared with 15% glycerol and preserved at -80°C.
The positive clones carrying the RGA fragments were sequenced using the T7/Sp6 Universal sequencing primer. RGA amplified fragment size of 450bp to 550bp range were selected and sequenced. Sequences were edited using DNA STAR/NCBI software for vector contamination.
Sequence submitted to NCBI nucleotide database
All RGA sequences identified have more than 200bp sequence length is submitted at NCBI database. The details are provided in Table 2.
| S.No. | RGA isolated from genomic DNA | NAME | Accession Number at Gene Bank Database |
|---|---|---|---|
| 1 | Yes | CBC 7 | JQ734556 |
| 2 | Yes | CBC 139 | JQ734557 |
| 3 | Yes | CBC 140 | JQ734558 |
| 4 | Yes | CBC 141 | JQ734559 |
| 5 | Yes | CBC 142 | JQ734560 |
| 6 | Yes | CBC 143 | JQ734561 |
| S.No. | RGA isolated from cDNA | NAME | Accession Number At Gene Bank Database |
| 1 | Yes | CBC 9 | JQ804950 |
| 2 | Yes | CBC 10 | JQ804951 |
| 3 | Yes | CBC 11 | JQ804952 |
| 4 | Yes | CBC 12 | JQ804953 |
| 5 | Yes | CBC 13 | JQ804954 |
| 6 | Yes | CBC 14 | JQ804955 |
| 7 | Yes | CBC 15 | JQ804956 |
| 8 | Yes | CBC 16 | JQ804957 |
| 9 | Yes | CBC 17 | JQ804958 |
| 10 | Yes | CBC 18 | JQ804959 |
| 11 | Yes | CBC 19 | JQ804960 |
| 12 | Yes | CBC 158 | JQ804961 |
| 13 | Yes | CBC 160 | JQ804962 |
| 14 | Yes | CBC 165 | JQ804963 |
| 15 | Yes | CBC 166 | JQ804964 |
| 16 | Yes | CBC 168 | JQ804965 |
| 17 | Yes | CBC 169 | JQ804966 |
| 18 | Yes | CBC 133 | JQ804967 |
| 19 | Yes | CBC 134 | JQ804968 |
| 20 | Yes | CBC 135 | JQ804969 |
| 21 | Yes | CBC 136 | JQ804970 |
Table 2: Coffea arabica RGA sequence (>200bp) submitted to NCBI Gene Bank database details.
Results
RGA cloning using degenerate primers
Genomic DNA Isolation
Genomic DNA isolated from S.288 using CTAB method has yielded 50 μg of genomic DNA from 100 mg of leaf tissue. CTAB method has yielded DNA with RNA contamination. RNAase treatment has successfully removed RNA contamination.
RGA amplified from genomic DNA using degenerate primers has yielded desire fragment of 500 bp with only one set of primer combination Ploop4 and GLPL4. Most of the other combination failed to amplify, only combination of Ploop2 with GLPL1, Ploop2 with GLPL2, Ploop2 with GLPL3, Ploop2 with GLPL4, Ploop4 with GLPL1, Ploop4 with GLPL2, Ploop4 with GLPL3 and Ploop4 and GLPL4. Out of these eight combinations only one set of primer Ploop4 and GLPL4 has yielded desire 500 bp fragments without any unspecific fragment therefore used for cloning. Although Ploop2 with GLPL4 and Ploop4 with GLPL2 has yielded desire 500 bp amplified fragment with some un-specific fragment, not used for cloning. Surprisingly as mentioned by Noir et al. (2001) work says that Ploop4 and GLPL4 combination has not yielded any RGA. Therefore it was interesting to clone and study these fragments.
Other RGA primer combination of Int-Cof primers and twelve degenerate primers F1 to F6 forward and HDR1 to HDR6 reverse tried with S.288 DNA has failed to amplify in any of its 36 combination tried. This may be the primer sequence did not have best annealing with coffee genomic DNA /cDNA sequence of S.288 at NBS region. Out of 16 degenerate primers along with Ploop 4 and GLPL 4 primers were also used on genomic DNA of S.288 to isolate RGAs. Out of 16 pairs of degenerate primer combinations used on genomic DNA, only one pair (Ploop 4 and GLPL 4) resulted into expected size amplified products.
