Comparative Effectiveness of Activated Soil in Bioremediation of a Farmland Polluted Soil by Polyaromatic Hydrocarbon in the Niger Delta
Received: 11-Oct-2018 / Accepted Date: 29-Nov-2018 / Published Date: 30-Nov-2018 DOI: 10.4172/2155-6199.1000456
Keywords: Activated soil; Bioremediation; Polyaromatic hydrocarbons; PAH-degrading bacteria; Polymerase chain reaction
Introduction
Background
Niger Delta Region of Nigeria is one of the top countries in the world with huge oil and gas reserve. The exploration of this crude resource gives rise to hydrocarbon pollution through routine oil operation, vehicular accident and illegal petroleum refining activities. The latter is currently the leading source of oil pollution in the region and the likelihood of putting a stop to this act is very slim because of the economic benefits enjoyed by the perpetrators and their collaborators. The hydrocarbon wastes emanating from these refineries are of high molecular weight including polyaromatic hydrocarbons (PAHs). The eco-toxicological impacts already in manifestation include air borne black soot, changes in soil physicochemical features, depletion in biodiversity, ground water contamination, bioaccumulation in environmental receptors and cancer related diseases and deaths [1].
Polyaromatic hydrocarbons (PAHs) are a group of organic pollutants that consist of two or more aromatic rings arranged in different configurations [2]. They are ubiquitous [3], persistent due to their inert nature [4], and are highly insoluble in water [5]. According to Kumar et al. [6], there are not less than 30 parent PAHs in crude oil out of which 16 have been considered as priority pollutants by the United State Environmental Protection Agency (US EPA), World Health Organisation (WHO) and European Union (UN) [7] due to their mutagenic, carcinogenic and teratogenic nature.
Remediation of these contaminants is necessary for environmental safety, sustainable development and human health. The mechanistic principles of PAHs’ remediation are physical, chemical and biological methods [8]. Physical and chemical methods are preferred to biological method when exigency calls, however they are capital intensive and require high energy with huge consumption of synthetic chemicals which may pose additional environmental challenge [9]. These and other reasons such as simplicity of technology, minimal site disruption, flexibility to be combined with other physicochemical methods (treatment train) of remediation are reasons bioremediation has become an attractive technology these days. The most critical advantage is that the biodegradation agents are hugely unlimited, renewable, versatile, flexible and ubiquitous [10].
Bacteria stand out to be the most resourceful organisms when it comes to bioremediation. Bacteria can survive anaerobic and prohibitive environments such as acid mines, heavy metal and radioactive dumps. Bacterial adaptive physiological and degradative competence makes them more versatile than any other group of organisms. More so, they respond to selective pressure [11] stimulation and genetic recombination far more than any other microorganism, thereby producing requisite biomass, biosurfactants, exopolysacharrides, enzymes and catabolic genes to degrade bioavailable pollutants [12].
These aforementioned biotic factors are expressed in basic forms of bioremediation such as bioattenuation, biostimulation and bioaugementation. Bioattenuation relies on natural processes to deplete contaminants through biological transformation [9]. Biostimulation requires the introduction of nutrients and/or oxygen to a polluted site as to encourage the growth of naturally occurring pollutant-degrading microorganisms. Bioaugmentation involves the seeding of allochthonous wide type or genetically modified microorganisms to polluted sites in order to accelerate the removal of hazardous compounds of interest [13].
Biodegradation of PAHs can be carried out aerobically and anaerobically [14] via metabolic and co-metabolic pathways [10]. Aerobic biodegradation of PAHs are well studied with established pathways and signatory metabolites for each PAH molecule [15]. Two and three ringed PAHs degrade faster than the high molecular weight counterparts. During aerobic metabolism, PAHs by dioxygenase action changes to cis-dihydrodiols, the latter is transformed into dihydroxycompounds through biochemical oxidation [16]. Catechol, gentisate, protocatachuate and phthalates are unique intermediates which are then subjected to ring cleavage (ortho or meta) pathways. The metabolites formed are then fed into the tricarboxylic acid cycle (TCA). Anaerobic biodegradation of PAHs is still at its developing stage. Thus its metabolic pathways are still on their nascent stage of study [17].
Biodegradation is the vehicle through which bioremediation is carried out. Bioattenuation, biostimulation and bioaugmentation are the three forms of bioremediations and has been employed in remediation of polluted sites in the Niger Delta. However, activated soil as an option of bioaugmentation is less reported even though it is arguably the cheapest method of bioremediation in the Niger Delta of Nigeria. The aim of this study is to carry out aerobic laboratory treatability study, under conditions that favour bacterial metabolism, to ascertain the effectiveness of activated soil in decontaminating PAHs against other bioremediation techniques.
Materials and Methods
Site description and collection
The soil samples for this study were collected from Ngia Ama (4.7947°N, 6.6831°E) Tombia in Degema Local Government Area of Rivers State where illegal refining history. The pollution in this island was only six months old with respect to the sampling time. The second sample was collected from Bomu (4.6340°N, 7. 3559°E) in Gokana Local Government Area of Rivers State. Pollution at this site dates back to 2007. Soil samples were collected in four different points at Bomu, merged to form a composite sample. At Ngia Ama, obviously polluted soil samples were collected at three spots to make a composite soil sample. The soil samples were collected with sterile hand trowel at 0-30 cm depth, put in a sterile polythene bag and transported to the laboratory within a time frame of 6 hours.
Soil samples preparation
The collected soil samples were processed, removing all non-soil matters. One kilogram of the contaminated soil samples were preserved at 4°C for later physicochemical and microbiological baseline studies [18]. The remaining soil samples were air-dried for a period of 16 h [19] in a clean ventilated laboratory. Each of the soil samples was pulverized and passed through (2 mm pore size) sieve [20]. The soil samples were thoroughly mixed to ensure proper mixing of the contaminant thereby achieving homogeneity. The processed soil samples were then kept in a sterile PVC bag at ambient temperature for later use.
Baseline study
The physicochemical and microbiological analysis of the soil samples, Bomu activated soil (ASB) and Ngia Ama polluted soil (PSN), were carried out according to Chikere et al. [3]. Both contaminated soil samples were obtained and analyzed to quantify THB, PAH, total petroleum hydrocarbon (TPH), organic carbon, nitrate, phosphate, nitrogen, total hydrocarbon content (TOC), pH, exchangeable ion conductivity and carbon-nitrogen ratio.
