Research Article Open Access
Genetic Diversity Studies on Selected Rice (Oryza Sativa L.) Genotypes Based on Amylose Content and Gelatinization Temperature
| Mawia A. Musyoki1*, Wambua F. Kioko1, Ngugi P. Mathew1, Karau G. Muriira2, Nyamai D. Wavinya1, Matheri Felix1, Langat R. Chemtai1, Njagi S. Mwenda1, Mworia J. Kiambi1 and Ngari L. Ngithi1 | |
| 1Department of Biochemistry and Biotechnology, School of Pure and Applied Sciences, Kenyatta University, P.O. Box 43844-00100, Nairobi, Kenya | |
| 2Molecular Laboratory, Kenya Bureau of Standards, Kenya | |
| Corresponding Author : | Mawia A. Musyoki Department of Biochemistry and Biotechnology School of Pure and Applied Sciences Kenyatta University, P.O. Box 43844-00100 Nairobi, Kenya E-mail: amosyokis@gmail.com |
| Received: September 19, 2015; Accepted: October 01, 2015; Published: October 07, 2015 | |
| Citation: Musyoki MA, Kioko WF, Mathew NP, Muriira KG, Wavinya ND, et al. (2015) Genetic Diversity Studies on Selected Rice (Oryza Sativa L.) Genotypes Based on Amylose Content and Gelatinization Temperature. Adv Crop Sci Tech 3:193. doi:10.4172/2329-8863.1000193 | |
| Copyright: © 2015 Musyoki MA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
| Related article at Pubmed, Scholar Google | |
Visit for more related articles at Advances in Crop Science and Technology
Abstract
Improving cooking and eating quality of rice is one of the important objectives of many breeding programs. The aim of the study was to carry out genetic diversity studies on selected rice (Oryza sativa L.) genotypes from Kenya and Tanzania based on amylose content and gelatinization temperature using microsatellite markers. Power marker version 3.25 and GenALEx version 6.5 softwares were used to analyze the data. The number of alleles per locus ranged from 2 to 4 alleles with an average of 2.75 alleles across 8 loci obtained in this study. The polymorphic information content (PIC) values ranged from 0.2920 (RM 202) to 0.6841 (RM 141) in all 8 loci with an average of 0.4697. Pair-wise genetic dissimilarity coefficients ranged from 0.9003 to 0.2201 with an average of 0.5627. Maximum genetic similarity was observed between R 2793 and BS 17, Supa and IR 64, R 2793 and ITA 310, Saro 5 and ITA 310, Saro 5 and R 2794. Minimum similarity of was observed between Wahiwahi and BW 196, IR 64 and BW 196. The dendogram based on cluster analysis by microsatellite polymorphism grouped the rice genotypes into 2 clusters effectively differentiating Kenyan and Tanzanian rice genotypes based on amylose content and gelatinization temperature. The results obtained suggested that use of microsatellite markers linked to Quantitative Trait Loci (QTLs) controlling these two traits could effectively be utilized for diversity analysis among diverse rice genotypes.