RGA cloning using non-degenerate primers
RGA amplified from genomic DNA
RGA amplified from genomic DNA using non-degenerate primers has yielded desire fragment of 500 bp. Some unspecific DNA fragment also amplified which was not used for cloning.
RGA amplified from cDNA
Total RNA Isolation
Approximately 35 to 40 μg of RNA was estimated from 100 mg of S.288 leaf tissue. The RNA quality was found good as indicated by agarose gel separation. mRNA was purified and quality was checked on agarose gel.
RGA amplified from cDNA using non-degenerate primers has yielded desire fragment of 500 bp. Some unspecific DNA fragment also amplified which was not used for cloning.
Cloning
Cloning of above amplified RGA fragments were electroeluted from the agarose gel and quantified small quantity on agarose gel. Cloning was performed and positive clones were screened first on blue white selection plates with high transformation efficiency 8 x 107 indicates the successive ligation reaction, second screening with re-patching on selection plates, and third screening by isolation of plasmid from selected clones and amplification with M13 universal primer which amplify the region of plasmid containing cloned insert.
Sequence analysis
Sequences of these clones were analyzed as mentioned above and discussed in the next section (Table 3, 4, 5).
| S.No | RGA Clone name CBC |
Insert size (bp) | NCBI BLAST best hit | |||
|---|---|---|---|---|---|---|
| Score | E value | Accession No. | Details of hit | |||
| 1 | CBC 7 | 500 | 924 | 0.0 | AJ298884.1 | Coffea arabica partial ORF for disease resistance-like protein |
| 2 | CBC8 | 340 | 182 | 1e-42 | AJ308824.1 | Coffea arabica microsatellite DNA |
| 3 | CBC139 | 486 | 41.0 | 5.5 | AJ298884.1 | Coffea arabica partial ORF for disease resistance-like protein |
| 4 | CBC140 | 528 | 926 | 0.0 | AY606825.1 | Coffea arabica clone P5RANDF disease resistance-like protein gene |
| 5 | CBC141 | 523 | 911 | 0.0 | AJ298884.1 | Coffea arabica partial ORF for disease resistance-like protein |
| 6 | CBC142 | 527 | 911 | 0.0 | AY606825.1 | Coffea arabica clone P5RANDF disease resistance-like protein |
| 7 | CBC143 | 486 | 41.0 | 5.5 | AJ298884.1 | Coffea arabica partial ORF for disease resistance-like protein |
Total RGAs isolated from genomic DNA using Ploop-Cof and GLPL-Cof and sequenced = 7 Number of clones sequence matches with disease resistance-like protein = 6 Number of clones sequence not matches with disease resistance-like protein = 1
Table 3: Details of NCBI BLASTN sequence results of RGAs isolated from genomic DNA using non degenerate primers.
| S.No | RGA Clone name CBC |
Insert size (bp) | NCBI BLAST best hit | |||
|---|---|---|---|---|---|---|
| Score | E value | Accession No. | Details of hit | |||
| 1 | CBC183 | 514 | 857 | 0.0 | EF566662.1 | Coffea spp. RGA putative NBS domain resistance protein gene |
| 2 | CBC184 | 592 | No Hit | |||
| 3 | CBC185 | 528 | 867 | 0.0 | EF566686.1 | Coffea spp. RGA putative NBS domain resistance protein gene |
| 4 | CBC186 | 525 | 852 | 0.0 | AJ298884.1 | Coffea arabica partial ORF for disease resistance-like protein |
| 5 | CBC187 | 507 | 144 | 4e-36 | ABU51609.1 | Putative NBS domain resistance protein [Coffea spp.] |
| 6 | CBC188 | 531 | 880 | 0.0 | EF566686.1 | Coffea spp. RGA putative NBS domain resistance protein gene |
| 7 | CBC189 | 530 | 880 | 0.0 | EF566686.1 | Coffea spp. RGA putative NBS domain resistance protein gene |
| 8 | CBC190 | 454 | No Hit | |||
| 9 | CBC191 | 530 | 880 | 0.0 | EF566686.1 | Coffea spp. RGA putative NBS domain resistance protein gene |
| 10 | CBC192 | 600 | No Hit | |||
| 11 | CBC193 | 527 | 915 | 0.0 | AJ298884.1 | Coffea arabica partial ORF for disease resistance-like protein, |
Total RGAs isolated from genomic DNA using Ploop-4 and GLPL-4 and sequenced = 11 Number of clones sequence matches with disease resistance-like protein =8 Number of clones sequence not have significant match = 3
Table 4: Details of NCBI BLASTN sequence results of RGAs isolated from genomic DNA using degenerate primer.