Physicochemical analyses
Total organic carbon (TOC) was determined according to Avramidis et al. [21]. To determine the total hydrocarbon content, ten grams of soil was added into an extracting flask followed by addition of 10 ml of n-hexane. The suspension was shaken for half an hour and filtered. The filtrate’s absorbance reading was taken at 420 nm. The amount of total hydrocarbon content in the soil was ascertained through extrapolation of plotted graph using a reference curve prepared with Bonny light. Determination of total petroleum hydrocarbon (TPH) was carried out according to Romanus et al. [22]. Determination of polyaromatic hydrocarbons (PAHs) was analyzed according to Rasdy et al. [23]. The GC-FID analysis was carried out on baseline, day 0, 15 and 30.
Total (Kjeldahl) nitrogen was conducted according to Diekow et al. [24]; phosphorus was ascertained according to APHA (1998). Soil pH was carried out by adding 10 g of each sample into a 100 ml of clean beakers. Deionized water (20 ml) was added and the suspension was thoroughly stirred with glass rod for 30 minutes to obtain homogenous mixture, after which calibrated pH meter (pH tester 20) was dipped into the beaker containing the suspension and the pH value was recorded after a steady reading. Average pH value was taken from triplicate readings. Soil conductivity test was done by dissolving 10 g of soil sample in 20 ml of deionized water and left standing for half an hour. The slurry was then filtered. The conductivity of the filtrate was determined by Hanna digital conductivity meter. Nitrate was determined according to Igwo-Ezikpe et al.
Bioremediation experimental set-up
Five microcosms were set-up with Ngia Ama polluted soil. Microcosm for biostimulation was defined by an inorganic fertilizer (NPK 20:10:10). NPK 20:10:10 is an inorganic fertilizer that has 20% nitrogen, 10% phosphorus (P2O5) and 10% potassium (K2O). For bioaugmrntation/biostimulation, same fertilizer and activated soil from Bomu was used. To simulate bioaugmentation microcosm the activated soil was mixed with sterilized Ngia Ama sample. Sample of Ngia Ama with no amendment was used as bioattenuation microcosm while sterilized Ngia Ama sample was made as control. Plastic container was used in all the set-ups. The set-up is summarized in Table 1.
| Microcosm code | Description |
|---|---|
| BST | 975 g of PSN+25 g of NPK (20:10:10)+5 ml of distilled water per day |
| BSAG | 950 g PSN+25 g of NPK+25 g of activated ASB+5 ml of distilled water per day |
| AGN | 975 g of PSN (sterilized)+25 g of activated ASB+5 ml of distilled water per day |
| BAT | 1000 g of PSN+5 ml of distilled water per day |
| STL | 1000 g of PSN (sterilized)+5 ml of distilled water per day |
Table 1: Experimental set-up for soil bioremediation with monitoring on day 0, 15 and 30.
Microbiological analysis
Isolation and enumeration of total heterotrophic bacteria (THB) was done according to Agamuthu et al. [25] with some modifications. Hydrocarbon utilizing bacteria (HUB) was quantified using Bushnell Haas (BH) solid media supplemented with 0.05 g/ml of nystatin and 1% Bonny Light oil [26]. Aliquot of 0.1 ml from 10-4 and 10-5 dilutions were plated out in triplicates. The plates were incubated at 27°C for 7 days before colonies were counted. For the enumeration of PAHdegrading bacteria, one gram of soil sample from each treatment cell was added to 9 ml of BH broth supplemented with 1% mixed PAH (0.3 g/l) solution. An aliquot from 10-3 and 10-4 dilutions were spread on BH agar plate with 0.05 g/ml of nystatin in triplicates, incubated at room temperature for 7 days and enumeration done in colony forming unit per gram [27]. This analysis was carried out on each of the samples from the different treatment in time-course.
Tentative identification of isolates
Colonies formed on BH solid agar plates were sub-cultured on nutrient agar. Pure isolates retrieved from the microbial analyses were tentatively assigned identity based on their phenotypic and biochemical characterization [28].
Molecular analyses
DNA extraction: DNA extraction was carried out on pure isolates from ASB and PSN samples (baseline and day 30 respectively) using the ZP Fungi/Bacteria DNA MiniPrepTM supplied by Inqaba BiotecTM South Africa following the manufacturer’s instructions. The DNA quantity and purity were ascertained with a NanodropR ND-1000 UV-Vis Spectrophotometer. The genomic DNA was stored at -20°C for PCR analysis [29].
PCR amplification of the 16S rRNA genes: The 16S rRNA regions of the isolates’ rRNA genes were amplified using the 27 F and 1492 R primers on a thermal cycler for 35 cycles at a final volume of 50 microlitres. The PCR-mix contained X2 Dream taq Master mix (DNTPs, taq polymerase and MgCl), primers (at a concentration of 0.4 M) and extracted DNA (template). The thermocycling parameters followed thus: initial denaturation at 95°C for 5 minutes; denaturation at 95°C for 30 s; annealing at 52°C for 30 s; extension at 72°C for 30 s with 35 cycles and final extention at 72°C for 5 minutes before holding and cooling to 4°C [30]. Agarose gel electrophoresis was conducted according to Ding et al. [31].
Small subunit ribosome (16S rRNA) Gene Sequencing and Phylogenetic Analyses: The PCR amplicons from isolates of ASB and PSN samples were sequenced using a 3500 genetic analyzer. Bioedit algorithm was use to edit the resulting 16S rRNA sequence. BlastN was used to download similar sequences from the database of National Biotechnology Information Center (NCBI) to determine similarity between sequences and percentage similarities between matches [32]. ClustalX was employed to align the sequences. Neighborhood-Joining method (in MEGA 6.0) was used to generate the evolutionary tree [33] and analyzed by the bootstrap method [34]. The evolutionary distance computed by Jukes and Cantor [35] which are in the units of the number of base substitutions per site.
Statistical analyses
Data originating from this study was statistically analyzed using IBM SPSS Version 20 to determine the level of significance at p ≤ 0.05. Chi-square and Post Hoc test were respectively tested for homogeneity and mean differences. A two-way analysis of variance (ANOVA) was used to analyze data from the bioremediation study taking into consideration the three different factors such as bacterial types, time and treatment and the interaction amongst these factors. For the loss of PAHs, a one-way ANOVA was used. Excel inbuilt statistical package (XLSTAT Ecology Version 2016.04.32525) was used for the graphical presentation of data.