| Keywords |
| Oryza sativa; Oryza glaberrima; Heterozygosity; Germplasm; Genotypes |
| Introduction |
| Rice (Oryza spices) is a monocotyledonous plant belonging to the family Granineae and subfamily Oryzoidea. It is cultivated under diverse eco-geographical conditions in various tropical and subtropical countries [1]. Due to its importance as a food crop, rice is being planted on approximately 11% of the Earth’s cultivated land area [2]. It is the grain with the third highest production globally after sugarcane and maize (FAOSTAT, 2012). Oryza sativa and Oryza glaberrima are the only two cultivated species of rice while the other species are wild. Oryza sativa is commonly grown in Asia, North and South America, Europe and Africa. Oryza glaberrima is highly grown in West African but due to higher yields of O.sativa and O. glaberrima-sativa varieties; it is being replaced in most parts of Africa [3]. |
| There are two classes of rice based on starch content, that is, waxy and non waxy rice. Glutinous or waxy rice in which endosperm starch lacks or has very little amylose content consists mainly of amylopectin starch [4]. The ratio of amylose to amylopectin has a major effect on the physical properties of starch. When cooked, the semi-crystalline structure of rice starch is disrupted thus transforming the starch into a softer, edible, and gel-like material [5]. Generally, the amylose content of milled rice is classified into five classes: waxy (0-2%), very low amylose (3-9%), low amylose (10-19%), intermediate amylose (20-24%) and high amylose (above 24%) [6].The cooking temperature at which water is absorbed and the endosperm starch granule swell irreversibly with subsequent loss of crystalline structure is referred to as gelatinization temperature (GT) [7].Gelatinization temperature is an important component of rice cooking quality. Rice grain with low gelatinization temperature takes shorter cooking times leading to significant potential savings in fuel costs [8]. Three classes of GT are recognized in rice breeding programs: high (>74°C), intermediate (70- 74°C), and low (<70°C) [9,10]. Besides waxy and alk genes that controls amylose content and gelatinization temperature in rice, there are other several QTLs within the rice genome that are linked to these two genes. |
| There is a wide range of rice varieties grown both in Kenya and Tanzania. These rice cultivars are either local landraces or improved varieties and they express different levels of amylose and amylopectin that influences amylose content and gelatinization temperature in rice respectively. Since these two traits are key determinant in cooking and eating qualities of rice, unscrupulous traders often blend rice grains which have good cooking and eating quality traits with grains which have poor cooking and eating quality traits based on amylose content and gelatinization temperature to make more profit from their trade. This causes a negative impact on rice trade and consumption. Accurate evaluation of these two traits is difficult and has hindered development of better varieties with good eating and cooking qualities by rice breeders both in Kenya and Tanzania. The various physicochemical methods commonly used to determine amylose content and gelatinization temperature in rice are often inaccurate and time consuming. However, genetic diversity analysis on these selected rice genotypes from Kenya and Tanzania based on amylose content and gelatinization temperature using microsatellite markers has not yet been studied. |
| Molecular markers can have a number of applications in agriculture, and their application in rice improvement has been reviewed [11]. Simple Sequence Repeat (SSR) markers are easily available for any region of the genome. In rice, SSR markers have been effectively utilized for many purposes such as genetic diversity and relatedness [12], QTL mapping [13], mutation analysis [14] and maker assisted selection [15]. This study was carried out to estimate the pattern and level of genetic diversity and relatedness among selected rice genotypes from Kenya and Tanzania along with other germplasm from Philippine based on amylose content and gelatinization temperature. |
| Materials and Methods |
| Plant materials |