| S.No | RGA Clone name CBC |
Insert size (bp) | NCBI BLAST best hit | |||
|---|---|---|---|---|---|---|
| Score | E value | Accession No. | Details of hit | |||
| 1 | CBC9 | 504 | 918 | 0.0 | AJ298884.1 | Disease resistance like protein Coffea arabica |
| 2 | CBC10 | 523 | 936 | 0.0 | AJ298884.1 | Disease resistance like protein Coffea arabica |
| 3 | CBC11 | 522 | 287 | 2e-82 | CAC82597.1 | Disease resistance like protein Coffea arabica |
| 4 | CBC12 | 521 | 289 | 5e-83 | CAC82597.1 | Disease resistance like protein Coffea arabica |
| 5 | CBC13 | 521 | 286 | 2e-82 | CAC82597.1 | Disease resistance like protein Coffea arabica |
| 6 | CBC14 | 517 | 284 | 2e-85 | CAC82597.1 | Disease resistance like protein Coffea arabica |
| 7 | CBC15 | 522 | 289 | 4e-83 | CAC82597.1 | Disease resistance like protein Coffea arabica |
| 8 | CBC16 | 522 | 287 | 2e-82 | CAC82597.1 | Disease resistance like protein Coffea arabica |
| 9 | CBC17 | 522 | 287 | 2e-82 | CAC82597.1 | Disease resistance like protein Coffea arabica |
| 10 | CBC18 | 511 | 289 | 3e-83 | CAC82597.1 | Disease resistance like protein Coffea arabica |
| 11 | CBC19 | 522 | 289 | 5e-83 | CAC82597.1 | Disease resistance like protein Coffea arabica |
| 12 | CBC133 | 330 | 41 | 3.6 | AB191232.1 | Cucumis meto MRGA-3 gene for NBS-LRR type resistance protein |
| 13 | CBC134 | 330 | 41 | 3.6 | AB191232.1 | Cucumis meto MRGA-3 gene for NBS-LRR type resistance protein |
| 14 | CBC135 | 362 | 42.8 | 1.1 | AY739270.1 | Musa acuminate NBS-LRR resistance protein-like mRNA, partial cds |
| 15 | CBC136 | 330 | 41.0 | 3.6 | AB191232.1 | Cucumis melo mRNA-3 gene for NBS-LRR type resistance protein |
| 16 | CBC137 | 410 | 53.6 | 6e-04 | CP000113.1 | Myxococcus xanthus DK 1622, complete genome |
| 17 | CBC138 | 243 | - | - | - | No hit |
| 18 | CBC158 | 513 | 789 | 0.0 | AY606826.1 | Coffea canephora disease resistance like protein |
| 19 | CBC 159 | 306 | - | - | - | No hit |
| 20 | CBC160 | 523 | 929 | 0.0 | AJ298884.1 | Disease resistance like protein Coffea arabica |
| 21 | CBC161 | 95 | - | - | - | No hit |
| 22 | CBC162 | 5 | - | - | - | No hit |
| 23 | CBC163 | 427 | 87.7 | 3e-16 | SGN-U353589 | C.canephora |
| 24 | CBC164 | 427 | 180 | 7e-42 | AY102435.1 | C.arabica microsatellite |
| 25 | CBC165 | 526 | 926 | 0.0 | AJ298884.1 | Disease resistance like protein Coffea arabica |
| 26 | CBC166 | 248 | 455 | 5e-125 | AJ298885.1 | Disease resistance like protein Coffea arabica |
| 27 | CBC167 | 590 | 77.8 | 3e-13 | SGN-U353389 | C.canephora |
| 28 | CBC168 | 263 | 427 | 1e-116 | AY606825.1 | Disease resistance like protein Coffea arabica |
| 29 | CBC169 | 425 | 710 | 0.0 | AY606825.1 | Disease resistance like protein Coffea arabica |
| 30 | CBC170 | 384 | 71.9 | 1e-11 | SGN-U353192 | C.canephora |
| 31 | CBC171 | 389 | 105 | 1e-21 | SGN-U353589 | C.canephora |
| 32 | CBC172 | 93 | 102 | 2e-19 | AJ298884.1 | Disease resistance like protein Coffea arabica |
Total RGAs isolated from cDNA using Ploop-Cof and GLPL-Cof and sequenced = 32
(CBC 9 to 19. CBC 133 to 138, CBC 158 to 172)
Number of clones sequence matches with disease resistance-like protein = 18
(CBC 9 to 19, CBC 158, 160, 165, 166, 168, 169 and 172)
Matched with RGA but not from coffee = 4 (CBC 133,134,135,136)
Matched with coffee but not RGA = 5 (CBC 163, 164, 167, 170,171)
Matched with other species =1 (CBC 137)
Not matched or no sequence = 4 (CBC 138, 159, 161,162)
Table 5: Details of NCBI BLASTN sequence results of RGAs isolated from cDNA using non-degenerate primer.
