Research Article Open Access
Identification of an Alpha-Tubulin Gene from the Chinese Mitten Crab Eriocheir sinensis: Expression Profiles under Immune Challenge and during Larval Development, Autoimmune hepatitis, Immune Cell Therapy, Auto immune Disease
Peng Li*, Xianfeng Jiang, Bin Sun, Yubo Lin, Jie Yan, Kaiya Zhou*Jiangsu Key Laboratory for Biodiversity and Biotechnology and Jiangsu Key Laboratory for Aquatic Crustacean Diseases, College of Life Sciences, Nanjing Normal University, Nanjing 210023, China
- Corresponding Author:
- Peng Li
Jiangsu Key Laboratory for Biodiversity and Biotechnology and Jiangsu Key Laboratory for Aquatic Crustacean Diseases
College of Life Sciences, Nanjing Normal University, Nanjing 210023, China
Tel: +86-25-85891667
Fax: +86-25-85891786
E-mail: biolipeng@163.com - Kaiya Zhou
Jiangsu Key Laboratory for Biodiversity and Biotechnology and Jiangsu Key Laboratory for Aquatic Crustacean Diseases
College of Life Sciences, Nanjing Normal University, Nanjing 210023, China
Tel: +86-25-85891667
Fax: +86-25-85891786
E-mail: kyzhounj@126.com
Received date: August 31, 2015; Accepted date: September 23, 2015; Published date: September 30, 2015
Citation: Li P, Jiang X, Sun B, Lin Y, Yan J, et al. (2015) Identification of an Alpha- Tubulin Gene from the Chinese Mitten Crab Eriocheir sinensis: Expression Profiles under Immune Challenge and during Larval Development. J Biotechnol Biomater 5:201. doi:10.4172/2155-952X.1000201
Copyright: © 2015 Li P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
In this study, an alpha-tubulin gene (EsTUBA, GenBank: KJ509188) was isolated and identified from the Chinese mitten crab Eriocheir sinensis. The EsTUBA mRNA expression patterns under larval brachyurization development and immune challenge stress by Vibrio parahaemolyticus injection in adult crabs were evaluated. EsTUBA cDNA was 1,695 base pairs (bp) with an open-reading frame of 1,356 bp encoding 451 amino acids (49.95 kDa) and containing GTPase domain and C-terminal Tubulin domain. Sequence alignment showed that EsTUBA shared 95-98% identity and exactly similar structural features with its counterparts reported in other animals. Phylogenetic analysis indicated that EsTUBA clustered into one group together with Tubulins from other crustaceans. Sequence alignment, structure comparison and bioinformatic analyses revealed that EsTUBA is member of the Tubulin family. EsTUBA was fluctuant expressed in all tested larval development stages and various organs, and the levels of EsTUBA mRNA expressed in hepatopancreas, gill and intestine, are significantly expression fluctuated and induced after bacteria V. parahaemolyticus injection, while in heart, is almost consistently expressed at 0, 6, 12, 24, 36 and 48 h post-injection compared to control. The molecular cloning of EsTUBA gene is important for further study on the function of this gene.
Keywords
Chinese mitten crab; Alpha-tubulin; cDNA cloning; Sequence analysis; mRNA expression
Introduction
The cytoskeleton of eukaryotic cells is comprised of microtubules (MTs), intermediate filaments and microfilaments. MTs are hollow cylindrical polymers built through the lateral association of protofilaments composed of longitudinally aligned head-to-tail αβ- tubulin dimmers [1,2]. Tubulin is globular protein and consists of several distinct families, including the alpha (α), beta (β), gamma (γ), delta (δ), epsilon (ε), zeta (ζ), iota (ι), eta (η), theta (θ) and kappa (κ) tubulin [3,4]. Tubulins as important component of the MTs in the cytoskeleton have been well known for a long time [5-8]. The tubulin family builds the MTs that play highly diverse and essential roles in eukaryotic cells, including cell division and elongation [9-11], maintaining cell structure [9,12], vesicle transport [11], signal transduction [4], etc. During the last decade, many studies on the structure and function of tubulins and their interaction with other proteins have been reported [8,13]. Functional conservation of tubulins across kingdoms is supported by copolymerization of heterologous or chimeric α-tubulin and β-tubulin, either in vitro [14] or in vivo [15,16]. Study on α-tubulin in Japanese pine sawyer Monochamus alternatus indicated that α-tubulin is also very important in the animal development [17]. And in recent years, tubulins have been shown associated with post-transcriptional regulation [18], mitochondrial metabolism and arrangement [7,8], ependymal cells formation and axonic elongation of neuroblasts [19], knowledge about the role of tubulins have widened.