| A total of 13 rice genotypes comprising of local landraces and improved rice genotypes were collected from Mwea Irrigation Agricultural Development Centre (MIAD) in Mwea, Kenya and Kilimanjaro Agricultural Research Institute in Moshi, Tanzania. The name, country of origin and category of the rice genotype chosen for the study are given in (Table 1). Genotype IR 64 was used as the check variety since it is known to have both intermediate amylose content and gelatinization temperature [16]. |
| DNA extraction and SSR marker analysis |
| Genomic DNA was extracted from seed samples using a modified CTAB method [17]. The quality of the genomic DNA was determined in a 1% agarose gel in 100ml TBE electrophoresis by running 10 μl of genomic DNA at a voltage of 75 for 45 minutes. DNA purity for each sample solution was evaluated using a spectrophotometer which employed the Thermo Scientific Nano drop 2000 system (Wilmington, USA). A set of 8 microsatellite markers shown in (Table 3) covering different genomic regions of rice were selected from published databased search for rice Simple Sequence Repeats (SSR) markers [18,19]. |
| The PCR reactions were carried out in Thermal cycler (Bio Rad Inc. USA) with the total reaction volume of 25μl containing, 5 μl of genomic DNA, 1X assay buffer, 200 μM of dNTPs, 2 μM MgCl2, 0.2μM of forward and reverse primer and 1 unit of Taq DNA polymerase (Fermentas Life Sciences). The PCR cycles were programmed as 95°C for 2 min, 94oC for 1 min, 55°C 72°C for 2 min for 35 cycles and an additional temperature of 72°C for 10 min for final extension. The amplified products were separated on 1.0 percent agarose gel prepared in 0.5X TBE buffer prestained with 10 μl of ethidium bromide then electrophorized at 100V for 1 hour. The gel was then visualized under UV trans-illuminator and photographs were taken using gel documentation instrument. The PCR products were sized against l00bp DNA ladder (Life sciences-USA). Clearly resolved, unambiguous bands were scored visually for their presence or absence with each primer. The scores were obtained in the form of matrix with ‘1’ and ‘0’, indicating the presence and absence of bands in each genotype respectively. |
| Data management and analysis |
| Using the Power Marker version 3.25 software package [20], the diversity of each accession was analysed on the basis of three statistical parameters: allele number, gene diversity and polymorphism information content (PIC), which measures the genic diversity [21]. Genetic distance was calculated using ‘’C.S Cord 1967’’ distance [22] followed by phylogeny reconstruction using rooted UPGMA as implemented in Power Marker with the tree viewed using Treeview. To visualize the relationship between the sample genotypes among the 13 rice varieties, principle coordinate analysis (PCoA) was conducted using GenALEx 6.5 software. It was chosen to complement the UPGMA cluster analysis. Furthermore, to reveal the partition and variation within and among the genotypes, analysis of molecular variance (AMOVA) was carried out using GenALEx 6.5 statistical software [23]. |
| Results and Discussion |
| Assessment of polymorphism from SSR profiles |
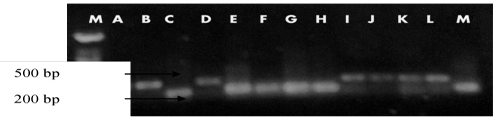
| Out of 12 SSR markers used, only 8 markers were polymorphic and showed consistent banding patterns and amplification of each genotype and were ultimately chosen for assessing genetic diversity among the rice genotypes studied. A total of 22 alleles were detected at the loci of 8 microsatellite markers across 13 rice genotypes. The allelic richness per locus varied from 2 to 4, with an average of 2.75 alleles. RM 141 produced the highest number of polymorphic alleles while RM 125, RM 202, and RM 253 produced the least number of polymorphic alleles as shown in (Table 2). |
| Occurrence of null allele was also observed among some genotypes whenever an amplification product could not be detected in their combination. Experiments detecting null alleles were all repeated at least once to ensure that the absence of an amplified product was not as a result of experimental error. Nineteen SSR loci showed null alleles in two to eight of the 13 rice genotypes. The genotypes having the largest proportion of null alleles were Kahogo and IR 54 (null alleles at four loci) and IR 64 and Supa (null alleles at two loci). The frequency of null allele was not included in the genetic diversity calculations for each SSR locus since they might decrease the apparent heterozygosity in a population leading to deviation of genotypes in a sample from Hardy- Weinberg expectations. |