Comparison of RGAs with known resistance genes
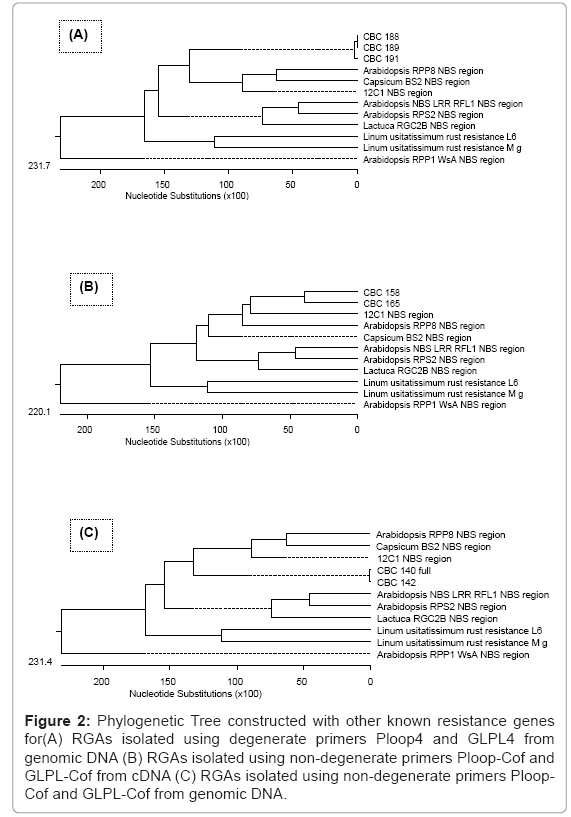
Only those sequences which have respective domains of Ploop (GVGKTT) and GLPL (GLPLAL) at both sides, compared with Arabidopsis thaliana disease resistance protein RPP8 (Accession No. AF089710.1), Capsicum chacoense disease resistance protein BS2 (Accession No. AF202179.1), Lycopersicon esculentum resistance complex protein I2C-1 (Accession No. AF004878.1), Arabidopsis thaliana NBS/LRR disease resistance protein RFL1 (Accession No. AF074916.1), Arabidopsis thaliana Col-0 resistance to Pseudomonas syringae RPS2 (Accession No. U12860.1), Lactuca sativa resistance protein candidate RGC2B (Accession No. AF113948.1), Linum usitatissimum alternatively spliced rust resistance L6 gene (Accession No. U27081.1), Linum usitatissimum rust resistance protein M gene (Accession No. U73916.1), Arabidopsis thaliana disease resistance protein RPP1-WsA gene (Accession No. AF098962.1) and resistance genes NBS region. Phylogenetic tree was constructed to observe the distance with existing R genes (Figure 2 ).
Figure 2: Phylogenetic Tree constructed with other known resistance genes for(A) RGAs isolated using degenerate primers Ploop4 and GLPL4 from genomic DNA (B) RGAs isolated using non-degenerate primers Ploop Cof and GLPL-Cof from cDNA (C) RGAs isolated using non-degenerate primers Ploop- Cof and GLPL-Cof from genomic DNA.