The Chinese mitten crabs (Eriocheir sinensis: family Grapsidae) are limnic and intertidal crabs distributed in Asia and are one of the major groups of brachyuran crabs in the world. They undergo brachyurization metamorphosis during the early physiological stages characterized by a reduced abdomen, folded beneath the cephalothorax, and inserted between the pereiopods or in a special cavity. This crab produced in the coastal rivers and estuaries of the Yellow Sea is a traditional savory food, and therefore is one of the most important cultivated aquatic species in China [20]. Due to the rapid development of crab farming, the country's crab production reached about 650,000 tons in 2012. Various diseases caused by viruses, bacteria, etc., have also begun to emerge and have resulted in enormous losses. Up to now, some immune-related genes of E. sinensis, such as C-type lectin [21], Hsp70 [22], Hsp90 [22,23], and secreted ferritins [24] etc., has been reported. But remarkably little is known about crustacean tubulins and how they involved in innate immune responses. At this time it is still unclear whether the tubulin gene (EsTUBA) expression of the Chinese mitten crab is related to the physiological stages in which brachyurization occurs, and plays role in the immune system of E. sinensis. In this study we have cloned the full-length cDNA of EsTUBA from E. sinensis and examined expression profiles in association with larval development and immune challenge stress. Due to association of α-tubulin with cell division [9,11] and immunoprotection [25], studies on the expression regulation of this gene would be critical to understand its role during the brachyurization development and immune challenge stress in the economically important crabs.
Materials and Methods
Sample preparation and animal immune challenge
Embryo (stage O, embryo stage; heart beat is about 90 times/min) and nine stages of the larvae whole body tissue samples (stage Z1, the first zoeal stage; stage Z2, the second zoeal stage; stage Z3, the third zoeal stage; stage Z4, the fourth zoeal stage; stage Z5, the fifth zoeal stage; stage M, megalopa stage; stage J1, the first juvenile crab stage; stage J2, the second juvenile crab stage; stage J3, the third juvenile crab stage) of E. sinensis were collected from an aquaculture hatchery. The samples (Supplementary Table S1) were dissected and preserved in RNAlater RNA Stabilization Reagent (QIAGEN, Germany), cut into tiny particles (less than 0.2 mm thick) in separate tubes with small size surgical scissors, kept overnight at +4ºC, and finally stored at −20ºC before total RNA isolation.
Healthy adult Chinese mitten crabs (n=120; 90 ± 10 g wet weight) were collected from an aquaculture farm. The crabs were acclimated for two weeks at 22 ± 3ºC in filtered, aerated freshwater before bacterial infection experiment. Vibrio parahaemolyticus (Collector number: 1.1614) were purchased from China General Microbiological Culture Collection Center (CGMCCC). After rejuvenation, the strain was static cultured in Luria Bertani (LB) medium at 28ºC for 24 h. T?hen the bacteria were washed off and diluted into 1 × 107 colony-forming units (CFU)/ml in 0.1 M phosphate buffer solution (PBS, pH 7.4). One hundred and twenty crabs were divided into three groups. Forty-five crabs were injected into the arthrodial membrane of the last pair of walking legs with live V. parahaemolyticus (approximately 1.0 × 107 CFU/100 g body weight) and introduced into clean tanks. Meanwhile, forty-five crabs were injected with PBS (0.1 mol·L-1/100 g body weight) as control. Additionally, thirty crabs were normal feeding only and without any processing as blank control. Procedures adopted in this study were approved by the Institutional Animal Care and Use Committee of the Nanjing Normal University (SOXR (Jiangsu) 2012- -004). Five crabs were randomly sampled at each time interval of 0, 6, 12, 24, 36 and 48 h after injection of V. parahaemolyticus and PBS. Various tissues samples (heart, gill, hepatopancreas and intestine) were collected from infected and control crabs (Supplementary Table S1 online). The samples were harvested as described above and stored at −20ºC before total RNA isolation.
Total RNA extraction and cDNA synthesis
Total RNA was extracted separately from E. sinensis tissues using the RNeasy Mini Kit (QIAGEN, Germany) according to the manufacturer’s protocol. The total RNA concentration and quality were estimated using spectrophotometry at an absorbance at 260 nm in a biophotometer (Eppendorf, Germany) and agarose-gel electrophoresis, respectively. The cDNA used to obtain 5'- and 3'-end sequences of EsTUBA gene was synthesized using 5' RACE System for Rapid Amplification of cDNA Ends, Version 2.0 (Invitrogen, CA, USA) and SMART™ RACE cDNA Amplification Kit (Clontech, CA, USA) with SuperScript II RT (Invitrogen, CA, USA) according to the user’s manual. The synthesized cDNA was stored at −70ºC until used.