| Since gene diversity is the measure of expected heterozygosity, the average gene diversity among the 13 rice populations was 0.5503. The gene diversity values ranged from 0.3550 (RM 202) to 0.7337 (RM 141) as shown in Table 3 Polymorphism information content (PIC) value is a measure of polymorphism among varieties for a marker locus used in linkage analysis. The eight SSR markers used in this study produced polymorphic bands as shown in (Figure 1). The PIC value of each marker, which can be evaluated on the basis of its alleles, varied greatly for all tested SSR loci. The values ranged from 0.2920 (RM 202) to 0.6841 (RM 141) with an average of 0.4697 per locus as shown in (Table 2). |
| Pairwise genetic dissimilarity |
| A dissimilarity matrix based on the “C.S Cord 1967” shared SSR alleles was used to determine the level of relatedness among the rice genotypes based on amylose content and gelatinization temperature. The pairwise genetic dissimilarity values as shown in table 3 ranged from 0.9003 to 0.2201 with an average of 0.5627. Wahiwahi and BW 196; and IR 64 and BW 196 exhibited the highest genetic dissimilarity of 90.03% followed by IR 64 and BS 370; Wahiwahi and BS 217; and Kahogo and ITA 310 with dissimilarity of 78.78%. On the other hand, R 2793 and BS 217; Supa and IR 64; R 2793 and ITA 310; Saro 5 and ITA 310; and Saro 5 and R 2794 depicted the lowest genetic dissimilarity of 22.51%. |
| Clustering of rice genotypes |
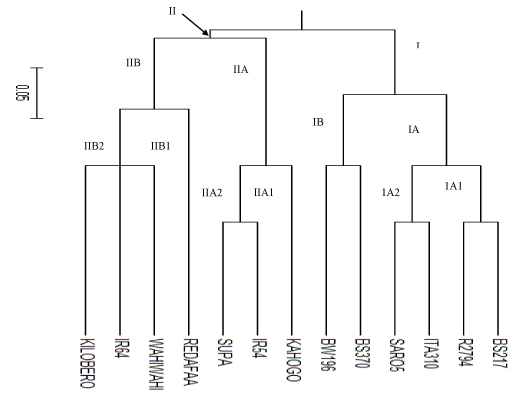
| A rooted UPGMA tree presented in (Figure 2) was constructed using the “C.S Chord 1967” distance values (Cavalli-Sforza and Edwards, 1967) in Power Marker with tree viewed using Treeview software. It revealed genetic relatedness among the 13 rice genotypes based on amylose content and gelatinization temperature using the 8 microsatellite markers. Genotypes that are derivatives of genetically similar types clustered together. The rice genotypes were clustered into two major groups; that is: group I and group II as shown in Figure 2. Group I consisted of the 5 improved genotypes form Kenya and one improved genotype from Tanzania. Group I was further divided into two subgroups; IA and IB. IA was further divided into two small groups; IA1 and IA2. Group IA1 comprised of R 2793 and BS 217 while group IA2 comprised of Saro 5 and ITA 310. Group IB comprised only of BW 196 and BS 370. |
| On the other hand, Group II was comparatively diverse since it consisted of 7 genotypes; 5 local landraces from Tanzania and 2 improved genotypes from Philippine. Group II was divided into two major subgroups; IIA and IIB. Major subgroup IIA was further divided into minor subgroups; IIA1 and IIA2. Group IIA1 consisted only of Kahogo while subgroup IIA2 consisted of Supa and IR 54. The other major subgroup IIB was further divided into two minor subgroups; IIB1 and IIB2. Subgroup IIB1 consisted only of Red Afaa while subgroup IIB2 comprised of Kilombero, IR 64 and Wahiwahi. |
| Principle coordinate analysis (PCoA) |
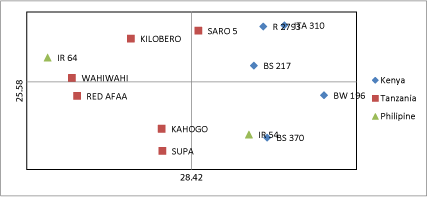
| Principle coordinate analysis (PCoA) based on Nei’s genetic distance (Nei 1972) was used to visualize the genetic relationship among the accessions as shown in (Figure 3). The PCoA supports the results obtained from UPGMA cluster analysis. The first two principle axes accounted for 28.42% and 25.58% variation respectively. The first principle axis comprised of Saro 5, R 2793, ITA 310, and BS 217. The second quadrant comprised of Kilobero, Wahiwahi and IR 64. The third quadrant comprised of Red Afaa, Kahogo and Supa while the forth quadrant comprised of IR 54, BS 370 and BW 196. |
| Analysis of molecular variance (AMOVA) |
| Analysis of molecular variance (AMOVA) was used to determine the proportion of genetic variation partitioned among and within the 13 rice genotypes. Twenty four percent (24%) (P<0.001) of genetic variation partitioned among genotypes and 76% (P<0.001) within the genotypes as shown in (Table 4). |
| Discussion |