Discussion
Sequence of all these RGA clones were analyzed at NCBI database for vector contamination at (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Edited sequence of clones were analyzed at National Centre for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov/) and sol genomic network (SOL) (http://solgenomics.net/) data bases for sequence homology of the isolated clones using the BLAST (Basic Local Algorithms Sequence Tools). BLASTN sequence analyzed data was tabulated, which explains the homology with resistance like proteins. Sequence analysis was performed using DNA Star software. Duplicate sequences, which have primers at both sides, were taken as single sequence for further analysis. Sequences which have primer sequence deletion for single nucleotide were corrected using DNA Star software. All sequences were analyzed at SeqMan (DNA Star) to find the contig and SNP’s present.
RGA isolation from genomic DNA using degenerate primer
RGAs were tried to isolate using 16 combination of degenerate primers used on genomic DNA, only one pair (Ploop4 with GLPL4) resulted in the amplified product of expected size. Eleven randomly selected clones were sequenced and analyzed. Out of these 11 clones, 8 showed homology to RGAs. Size of these clones varied from 507 to 531 bp, indicating that they are related to disease resistance genes. The remaining 3 clones did not have significant homology to any RGA or any known sequence in both data bases searched. The BLASTN result details were shown at (Table 4). CBC 185 which is 528 bp, CBC 188 which is 531 bp, CBC 191 which is 530 bp has 99% matches with Coffea RGA clone accession number EF566686.1. CBC 186 which is 525 bp clone has 96% homology with Coffea arabica partial ORF for disease resistance like protein accession number AJ298884.1. CBC 189 which is 530 bp clone has 99% homology with Coffee RGA clone putative for NBS domain resistance protein gene accession number EF566680.1. Similarly CBC 193 which is also a 527 bp clone has 98% homology with Coffea arabica partial ORF for disease resistance like protein (NBS clone) accession number AJ298884.1. These sequence multiple alignment comparison suggest different range of RGA class.
These clones sequence were analysed for duplication and to search SNP present in their sequence. CBC 188 and CBC 189 are duplicate clones having identical sequence data, therefore CBC 188 taken for the studies. After assembled these sequences in SeqMan DNA Star six contig found. First contig containing 4 sequences CBC 183, CBC 185, CBC 191 and CBC 188. CBC 185, CBC 188 and CBC 191 have difference in first and last 25 bp positions. CBC 185 has single nucleotide polymorphism (SNP) at sequence position 228. Similarly CBC 188 has SNP at position 249 bp and 462 positions. CBC 191 has SNP present at sequence position 251 bp. Second contig contain two sequences CBC 186 and CBC 193. The other contigs have one sequence each, Contig 3 has CBC 187, Contig 4 has CBC 184, Contig 5 has CBC 192 and Contig 6 have CBC 190.
The other set of degenerate primer used, failed to give any amplification with coffee DNA suggests there is difference in the sequence of primer binding region. This made the primers all 36 combinations un-usable with coffee samples.
RGAs isolated from genomic DNA using non-degenerate primer
RGAs isolated from genomic DNA using Ploop-Cof and GLPLCof primer were sequenced and analyzed. Out of seven clones (CBC 7, CBC8, CBC 138, CBC 139, CBC140, CBC 141, CBC142 and CBC 143) searched; five clones sequence has a match with RGAs. BLASTX for protein database search indicate all these clones have one class of RGA. NCBI BLASTN data were tabulated (Table 3), which indicates that most among the five have 98% match with C.arabica/C.canephora disease resistance like protein. Clones CBC7 which is 500bp, CBC 139 which is 486bp , CBC141 which is 523 and CBC 143 which is 486bp has 98% match with the Coffea arabica partial ORF for disease resistance-like protein, accession number AJ298884.1. These clones were BLASTX at Gene bank protein database had a match with disease resistance like protein accession number CAC82597.1. CBC 142 the only sequence has 98% identity with accession number AY606825.1 at nucleotide database and accession number CAC82609.1 at protein data base which is disease resistance-like protein identified from Coffea canephora. Although CBC 8 which is 340bp a short fragment clone has matched with Coffea arabica microsatellite DNA. Multiple alignment comparison of these RGA sequences indicates that they share similarity to class A of disease resistance NBS-LRR genes. CBC 140 clone sequence which has expected size DNA fragment insert of 528 bp but failed to match with any known sequence at both database. Only those sequence has complete sequence were included for further studies.