Nucleotide sequence accession numbers
The draft nucleotide sequences described in this study have been submitted to GenBank under accession numbers KX754444 to KX7554459
Results
Baseline characteristics of ASB and PSN polluted soil samples
The values of the baseline physicochemical (pH, electrical conductivity, nitrate, phosphate, total nitrogen, total organic carbon, total petroleum hydrocarbon and polyaromatic hydrocarbon) and microbiological analysis of sample ASB and PSN are all shown in Table 2. All the physicochemical characteristics of ASB are appreciably lower in concentration and in value when compared to sample PSN.
| Physicochemical parameter | Unit | ASB sample | PSN sample |
|---|---|---|---|
| Feelings to the hand | Grainy | Smooth | |
| Colour | Dark | Light brown | |
| pH | - | 4.5 | 6.86 |
| Electrical conductivity | µS/cm | 12 | 174 |
| Nitrate | mg/kg | 0.9 | 4.36 |
| Phosphate | mg/kg | 0.1 | 0.76 |
| Total nitrogen | mg/kg | - | 0.241 |
| Total organic carbon | % | 0.15 | 0.95 |
| Total petroleum hydrocarbon (TPH) | mg/kg | 858 (C9) | 2,067.72 |
| Polyaromatic hydrocarbon (PAH) | ppm | Total: 5.84 | Total: 193 |
| Bacterial counts THB | CFU/g | 7.1 × 109 ± 8.2 × 107 | 2.2 × 107 ± 8.2 × 105 |
| HUB | CFU/g | 1.9 × 106 ± 8.2 × 104 | 3.6 × 105 ± 8.2 × 103 |
| PDB | CFU/g | 4.6 × 105 ± 8.2 × 103 | 4.9 × 104 ± 8.2 × 102 |
Table 2: Baseline characteristics of activated soil from Bomu (ASB) and polluted soil from Ngia Ama (PSN).
On the other hand the bacterial count of total heterotrophic bacteria (THB), hydrocarbon utilizing bacteria (HUB) and PAH-degrading bacteria (PDB) of sample ASB are higher than the PSN sample with a value between 1 to 2 order of magnitude.
Isolation and enumeration of total heterotrophic bacteria (THB), hydrocarbon degrading bacteria (HUB) and PAH degrading bacteria (PDB) counts in treated sample and the control
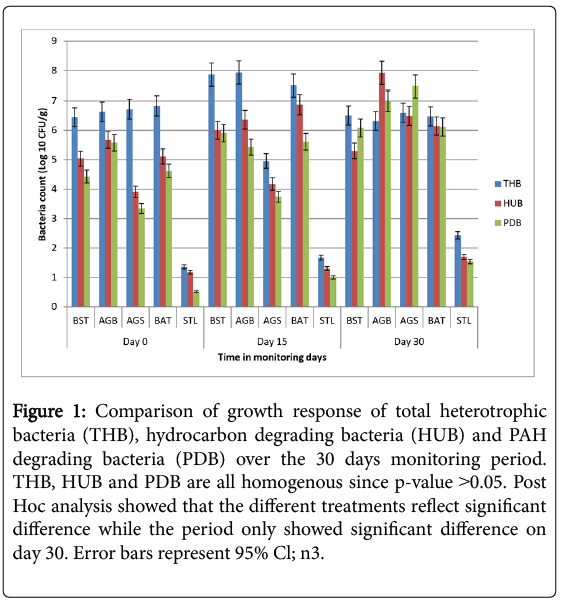
Microbiological counts were carried out in tandem with bioremediation monitoring over a period of 30 days in a laboratory condition. Evidence of microbial activities was ascertained by pure isolate and quantification of total heterotrophic bacteria (THB) counts, hydrocarbon utilizing bacteria (HUB) counts and PAH-degrading bacteria (PDB) counts in the various microcosms such as biostimulation (BST), bioaugmentation/biostimulation (BSAG), bioaugmentation (AGN) and a sterilized control (STL). Figure 1 shows the graphical presentation, comparatively.
Figure 1: Comparison of growth response of total heterotrophic bacteria (THB), hydrocarbon degrading bacteria (HUB) and PAH degrading bacteria (PDB) over the 30 days monitoring period. THB, HUB and PDB are all homogenous since p-value ˃0.05. Post Hoc analysis showed that the different treatments reflect significant difference while the period only showed significant difference on day 30. Error bars represent 95% Cl; n3.
Total heterotrophic bacteria (THB) population ranged from 5.2 × 103 CFU/g to 8.6 × 107 CFU/g across the microcosms. The highest THB value was recorded in BSAG at day 15 with a value of 8.6 × 107 CFU/g and highest in AGN at day 30 with a value of 4.0 × 106 CFU/g. The least count was recorded in STL at day 0. Hydrocarbon utilizing bacteria (HUB) count ranged from 8.2 × 103 CFU/g to 7.3 × 107 CFU/g across the different microcosms. The highest value was from BSAG (8.4 × 107 CFU/g) and least in STL (15 CFU/g) at day 30 and day 0 respectively. PDB count ranged from 2.2 × 103 CFU/g to 3.0 × 107 CFU/g across the microcosms. The highest and the lowest values were from AGN (3.0 × 107 CFU/g) at day 30 and STL (3.3 CFU/g) at day 0 respectively.
Biochemical characterization of PAH degrading bacteria (PDB)
Biochemical characterization of isolated PAH-degrading bacteria is shown in Tables 3 and 4.
| ASB1 | ASB2 | ASB3 | ASB4 | ASB5 | |
|---|---|---|---|---|---|
| Gram reaction | - | - | - | - | + |
| Cell morphology | Rods | Rods | Rods | Rods | Rods |
| Pink | Creamy | Greenish | Pink | Creamy | |
| Circular | Circular | Circular | Circular | Irregular | |
| Biochemical test | |||||
| Oxidase | - | + | + | + | - |
| Catalase | + | + | + | + | - |
| Citrate | + | + | + | + | + |
| Urease | + | + | - | + | - |
| Indole | - | - | - | - | - |
| Methyl red | - | - | - | - | + |
| Voges Proskauer | + | - | - | - | + |
| Motility | + | + | + | + | + |
| Lactose fermentation | + | - | - | - | + |
| H2S production | - | + | - | - | - |
| Gas production | + | + | + | + | + |
| Spore test | - | - | - | - | + |
| Tentative identification | Enterobacter sp | Klebsiella sp | Pseudomonas sp | Serratia sp | Bacillus sp |
Table 3: Biochemical characteristics of bacterial isolates from the Bomu activated soil (ASB).