| Assessment of genetic diversity is a key factor for germplasm conservation, characterization and breeding. Classical breeding affects genetic diversity by selection of combination of outcomes from diverse allele frequencies and leads to favorable effects and loss of diversity [24]. Little was known about genetic diversity among Kenyan and Tanzanian rice varieties compared to those from Philippine. In the present study, a total of 8 SSR markers were used to assess genetic diversity of 13 rice genotypes. The results indicated a considerable level of genetic diversity among the genotypes used. Different numbers of alleles per locus were detected in all the 8 SSR markers used. This could have been attributed by remnant heterozygosity in some varieties and expected varietal heterogeneity where landrace varieties consist of mixtures of pure lines that contribute to their broad adaptation in traditional farming systems. These results were similar to those reported by Shahid MS et al. [25] using a different group of rice genotypes. In contrast, the average number of alleles per locus obtained in the present study was smaller than that reported in previous studies [26,27]. This difference in average allele per locus might be due to diverse nature of genotypes used by these authors and selection of SSR markers with different scorable alleles. |
| A null allele is a mutant copy of a gene at a locus that completely lacks that gene’s normal function. Occurrence of null alleles in some rice genotypes could have been attributed by chromosomal mutation within a particular gene locus. As a result, it was difficult to amplify the targeted gene sequence using the SSR primer pairs. Similar null allele occurrence was observed by Lapitan VC et al. [28]. |
| Gene diversity is the measure of expected heterozygosity. The genetic diversity mean value of 0.5503 depicted relative heterezygosity based on amylose content and gelatinization temperature among the 13 rice genotypes studied. The average gene diversity value was lower than what was reported by Lapitan VC et al. [28] whom obtained a value of 0.71 but slightly higher than the one obtained by Shahid MS et al. [25].The high mean gene diversity value reported by Lapitan VC et al. [28]. Could be as a result of high rate of exchange of genetic materials among the rice genotypes studied mostly during their genetic improvement. The relative heterozygozity reported in our study could have been attributed by mixing or exchange of genetic materials from different parental lines especially during rice improvement strategies. Conventional breeding of rice by rice breeders is one of major contributory factor to the exchange of genetic material between two lines resulting into a hybrid variety. |
| The level of polymorphism was determined by calculating polymorphism information content (PIC). Polymorphism information content is a measure of allele diversity at a locus. Loci which are highly informative have PIC value > 0.5), reasonably informative loci have PIC values between 0.25 and 0.5, and slightly informative loci have PIC value <0.25. Out of the 8 SSR markers used in this study, only 3 markers (RM 141, RM 225, and RM 434) had a PIC value greater than 0.5. These markers appeared to be highly informative and could therefore be utilized in marker assisted selection of rice genotypes because they are capable of distinguishing between genotypes. The mean PIC value observed in this study was higher than the PIC value of 0.31 recorded by Sivaranjani AKP et al. [29].This indicated that the genotypes used in the present study were more diverse due to differences in origin. |
| Maximum genetic distance values observed in our study between Wahiwahi and BW 196; IR 64 and BW 196 indicated high genetic dissimilarity between them and they showed more divergence. Chromosomal mutation and diverse geographical origin could be the contributory factor for the observed genetic dissimilarity between these rice genotypes. The minimum genetic distance values observed between R 2793 and BS 217; Supa and IR 64; R 2793 and ITA 310; Saro 5 and ITA 310; Saro 5 and R 2794 portrayed less genetic dissimilarity and having a very close relationship. The observed genetic dissimilarity is an indication of a common ancestral origin, or perhaps high rate interbreeding which results to sharing of similar alleles in their genome. This result was comparable with what was reported by Islam MM et al. [30] But slightly higher than what was observed by Shahid MS et al. [25]. A different value of average genetic similarity of 0.79 between 40 rice genotypes was reported by Ravi M et al. [31]. this genetic difference could be due to different group of rice genotypes used. The high level of similarity recorded by this author could be due to the intra specific variation in the germplasm used. |