To look for contigs among these sequences were analyze with SeqMan DNA Star. After removing the duplicate clone sequences, which have both primers sequences were analyze for contigs. CBC 140 and CBC 142 have identical sequence therefore CBC 140 taken for study which will represent for CBC 142. Similarly out of CBC 139 and CBC 143 which are identical sequence, only CBC 139 taken for studies which will represent for CBC 143. CBC 7 and CBC 141 have difference at sequence at position 199, and they also have single nucleotide addition at position 502. Therefore CBC7, CBC 139, CBC 140 and CBC 141 were assembled and found two contigs. First contig have three sequences CBC 7, CBC 140 and CBC 141. CBC7 and CBC 141 has almost identical sequence except for SNP present at position 199, and one nucleotide addition present in CBC 141 which is missing in CBC 7 at sequence position 506. Second contig has only one sequence CBC 139.
RGAs isolated from cDNA using non-degenerate primer
RGAs isolated from cDNA prepared with RNA of Coffea arabica Cv S.288 to understand the expression profile. The results were shown with best NCBI/ SGN nucleotide BLAST at (Table 5). RGAs were isolated using Ploop-Cof and GLPL-Cof primers from cDNA first strand. Amplified product of desire range were sequenced and analyzed at NCBI/SGN database. Thirty two randomly picked clones were sequenced and analyzed. Out of 32 clones, 22 clones showed homology with RGAs, size of clones varied in the range of 480 bp to 525 bp. Out of these 22 clones, 18 (CBC 9 to 19, CBC 158, 160, 165, 166, 168, 169 and 172) clones sequence have homology with Coffea species and other 4 (CBC 133, 134, 135, 136) clones sequence have homology with other plant species RGAs. The remaining clones among them 5 (CBC 163, 164, 167, 170,171) have homology with coffee sequences but they are not shown any homology with RGAs. Four clones sequence (CBC 138, 159, 161,162) data is short read therefore not included in studies. One clone (CBC 137) has homology with other plant species which is also not included in studies. CBC 9, CBC 10, CBC 160, CBC 165, CBC 166 and CBC 172 have 98% homology with Coffea arabica, disease resistance like protein accession number AJ 298884.1. CBC 11, CBC 12, CBC 13, CBC 14, CBC 15, CBC 16, CBC 17, CBC 18 and CBC 19 have 92% homology with Coffea arabica, disease resistance like protein accession number CAC82597.1. CBC 133, CBC 134 and CBC 136 which are 330 bp clones have 100% homology with Cucumis melo mRNA-3 gene for NBS-LRR type resistance protein accession number AB191232.1. CBC 168 and CBC 169 which are 263 and 425 bp respectively have 96% homology with Coffea arabica disease resistance like protein accession number AY606825.1. CBC163, CBC 167 and CBC 171 which are clones of 427 bp, 590 bp and 389 bp sizes respectively have 92% homology with Coffea canephora sequence accession number SGN-U353589. These clones have primer sequence in the both side which indicate the full clone sequence but not the expected size. CBC 135 which is 362 bp clone has 100% homology with Musa acuminata NBS-LRR resistance protein-like accession number AY739270.1. CBC 137 which is 410 bp clone has 78% homology with Myxococcus xanthus DK 1622, complete genome accession number CP000113.1. CBC 164 which is 427 bp clone has 98% homology with Coffea arabica microsatellite accession number AY102435.1. CBC 136 which is 330 bp clone has 100% homology with Cucumis melo mRNA-3 gene for NBSLRR type resistance protein accession number AB191232.1. CBC 158 which is 513 bp clone has 93% homology with Coffea canephora disease resistance like protein accession number AY 606826.1. CBC 167 which is 590 bp clone has 92% homology with Coffea canephora accession number SGN-U353389.
These clone sequences were analyze at SeqMan DNA Star and found eight contigs. First contig contains nine sequences CBC 10, CBC 11, CBC 14, CBC 15, CBC 16, CBC 17, CBC 19, CBC 160 and CBC 165. These sequences have SNP’s at position 66, 91, 123, 139, 144, 200, 236, 246, 277, 285, 350, 351, 368, 375, 376, 468 and 495 sequence positions.