| PSN-1 | PSN-2 | PSN-3 | PSN-4 | PSN-5 | PSN-6 | PSN-7 | |
|---|---|---|---|---|---|---|---|
| Gram reaction | - | - | + | - | - | - | - |
| Cell morphology | Rods | Rods | Rods | Rods | Rods | Rods | Rods |
| Creamy | Greenish | Whitish | Greenish | Greenish | Greenish | Greenish | |
| Pin point | Medium | Small | Medium | Large | Large | Medium | |
| Dry | Mucoid | Dry | Mucoid | Mucoid | Mucoid | Mucoid | |
| Circular | irregular | Circular | Circular | Irregular | Circular | Irregular | |
| Biochemical test | |||||||
| Oxidase | + | + | + | + | + | + | - |
| Catalase | + | - | + | + | + | + | + |
| Citrate | + | + | + | + | + | - | + |
| Urease | |||||||
| Indole | - | - | - | - | - | - | - |
| Methyl red | + | - | - | - | + | - | - |
| Voges Proskauer | - | + | - | + | + | + | - |
| Motility | + | + | + | + | + | + | + |
| Lactose fermentation | - | - | + | - | - | - | - |
| H2S production | - | - | - | - | - | - | - |
| Gas production | - | - | - | - | - | - | - |
| Spore test | - | - | - | - | - | - | - |
| Tentati identification | Alc. sp | Ps. sp | Artr. sp | Ps. sp | Ps. sp | Ps. sp | Ps. sp |
Table 4: Biochemical characteristics of bacterial isolates from Ngia Ama polluted soil (PSN). Key: Alc.: Alcaligenes , Ps.: Pseudomonas , Artr.: Arthrobacter , sp.: species; +=positive to biochemical test; -=not positive biochemical test.
Physicochemical analysis of PSN sample
The pH highest value was 6.48 in AGN microcosm on day 0 and gradually declined to its lowest level (4.75) on day 30 in BST microcosm. Nitrogen content started with a peak of 4.27 mg/kg at day 0 in BSAG, got to its lowest level of 2.24 mg/kg on day 30 in BAT. Phosphate utilization started with its highest value (0.82 mg/kg) on day 0 in BSAG and got its lowest value (0.49 mg/kg) on day 30 in BAT. The trends of conductivity, total nitrogen, total hydrocarbon content, total organic carbon and carbon-nitrogen ratio are shown in Tables 5-7.
| Physicochemical parameter | BST | BSAG | AGN | BAT | STL |
|---|---|---|---|---|---|
| pH | 6.07 | 6.08 | 6.48 | 6.10 | 5.40 |
| Conductivity (µS/cm) | 186 | 168 | 160 | 166 | 154 |
| Nitrate (mg/kg) | 0.632 | 0.840 | 0.610 | 0.488 | 0.365 |
| Phosphate (mg/kg) | 0.78 | 0.82 | 0.76 | 0.66 | 0.62 |
| Total nitrogen (mg/kg) | 3.98 | 4.27 | 3.85 | 3.68 | 3.34 |
| Total hydrocarbon content (%) | 4.49 | 5.15 | 3.42 | 5.07 | 3.86 |
| Total organic carbon (%) | 1.39 | 2.20 | 0.87 | 2.84 | 1.54 |
| Carbon-nitrogen ratio | 5.345 | 6.215 | 3.152 | 9.896 | 5.811 |
| TPH (mg/kg) | 13.88 | 13.55 | 10.98 | 11.65 | 7.23 |
| ∑PAH (ppm) | 99.268 | 108.933 | 98.425 | 78.010 | 77.908 |
| Total PAH (ppm) | 8.25843 | 10.23276 | 6.72978 | 3.54782 | 4.20712 |
Table 5: Day 0 physicochemical characteristics of treated Ngia Ama soil sample undergoing bioremediation.
| Physicochemical parameter | BST | BSAG | AGN | BAT | STL |
|---|---|---|---|---|---|
| pH | 5.36 | 5.45 | 5.10 | 4.80 | 5.46 |
| Conductivity (µS/cm) | 136 | 130 | 128 | 145 | 157 |
| Nitrate (mg/kg) | 0.287 | 0.354 | 0.276 | 0.265 | 0.345 |
| Phosphate (mg/kg) | 0.72 | 0.80 | 0.70 | 0.59 | 0.55 |
| Total nitrogen (mg/kg) | 2.85 | 3.56 | 2.84 | 2.35 | 3.00 |
| Total hydrocarbon content (%) | 4.09 | 4.70 | 3.94 | 3.77 | 3.05 |
| Total organic carbon (%) | 1.44 | 3.40 | 2.80 | 1.34 | 2.18 |
| Carbon-nitrogen ratio | 2.279 | 4.048 | 4.590 | 2.746 | 5.973 |
| TPH (mg/kg) | 9.37 | 8.00 | 7.54 | 6.10 | 5.10 |
| ∑PAH (ppm) | 79.537 | 85.591 | 80.889 | 58.979 | 54.324 |
| Total (ppm) | 5.78818 | 8.77706 | 5.44378 | 1.93491 | 4.01295 |
Table 6: Day 15 physicochemical characteristics of treated Ngia Ama soil sample undergoing bioremediation.
| Physicochemical parameter (ppm) | BST | BSAG | AGN | BAT | STL |
|---|---|---|---|---|---|
| pH | 4.75 | 5.43 | 5.39 | 4.81 | 5.38 |
| Conductivity (µS/cm) | 255 | 177 | 224 | 185 | 188 |
| Nitrate (mg/kg) | 0.233 | 0.308 | 0.251 | 0.243 | 0.319 |
| Phosphate (mg/kg) | 0.69 | 0.76 | 0.62 | 0.49 | 0.50 |
| Total nitrogen (mg/kg) | 2.77 | 3.02 | 2.58 | 2.24 | 2.78 |
| Total hydrocarbon content (%) | 4.38 | 4.20 | 3.52 | 4.38 | 3.82 |
| Total organic carbon (%) | 1.62 | 1.60 | 2.00 | 2.40 | 1.80 |
| Carbon-nitrogen ratio | 3.857 | 3.704 | 3.846 | 3.857 | 4.390 |
| TPH (mg/kg) | 3.20 | 2.72 | 3.89 | 3.84 | 3.81 |
| ∑PAH (ppm) | 62.575 | 54.907 | 60.213 | 45.609 | 48.019 |
| Total PAH (ppm) | 2.5894 | 3.4336 | 2.1293 | 1.3021 | 2.3786 |
Table 7: Day 30 physicochemical characteristics of treated Ngia Ama soil sample undergoing bioremediation.