| On comparing Kenyan and Tanzanian rice genotypes, it was found that Kenyan genotypes were closely related compared to Tanzanian genotypes, with a mean genetic dissimilarity coefficient of 0.3939 against that of 0.5064 for the Tanzanian genotypes. The genetic closeness of Kenyan rice varieties could be as a result of high intra specific variation, evolution from a common ancestry and introgression of similar traits during the time of genetic improvement. On the other hand, the relatively high genetic dissimilarity witnessed among the Tanzanian varieties could be as a result of having diverse ancestral origins, high gene flow caused by cross pollination among these varieties and chromosomal mutations in their genome. |
| Clustering of these genotypes together, for example, BW 196 and BS 370 could be as a result of sharing common ancestry or similar genes were introgressed into their genome during their breeding. Surprisingly, group I genotypes which were all improved varieties were genetically distinct when compared with the IR 64 that was used as a check variety. Therefore, based on these results, it is evident that the six improved varieties studied have different levels of amylose content and gelatinization temperature. Thus further breeding on these genotypes should be carried out so as to introgress favorable genes conferring intermediate amylose content and gelatinization temperature in their genome so as to make them highly competitive in rice market. |
| Supa, a local landrace from Tanzania and IR 54, an improved cultivar with low amylose content from Philippine clustered together. Based on these results, these two genotypes share common alleles for waxy gene responsible for high amylose content and alk alleles associated with low gelatinization temperature. Therefore, Supa genotype does not have good cooking and eating quality characteristics. Genes expressing good quality traits should be introgressed into genome of this genotype. Kilombero, Wahiwahi and IR 64 were clustered together. Factors such as sharing of common ancestry and gene flow caused by interspecific gene transfer could be reason behind clustering together. Clustering of Kilombero and Wahiwahi together with IR 64 is an indication that these two Tanzanian local landraces have good cooking and eating qualities like those of IR 64. |
| Principle coordinate analysis is a method that visualizes similar and dissimilar data. It assigns similar or dissimilar matrix a location in a three dimensional space. It was chosen to complement the UPGMA cluster analysis by visualizing the relationship between the sample genotypes using genetic distances. The Kenyan improved genotypes were clearly separated from Tanzanian local landraces. Genotypes that grouped together were interpreted to have similar characteristics (closely related) while those apart interpreted to be different or distantly related. |
| Analysis of molecular variance revealed percentage variation between and among the Kenyan and Tanzanian rice genotypes used in this study. The high genetic variation within the sample populations could be due to increased gene flow or mutations of a number of repeats of a given genotype for a given SSR. In addition, natural selection mechanism could be another source of this high genetic variation within the rice genotypes studied. On the other hand, the relatively low genetic variation among these rice genotypes could be attributed by sharing of same SSR profiles among themselves. The low genetic variation among these genotypes could explain the probability of sharing a common ancestry despite the fact that they are grown in different countries. Similar huge differences in percentage variation between and among a group of rice genotypes studied using SSR markers. |
| Conclusion |
| The present study provided an overview of genetic diversity based on amylose content and gelatinization temperature among the rice genotypes studied. The UPGMA cluster analysis showed that all 13 rice genotypes could be easily distinguished based on the information generated by the 8 polymorphic SSR markers. The PIC values revealed that RM 141, RM 225, and RM 434 might be the best markers for identification and diversity estimation of rice genotypes. The genetic distance revealed that the Kenyan genotypes had relatively narrow genetic base compared to the Tanzanian genotypes. Therefore, it is highly important not only to conserve these genotypes, but also to reveal their gene pool and unlock other valuable genes for breeding purposes. |
| References |
References
- id="Reference_Titile_Link" value="1">Causse MA1, Fulton TM, Cho YG, Ahn SN, Chunwongse J, et al. (1994) Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138: 1251-1274.