Contig 2 has three sequences CBC 134, CBC 133 and CBC 136. These sequences have 100 % identity with each other, therefore CBC 133 sequence taken for studies. Now contig 2 has only one sequence as CBC 133. Contig 3 and contig 4 has one sequence each as CBC 158 and CBC 171 respectively. On the other hand Contig 5 has two sequences as CBC 163 and CBC 164. Contig 5 sequences have some insertion/ deletion of sequences. Contig 6, 7, 8 have one sequence each as CBC 167, CBC 137 and CBC 135 respectively.
Comparison of RGAs with known resistance genes
RGAs sequences were compared with existing R genes. RGAs phylogenetic tree analysis suggest that these RGAs are close to existing Arabidopsis thaliana disease resistance protein RPP8, Capsicum chacoense disease resistance protein BS2 and Lycopersicon esculentum resistance complex protein I2C-1. TIR and non TIR-NBSLRR sequences are distinguishable by amino acid motifs internal to their NBS domains. While motifs such as the Ploop, Kin-1a, and GLPLA signature are present in both classes, motifs RNBS-A-TIR (LQKKLLSKLL) and RNBS-D-TIR (FLHIACFF) are found exclusively in TIR class, while RNBS-A-non TIR (FDLxAWVCVSQxF) and RNBSD- nonTIR (CFLYCALFPED) are found in the non-TIR class (Meyers et al. 1999). The RGAs sequences obtained in current study were analyzed for the respective domains for identity of TIR and non-TIR class. Based on these, it may be concluded that all the RGAs isolated in this study belongs to nonTIR sub class. Sreenath et al. 2002 and Noir et al. 2001 also reported in earlier studies for nonTIR RGAs from coffee.
Acknowledgements
The authors are grateful to Dr. Jayarama, Director of Research, Central Coffee Research Institute, for carrying out this work. Author is grateful to U.G.C. for providing the fellowship.
References
- http://www.ico.org/.
- Aarts MG, te Lintel Hekkert B, Holub EB, Beynon JL, Stiekema WJ, et al (1998) Identification of R-gene homologous DNA fragments genetically linked to disease resistance loci in Arabidopsis thaliana. Mol Plant Microbe Interact 11: 251-258.
- Noir S, Combes MC, Anthony F, Lashermes P (2001) Origin, diversity and evolution of NBS type disease-resistance gene homologues in coffee trees (CoffeaL.) Mol Genet Genomics 265: 654-662.
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, et al. (1994) RPS2 of Arabidopsisthaliana: a leucine-rich repeat class of plant disease resistance genes. Science265: 1856-1860.
- Mindrinos M, Katagiri F, Yugl, Ausubel FM (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide binding site and leucine-rich repeats. Cell78: 1089-1099.
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, et al. (1995) Structure of the ArabidopsisRPM1 gene enabling dual specificity disease resistance.Science 269: 843-846.
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, et al. (1997) Different requirements for EDS1, and NDR1, by disease resistance genes define at least two R-gene-medicated signaling pathways in Arabidopis. Proc Natl Acad Sci U S A. 95: 10306-10311.
- Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S.Sobral BW, et al. (1999) Plant disease resistance genes encode members of an ancient and diverse family within the nucleotide-binding superfamily. Plant J20: 317-332.
- Pan Q, Liu Y-S, Budai-Hadrian O, Sela M, Carmel-Goren L, et al. (2000) Comparative genetics of nucleotide binding site-leucine rich repeat resistance gene homologues in the genomes of two dicotyledons: tomato and Arabidopsis. Genetics 155: 309-322.
- Liu JJ, Ekramoddoullah AK (2003) Isolation, genetic variation and expression of TIR-NBS-LRR resistance gene analogs from western white pine (Pinus monticola Dougl. ex. D. Don.). Mol Genet Genomics 270: 432-441.
- Budak H, Kasap Z, Shearman RC, Dweikat I, Sezerman U, et al. (2006) Molecular characterization of cDNA encoding resistance gene-like sequences in Buchloe dactyloides. Mol. Biotechnol 34: 293-301.
- Sreenath HL, Finkers HJ, Van Heusden AW (2002) Isolation and analysis of resistance gene analogues (rgas) in Coffee. Proceeding of Placrosym XV: 185-194.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 15999
- [From(publication date):
August-2012 - Dec 22, 2025] - Breakdown by view type
- HTML page views : 11202
- PDF downloads : 4797