Stoichiometric reduction of PAHs from crude oil polluted (PSN) soil sample
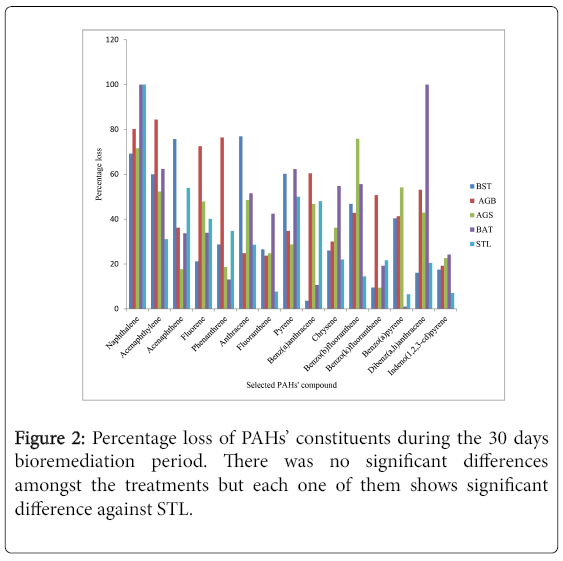
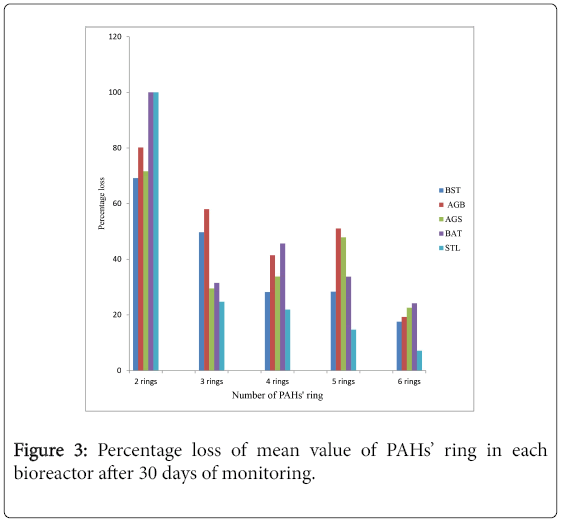
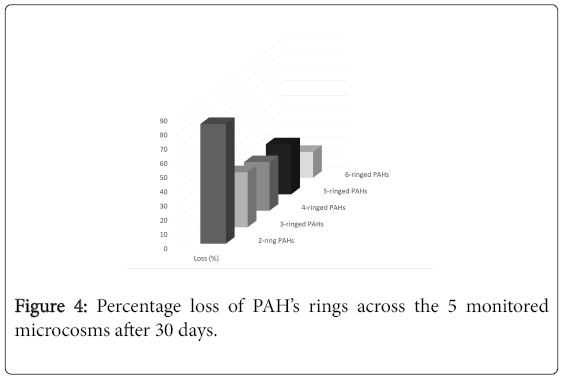
Chemical analysis of residual concentration of PAHs and TPHs using GC-FID revealed appreciable loss of PAHs and TPHs across all the microcosm setups within the prevailing laboratory conditions. ΣPAH concentration was between 103.878 ppm-68.8577 ppm on day 0 and reduced stoichiometrically to 60.1841 ppm-15.7733 ppm on day 30 across the microcosm setups. The percentage losses of total PAHs and TPHs as analyzed by GC-FID were 68.7 %, 70.0 % (BST), 66.4 %, 72.6 % (BSAG), 68.4 %, 64.7 % (AGN), 63.0 %, 67.0 % (BAT) and 43.4 %, 47.3 % (STL). Figure 2 shows the percentage loss of individual PAHs during the 30 days monitored period. The percentage degradation of the PAHs’ ring in each microcosm and across the 5 microcosms is illustrated in Figures 3 and 4 respectively.
Molecular characterization of bacteria isolate from ASB and PSI samples
The Megablast search for 16S rDNA sequence similarity gave an exact match from the NCBI database. The least percentage similarity was shown to be 99% with respect to other species. The computed evolutionary distances were in harmony with the 16S rRNA phylogenetic placement of the isolates within the genera and revealed a high similarity to the species than other genera with in Table 8 shows the homoloy analysis of the PDB, Figure 5 displays the gel electrophoresis result. The results are based on comparison of SSU rRNA gene sequences of the isolates to the sequence that shows the highest sequence similarity to the isolate.
| S/n. | Isolate code | Closest strain | Percentage similarity | *GenBank Feedback | GenBank Accession no. |
|---|---|---|---|---|---|
| 1 | B1 | Enterobacter xiangfanensis strain 9A | 100 | Enterobacter xiangfanensis | KX754444 |
| 2 | B2 | Shewanella haliotis strain 0315 | 100 | Shewanella haliotis strain | KX754445 |
| 3 | B3 | Pseudomonas denitrificans strain Y-16 | 100 | Pseudomonas denitrificans | KX754446 |
| 4 | B4 | Burkholderia terrestris strain R-233321 | 99 | Caballeronia terrestris | KX754447 |
| 6 | B6 | Acinetobacter calcoaceticus strain N7 | 100 | Acinetobacter calcoaceticus | KX754448 |
| 7 | B9 | Acinetobacter pittii strain AP_882 | 100 | Acinetobacter pittii strain | KX754449 |
| 8 | B11 | Pseudomonas nitroreducens strain VITWW2 | 99 | Pseudomonas nitroreducens | KX754450 |
| 9 | B12 | Pseudomonas otitidis strain IND2 | 100 | Pseudomonas otitidis | KX754451 |
| 10 | B13 | Stenotrophomonas maltophilia strain C_ | 100 | Stenotrophomonas maltophilia | KX754452 |
| 11 | B15 | Pseudomonas aeruginosa SJTD-1 | 100 | Pseudomonas aeruginosa | KX754453 |
| 12 | B16 | Unidentified | |||
| 13 | B37 | Pseudomonas fluorescens strain KRST 01 | 100 | Pseudomonas fluorescens | KX754454 |
| 14 | B38 | Pseudomonas aeruginosa SJTD-1 | 100 | Pseudomonas aeruginosa | KX754455 |
| 15 | B39 | Pseudomonas aeruginosa SJTD-1 | 100 | Pseudomonas aeruginosa | KX754456 |
| 16 | B40 | Pseudomonas fluorescens strain KRST 01 | 100 | Pseudomonas fluorescens | KX754457 |
| 17 | B41 | Exiguobacterium alkaliphilum strain 12/1 | 99 | Exiguobacterium alkaliphilum | KX754458 |
| 18 | B42 | Pseudomonas fluorescens strain KRST 01 | 100 | Pseudomonas fluorescens | KX754459 |
Table 8: Homology analysis (with BLAST) of PAH-degrading bacteria isolate.