- id="Reference_Titile_Link" value="2">Khush GS1 (2005) What it will take to feed 5.0 billion rice consumers in 2030.Plant Mol Biol 59: 1-6.
- id="Reference_Titile_Link" value="3">Linares OF (2002). African rice (Oryzaglaberrima): History and the future potential. Proceedings of the National Academy of Science of United States of America 99: 16360-16365.
- id="Reference_Titile_Link" value="4">Bertoft E, Koch K (2000) Composition of chains in waxy-rice starch and its structural units. Carbohydrate Polymers 41:121-132.
- id="Reference_Titile_Link" value="5">Ramesh M, Ali SZ, Bhattacharya KR (1999) Structure of rice starch and its relation to cooked-rice texture. Carbohydrate Polymers 38: 337-347.
- id="Reference_Titile_Link" value="6">Suwannaporn P, Pitiphunpong S, Champangern S (2007. Classification of rice amylose content by discriminant analysis of physicochemical properties. Starch-Stärke 59: 171-177.
- id="Reference_Titile_Link" value="7">Cuevas RP, Daygon VD, Corpuz HM, Reinke RF, Waters DL, et al. (2010) Melting the secrets of gelatinization temperature in rice. Functional Plant Biology 37: 439-447.
- id="Reference_Titile_Link" value="8">Fitzgerald MA1, McCouch SR, Hall RD (2009) Not just a grain of rice: the quest for quality.Trends Plant Sci 14: 133-139.
- id="Reference_Titile_Link" value="9">Juliano BO (2003) Rice chemistry and quality. Philippine Rice Research Institute 8-21.
- id="Reference_Titile_Link" value="10">Waters DL1, Henry RJ, Reinke RF, Fitzgerald MA (2006) Gelatinization temperature of rice explained by polymorphisms in starch synthase.Plant Biotechnol J 4: 115-122.
- id="Reference_Titile_Link" value="11">Collard BC1, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century.Philos Trans R SocLond B Biol Sci 363: 557-572.
- id="Reference_Titile_Link" value="12">PervaizZH, Rabbani MA, Khaliq I, Pearce SR, Malik SA (2010) Genetic diversity associated with agronomic traits using microsatellite markers in Pakistani rice landraces. Electronic Journal of Biotechnology 13: 4-5.
- id="Reference_Titile_Link" value="13">Guo Y, Cheng B, Hong D (2010) Construction of SSR linkage map and analysis of QTLs for rolled leaf in Japonica Rice. Rice Science 17: 28-34.
- id="Reference_Titile_Link" value="14">Wang Q1, Sang X, Ling Y, Zhao F, Yang Z, et al. (2009) Genetic analysis and molecular mapping of a novel gene for zebra mutation in rice (Oryza sativa L.).J Genet Genomics 36: 679-684.
- id="Reference_Titile_Link" value="15">Thomson MJ (2009) Marker-assisted breeding for abiotic stress tolerance in rice: Progress and future perspectives. Metro: 43-55.
- id="Reference_Titile_Link" value="16">Khush GS, Virk PS (2005) International Rice varieties and their impact. International Rice Research Institute, Manilla 115-135.
- id="Reference_Titile_Link" value="17">Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13-15.
- id="Reference_Titile_Link" value="18">Panaud O1, Chen X, McCouch SR (1996) Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.).Mol Gen Genet 252: 597-607.