Discussion
This study focused on hydrocarbon pollution emanating from artisanal oil refining activities which is one major subset of humanly induced environmental stressors with heavy carbon fragment and polyaromatic hydrocarbons (PAHs). The latter class of hydrocarbon (PAHs) serves as the central focus for this study and justified by the fact that PAHs are recalcitrant in nature and chemically toxicogenic, thus required to be eliminated or reduced to harmless level to resuscitate stressed ecosystem.
The low physicochemical characteristics of ASB sample (especially pH of 4.5) reflect low physiological activity for bacterial biodegradation [36]. When added as a bioaugmentation agent into sample PSN (with a pH value of 6.8) it increased bacteria (from sample ASB) physiological activity to degrade TPHs and PAHs [37] in isolation or complementary to degradation role played by the indigenous bacteria. Positive results of microbiological analysis lend credence to metabolic adaptability of bacteria thereby utilizing the hydrocarbons as carbon and energy source for growth and replication [38] across microcosms, except the control. Darmayati et al. [39] had earlier used activated soil to remediate hydrocarbon polluted soil.
Serial dilution technique was employed to enumerate total heterotrophic bacteria (THB), hydrocarbon utilizing bacteria (HUB) and polyaromatic hydrocarbon degrading bacteria (PDB). Besides, the technique was also used provide an index which indicates the differing bacteria types from different treatments. Though serial dilution technique is a corner stone of quantitative microbiology [40], it causes differences in community structure and metabolic redundancy [41]. This implies that specialist genera hardly grow in higher diluents save the generalist microbes. Coupled to these limitations, 90 to 99% of constitute culturable but not viable bacteria [3].
The colonies per g of soil was calculated using the formula:
NB/g=MPC × DF × V1/V2 × M
Where MPC=mean plate count; DF=dilution factor; V2=volume of original suspension; V2=inoculated volume; M=mass of soil added to V1.
Supported by this work, most literatures and research works have shown that Gram-negative bacteria are better degraders of hydrocarbons and dominate in processes involving crude oil degradation and remediation. Eze et al. [42] noted that Gram-negative bacteria predominate over Gram-positive bacteria in test samples due to the complexity of Gram-negative bacteria cell wall which hinders the penetration of certain substances and their entry into the cytoplasm. For instance Gram-negative bacteria cell walls possess porins which help in the selective uptake of substances by the cell and extrusion of others which may be harmful. Hydrocarbon degrading bacteria often display physiological responses that makes them insensitive to toxic effects, access insoluble hydrocarbons, or transfer large substances into the cytoplasm through biochemical mechanisms such as alteration of cell surface hydrophobicity, gaining of protection from hydrophilic lipopolysaccharide (specifically for Gram-negative bacteria) constituents and or using of repair mechanisms to compensate for losses in membrane as a result of lipophilic compound intercalation [12]. Gram-negative bacteria are less sensitive to the toxic effects of hydrocarbons compared to Gram-positive bacteria [43], hence dominate oil contaminated sites [44]. Most Gram-positive bacteria cannot counter the alteration of membrane architecture resulting from changes in protein conformation and fluidity due to insertion of hydrocarbons with the ultimate consequences of altering membrane-bound enzyme activities and disruption of the barrier and energy transduction roles [12].
Gamma-protobacteria, was the most dominant bacteria in this study. Vinas et al. observed in their study that different bacteria phylotypes of hydrocarbon degraders responded to different phases of bioremediation of PAHs and also in different nutrient amendments. According to them alpha-proteobacteria dominate in the early phase of bioremediation but when biostimulation is put into effect, betaproteobacteria and alpha-proteobacteria co-dominated. Against these findings, it was shown in this study that the gamma-proteobacteria dominated in all phases of the bioremediation process and also in all amendment conditions which fully corroborates with Sara et al. [45] findings and partially to Fuentes et al. [46] which demonstrated gamma-proteobacteria bloom to support the ecological concept of “conditionally rare taxa” to mean rareness is a temporary state conditioned by environmental stressors.
Among the gamma-protobacteria, Pseudomonas species stand tall (as reflected in this work) when it comes to hydrocarbon degradation and bioremediation. This could be explained why Pseudomonas is accorded dominant reference in research field of Environmental Microbiology or in discuss involving bioremediation and biodegradation of crude oil or its products in the environment, be it in soil, aquatic media, sediments, mangroves or air. Apart from PAHs, Pseudomonas strains also degrade wide range of petroleum compounds, including S-and N-heteroatoms and resins [47,48]. Species that have been used for extensive studies in biodegradation and bioremediation are Pseudomonas putida, Pseudomonas cepacia, Pseudomonas saccharophila, Pseudomonas xanthomarina, Pseudomonas aeruginosa, Pseudomonas pickettii 56 [49,50]. They degrade wider spectrum of hydrocarbons by virtue of their catabolic genes for metabolic versatility [43], cell surface activities for improved uptake of hydrocarbon substrate and bioavailability (Johnsen et al.), enzymes for metabolic and co-metabolic functions [16], the flexibility to coexist with other organisms to enhance biodegradation [51] and the ability to survive and proliferate (as a r-strategists) in an environment stressed with constraints and take advantage of such opportunities [52].
The biodegradation activity of the bacteria reflects in the decrease of TPHs and PAHs residual concentrations across the monitored test microcosms. The TPHs and PAHs losses experienced in the sterilized control reflect abiotic influence such as volatilization [53]. Differences in the loss of total PAHs and TPHs was not so much pronounced across the microcosm setups. This implies that the rate of degradation is not statistically significant at p ≤ 0.05 using analysis of variance (ANOVA). The 100% loss of naphthalene could be as a result of its solubility and volatility [19]. Pronounced loss between each ring category of PAHs means statistical significance at p ≤ 0.05. This implies that the ease of degradation is inversely proportional to the number of rings in PAHs.
Phylogenetic analysis based on the 16S rRNA is very useful and widely used due to its simplicity and allows to determine the structure of the bacterial population in soil metagenomics. The phylogenetic analysis was able to determine the taxonomic position of each of the isolate. The representation of a strain for more than two or more isolates with morphological dissimilarity as shown in Table 8 is an indication of phenotypic innovation. For instance isolates ASB37, ASB40 and ASB42 share same identity as Pseudomonas fluorescence strain KRST 01. This phenomenon could be attributed to cell’s response to the adverse effects of contaminants.
Conclusion
Petroleum artisanal refining activities contribute to the spate of hydrocarbon pollution in the Niger Delta thus, unleash polyaromatic hydrocarbons to the ecosystem causing ecological constraints. The soil sample from Bomu has much reduced hydrocarbon content in comparison to the soil sample from Ngia Ama. The soil sample from Bomu has hydrocarbon degrading bacteria thus served as an activated soil which was used to remediate the Ngia Ama polluted soil. Comparatively, the activated soil proves effective against biostimulation, bioattenuation and a combination of biostimulation and bioaugmentation, though no evidence of statistical difference was evidenced. Analysis of bacterial SSU rRNA gene sequence revealed that Enterobacter Shewanella, Pseudomonas , Burkholderia, Bacillus spp, Acinetobacter, Exiguobacter and Stenotrophomonas are good candidate for PAH degradation.
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- Kalantary RR, Mohseni-Bandpi A, Esrafili A, Nasseri S, Ashmagh FR, et al. (2014) Effectiveness of biostimulation through nutrient content on the bioremediation of phenanthrene contaminated soil. Journal of Environmental Health Science and Engineering 12: 143.
- Yu KSH, Wong AHY, Yau KWY, Wong YS, Tam NFY (2005) Natural attenuation, biostimulation and bioaugmentation on biodegradation of polycyclic aromatic hydrocarbons (PAHs) in mangrove sediments. Marine Pollution Bulletin 51: 1071-1077.
- Chikere BO, Chijioke-Osuji C (2006) Microbial diversity and physicochemical properties of a crude oil polluted soil. Nigerian Journal of Microbiology 20: 1039-1046.
- Isaac P, Sanchez LA, Bourguignon N, Cabral ME, Ferrero MA (2013) Indigenous PAH-degrading bacteria from oil-polluted sediments in Caleta Cordova, Patagonia Argentina. International Biodeterioration and Biodegradation 82: 207-214.
- Nganje TN, Neji PA, Ibe KA, Adamu CI, Edet A (2014) Fate, Distribution and Sources of Polycyclic Aromatic Hydrocarbons (PAHs) in Contaminated Soils in Parts of Calabar Metropolis, South Eastern Nigeria. Journal of Applied Sciences and Environmental Management 18: 309-316.
- Kumar A, Bisht BS, Joshi VD, Dhewa T (2011) Review on bioremediation of polluted environment: A management tool. International Journal of Environmental Sciences 1: 1079-1093.
- Zrafi-Nouira I, Saidane-Mosbahi D, Abdelghani S, Bakhrouf A, Rouabhia M (2012) Crude Oil Metagenomics for Better Bioremediation of Contaminated Environments. In Introduction to Enhanced Oil Recovery (EOR) Processes and Bioremediation of Oil-Contaminated Sites. InTech.
- Tomei MC, Daugulis AJ (2013) Ex situ bioremediation of contaminated soils: an overview of conventional and innovative technologies. Critical Reviews in Environmental Science and Technology 43: 2107-2139.
- Mrozik A, Piotrowska-Seget Z (2010) Bioaugmentation as a strategy for cleaning up of soils contaminated with aromatic compounds. Microbiological Research 165: 363-375.
- Kumar A, Ashok M, Rajesh S (2011) Crude oil PAH constitution, degradation pathway and associated bioremediation microflora: An overview. International Journal of Environmental Sciences 1: 1420-1439.
- Liang Y, Van Nostrand JD, Deng Y, He Z, Wu L, et al. (2011) Functional gene diversity of soil microbial communities from five oil-contaminated fields in China. The ISME Journal 5: 403-413.
- Van Hamme JD, Singh A, Ward OP (2003) Recent advances in petroleum microbiology. Microbiology and Molecular Biology Reviews 67: 503-549.
- Ravanipour M, Kalantary RR, Mohseni-Bandpi A, Esrafili A, Farzadkia M, et al. (2015) Experimental design approach to the optimization of PAHs bioremediation from artificially contaminated soil: application of variables screening development. Journal of Environmental Health Science and Engineering 13: 22.
- Peng RH, Xiong AS, Xue Y, Fu XY, Gao F, et al. (2008) Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiology Reviews 32: 927-955.
- Seo JC, Keum YS, Li QS (2009) Bacteria degradation of aromatic compounds. International Journal of Environmental Research and Public Health 6: 278-309.
- Al-Turki AI (2009) Microbial polycyclic aromatic hydrocarbons degradation in soil. Research Journal of Environmental Toxicology 3: 1-8.
- Foght J (2008) Anaerobic biodegradation of aromatic hydrocarbons: pathways and prospects. Journal of Molecular Microbiology and Biotechnology 15: 93-120.
- Akinbankole AS, Tunung R, Tennant AM (2015) Biochemical and molecular characterization of pyrene and anthracene metabolizing bacteria isolated from oil contaminated water and soil in Malaysia. Journal of Applied and Environmental Microbiology 3: 25-30.
- Luepromchai E, Lertthamrongsak W, Pinphanichakarn P, Thaniyavarn S, Pattaragulwanit K, et al. (2007) Biodegradation of PAHs in petroleum-contaminated soil using tamarind leaves as microbial inoculums. Biodegradation 29: 515-527.
- Scherr KE, Hasinger M, Mayer P, Loibner AP (2009) Effect of vegetable oil addition on bioaccessibility and biodegradation of polycyclic aromatic hydrocarbons in historically contaminated soils. Journal of Chemical Technology & Biotechnology 84: 827-835.
- Avramidis P, Nikolaou K, Bekiari V (2015) Total organic carbon and total nitrogen in sediments and soils: a comparison of the wet oxidation–titration method with the combustion-infrared method. Agriculture and Agricultural Science Procedia 4: 425-430.
- Akpe R, Ekundayo AO, Aigere SP, Okwu GI (2015) Bacterial degradation of petroleum hydrocarbons in crude oil polluted soil amended with cassava peels. American Journal of Research Communication 3: 99-118.
- Rasdy NFA, Sanagi MM, Ibrahim WAW, Naim AA (2008) Determination of polycyclic aromatic hydrocarbons in palm oil mill effluent by soxhlet extraction and gas chromatography-flame ionization detection. Malaysian J Anal Sci 12: 16-21.
- Dieckow J, Mielniczuk J, Knicker H, Bayer C, Dick DP, et al. (2007) Comparison of carbon and nitrogen determination methods for samples of a paleudult subjected to no-till cropping systems. Scientia Agricola 64: 532-540.