- id="Reference_Titile_Link" value="19">McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K et al. (2002). Development and Mapping of 2240 New SSR Markers for Rice (Oryza sativa L.). DNA Research 9: 199-207.
- id="Reference_Titile_Link" value="20">Liu K1, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis.Bioinformatics 21: 2128-2129.
- id="Reference_Titile_Link" value="21">Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms.Am J Hum Genet 32: 314-331.
- id="Reference_Titile_Link" value="22">Cavalli-Sforza LL, Edwards AW (1967) Phylogenetic analysis. Models and estimation procedures.Am J Hum Genet 19: 233-257.
- id="Reference_Titile_Link" value="23">Peakall R, Smouse P (2009) GenALEx6.5. Gene analyses in excel. Population genetic software for teaching and research. Molecular Ecology Notes 28: 2537-2539.
- id="Reference_Titile_Link" value="24">Singh RK, Sharma RK, Singh AK, Singh VP, Singh NK et al. (2004). Suitability of mapped sequence tagged microsatellite site markers for establishing distinctness, uniformity and stability in aromatic rice. Euphytica, 135: 135-143.
- id="Reference_Titile_Link" value="25">Shahid MS, Shahzad AN, Muhammad A (2013) Genetic diversity in basmati and non-basmati rice varieties based on microsatellite markers. Pakistan Journal of Botany 45: 423-431.
- id="Reference_Titile_Link" value="26">Kuroda Y, Sato YI, Bounphanousay C, Kono Y, Tanaka K (2007) Genetic structure of three Oryza AA genome species (O. rufipogon, O. nivara and O. sativa) as assessed by SSR analysis on the Vientiane Plain of Laos. Conservation Genetics 8: 149-158.
- id="Reference_Titile_Link" value="27">Shefatur RM, Rezwan MM, Samsul AM, Rahman L (2009) DNA fingerprinting of rice (Oryza sativa L.) cultivars using microsatellite markers. Australian Journal of Crop Science 3: 122-128.
- id="Reference_Titile_Link" value="28">Lapitan VC, Brar DS, Abe TE, Redoña ED (2007) Assessment of genetic diversity of Philippines rice cultivars carrying good quality traits using SSR markers. Breeding Science 57: 263-270.
- id="Reference_Titile_Link" value="29">Sivaranjani AKP, Manish K, Pandey I, Sudharshan G, Kumar GR, et al. (2010) Assessment of genetic diversity among basmati and non-basmati aromatic rice of India using SSR markers. Research communications 99: 2-25.
- id="Reference_Titile_Link" value="30">Islam MM, Ali MS, Prodhan SH (2012) SSR marker-based molecular characterization and genetic diversity analysis of aromatic landreces of rice (Oryza sativa L.). Bioscience and Biotechnology 1:107-116.
- id="Reference_Titile_Link" value="31">Ravi M, Geethanjali S, Sameeyafarheen F, Maheswaran M (2003). Molecular marker based genetic diversity analysis in rice (Oryza sativa L.) using RAPD and SSR markers. Euphytica 133: 243-252.
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 | Table 4 |
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 |
Relevant Topics
- Agricultural science
- Agronomy
- Climate impact on crops
- Crop Productivity
- Crop Sciences
- Crop Technology
- Field Crops Research
- Hybrid Seed Technology
- Irrigation Technology
- Organic Cover Crops
- Organic Crops
- Pest Management
- Plant Genetics
- Plant Breeding
- Plant Nutrition
- Seed Production
- Seed Science and Technology
- Soil Fertility
- Weed Control
Recommended Journals
Article Tools
Article Usage
- Total views: 15661
- [From(publication date):
December-2015 - Aug 29, 2025] - Breakdown by view type
- HTML page views : 10725
- PDF downloads : 4936
Peer Reviewed Journals
Make the best use of Scientific Research and information from our 700 + peer reviewed, Open Access Journals