- Agamuthu P, Tan YS, Fauziah SH (2013) Bioremediation of hydrocarbon contaminated soil using selected organic wastes. Procedia Environmental Sciences 18: 694-702.
- Stanley HO, Amakiri MA, Okerentugba PO (2015) Characterization of hydrocarbon utilizing bacteria in soil samples collected from various sites in Port Harcourt (Niger-Delta, Nigeria). Society for Science and Nature 4: 6-11.
- Ma L, Zhai Y, Feng D, Chan TC, Lu Y, et al. (2010) Identification of novel factors involved in or regulating initiation of DNA replication by a genome-wide phenotypic screen in Saccharomyces cerevisiae. Cell Cycle 9: 4399-4410.
- De la Maza LM, Pezzalo MN, Shigei JT, Tan GL, Peterson EM (2013) Colour Atlas of Medical Bacteriology. 2nd edn., American Society for Microbiology Press, Washinton, USA.
- Ren G, Ren W, Teng Y, Li Z (2015) Evident bacterial community changes but only slight degradation when polluted with pyrene in a red soil. Frontiers in Microbiology 6: 22.
- Hesham AEL, Mawad AM, Mostafa YM, Shoreit A (2014) Biodegradation ability and catabolic genes of petroleum-degrading Sphingomonas koreensis strain ASU-06 isolated from Egyptian oily soil. BioMed Research International 2014: 127674.
- Ding GC, Heuer H, Zuhlke S, Spiteller M, Pronk GJ, et al. (2010) Soil type-dependent responses to phenanthrene as revealed by determining the diversity and abundance of polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenase genes by using a novel PCR detection system. Applied and Environmental Microbiology 76: 4765-4771.
- Mahlen SD, Clarridge JE (2011) Evaluation of a selection strategy before use of 16S rRNA gene sequencing for the identification of clinically significant Gram-negative rods and coccobacilli. American Journal of Clinical Pathology 136: 381-388.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis. Molecular biology and evolution 30: 2725-2729.
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783-791.
- Jukes TH, Cantor CR (1969) Evolution of protein molecules. Mammalian Protein Metabolism 3: 132.
- Sihag S, Pathak H, Jaroli DP (2014) Factors Affecting the Rate of Biodegradation of Polyaromatic Hydrocarbons. Int J Pure App Biosci 2: 185-202.
- Alrumman SA, Standing DB, Paton GI (2015) Effects of hydrocarbon contamination on soil microbial community and enzyme activity. Journal of King Saud University-Science 27: 31-41.
- Kostka JE, Prakash O, Overholt WA, Green S, Freyer G, et al. (2011) Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Horizon oil spill. Applied and Environmental Microbiology 77: 7962-7974.
- Darmayati Y, Sanusi HS, Prartono T, Santosa DA, Nuchsin R (2015) The Effect of Biostimulation and Biostimulation-Bioaugmentation on Biodegradation of Oil-Pollution on Sandy Beaches Using Mesocosms. International Journal of Marine Science 5: 1-11.
- Ben-David A, Davidson CE (2014) Estimation method for serial dilution experiments. Journal of Microbiological Methods 107: 214-221.
- Franklin RB, Garland JL, Bolster CH, Mills AL (2001) Impact of dilution on microbial community structure and functional potential: comparison of numerical simulations and batch culture experiments. Applied and Environmental Microbiology 67: 702-712.
- Eze CN, Ogbonna JC, Anyanwu CU, Eze EA (2013) Determination of the relative abundance and distribution of bacteria and fungi in Bonny light crude oil-contaminated sandy loam soil. Scientific Research and Essays 8: 375-381.
- Lazaroaie MM (2010) Multiple responses of gram-positive and gram-negative bacteria to mixture of hydrocarbons. Brazilian Journal of Microbiology 41: 649-667.
- Kaplan CW, Kitts CL (2004) Bacterial succession in a petroleum land treatment unit. Applied and Environmental Microbiology 70: 1777-1786.
- Kleindienst S, Paul JH, Joye SB (2015) Using dispersants after oil spills: impacts on the composition and activity of microbial communities. Nature Reviews Microbiology 13: 388.
- Fuentes S, Barra B, Caporaso JG, Seeger M (2016) From rare to dominant: a fine-tuned soil bacterial bloom during petroleum hydrocarbon bioremediation. Appl Environ Microbiol 82: 888-896.
- Mansur AA, Adetutu EM, Makadia T, Morrison PD, Ball AS (2015) Assessment of the hydrocarbon degrading abilities of three Bioaugmentation agents for the bioremediation of crude oil tank bottom sludge Contaminated Libyan soil. Int J Environ Bioremed Biodegradation 3: 1-9.
- Wang Q, Zhang S, Li Y, Klassen W (2011) Potential approaches to improving biodegradation of hydrocarbons for bioremediation of crude oil pollution. Journal of Environmental Protection 2: 47.
- Rajaei S, Seyedi SM, Raiesi F, Shiran B, Raheb J (2012) Characteristic and potentials of indigenous oil-degrading bacteria inhabiting the rhizosphere of wild oat (Arena fatua) in South West of Iran. Iran Journal of Biotechnology 11: 32-40.
- Mandri T, Lin J (2007) Isolation and characterization of engine oil degrading indigenous microrganisms in Kwazulu-Natal, South Africa. African Journal of Biotechnology 6: 23-27.
- Romero MC, Urrutia MI, Reinoso HE, Kiernan MM (2010) Benzo [a] pyrene degradation by soil filamentous fungi. Journal of Yeast and Fungal Research 1: 025-029.
- Singer E, Webb EA, Nelson WC, Heidelberg JF, Ivanova N, et al. (2011) The genomic potential of Marinobacter aquaeolei–A biogeochemical opportunitroph. Applied and Environmental Microbiology 77: 2763-2771.
- Skupinska K, Misiewicz I, Kasprzycka-Guttman T (2004) Polycyclic aromatic hydrocarbons: physicochemical properties, environmental appearance and impact on living organisms. Acta Pol Pharm 61: 233-240.
Citation: Chikere CB, Fenibo EO, Akaranta O (2018) Comparative Effectiveness of Activated Soil in Bioremediation of a Farmland Polluted Soil by Polyaromatic Hydrocarbon in the Niger Delta. J Bioremediat Biodegrad 9:456. DOI: 10.4172/2155-6199.1000456
Copyright: © 2018 Chikere CB, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 5410
- [From(publication date): 0-2018 - Dec 20, 2025]
- Breakdown by view type
- HTML page views: 4427
- PDF downloads: 983