Cloning of full-length cDNA sequence
The full-length cDNA sequence of EsTUBA gene was made up of 5' RACE segments, expressed sequence tag (EST) segments and 3' RACE segments. Based on ESTs (355 base pairs (bp), dbEST_Id 62013217 and GenBank accession number GE329293; 167 bp, dbEST_Id 62013221 and GenBank accession number GE329297) sequence information obtained from a subtractive hybridization cDNA Library of E. sinensis [26]. New gene-specific primers (Table 1) were designed and synthesized to obtain full-length cDNA sequence. Primers including Abridged Anchor Primer (10 μM), Anchor Primer (10 μM), Abridged Universal Anchor Primer (AUAP, 10 μM) or Universal Amplification Primer (UAP, 10 μM), and EsTUBA-GSP1/EsTUBA-GSP2/EsTUBAGSP3 for EsTUBA were employed to amplify the 5'-ends of each cDNA, while 10 × Universal Primer A Mix (UPM) and EsTUBA-31/ EsTUBA-32 for EsTUBA were employed to obtain the 3'-ends of each cDNA. The PCRs for 5'-RACE reactions were carried out respectively in a 50 μl reaction volume: (1), the primary PCR containing 5 μl 10 × PCR buffer, 3 μl MgCl2 (25 mM), 1 μl dNTP Mix (10 mM), 2 μl Abridged Anchor Primer (10 μM), 31.5 μl sterilized water, 0.5 μl Taq DNA polymerase (5 units/μl), 2 μl gene-specific primer (10 μM) and 5 μl of RACE-Ready cDNA, and finally overlay with 50 to 100 μl of mineral oil. (2), the nested PCR containing 5 μl 10 × PCR buffer, 3 μl MgCl2 (25 mM), 1 μl dNTP Mix (10 mM), 1 μl AUAP or UAP (10 μM), 33.5 μl sterilized water, 0.5 μl Taq DNA polymerase (5 units/μl), 1 μl genespecific primer (10 μM) and 5 μl dilution of primary PCR product, and finally overlay with 50 to 100 μl of mineral oil. PCR conditions for RACE were as follows: initial denaturation at 94ºC for 1-2 min, 30- 35 cycles of 94ºC for 0.5-1 min, 55ºC for 0.5-1 min and 72ºC for 1-2 min. Extra extension at 72ºC for 5-7 min, indefinite hold at 5ºC until products are removed. The PCRs reactions with QC304F/QC304R were employed respectively to amplify the open reading frame (ORF) of EsTUBA cDNA. The PCR products were separated on a 2% agarose gel, and bands with expected sizes were cut from the gel and purified using a gel purification kit (Ayxgen, China). The freshly purified PCR products were cloned into the pMD® 18-T Vector (TaKaRa, China), and transformed into competent Escherichia coli cells DH5α (TaKaRa, China). The positive transformants were picked using a blue-white screen, and inserts were verified by PCR. Three of the positive clones were sequenced on an ABI PRISM 3730 Automated Sequencer using BigDye terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). The sequences obtained after 5'- and 3'-RACE were assembled using DNAStar Lasergene 7.1 software to generate full length cDNA.
| Application | Primer | Primer sequence (5’→3’) | Amplicon length (bp) |
| 5'-RACE | 5'RACE Abridged Anchor Primer AUAP UAP 5'RACE Anchor Primer |
5'-GGCCACGCGTCGACTAGTACGGGGGGGGGG-3' CUACUACUACUAGGCCACGCGTCGACTAGTAC-3' 5'-CUACUACUACUAGGCCACGCGTCGACTAGTACGGGGGGGGGG-3' |
|
| EsTUBA-GSP1 | 5'-ATCTGGTTTGCGGGCT-3' | 1166 bp | |
| EsTUBA-GSP2 | 5'-AAGCAGGCGTTGGTGATCTCA-3' | 1148 bp | |
| EsTUBA-GSP3 | 5'-GATAACGGGAGCGTAGGTCAC-3' | 1089 bp | |
| 3'-RACE | 3'-RACE 10 × Universal Primer A Mix (UPM) | 5'-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3' 5'-CTAATACGACTCACTATAGGGC-3' |
|
| EsTUBA-31 EsTUBA-32 | 5'-CATCCACTTCCCTCTGGTGACCTACGCT-3' 5'-GCTGTCTGTGTCTGAGATCACCAACGCC-3' |
643 bp 580 bp |
|
| ORF full-length confirmation semi-quantitative RT-PCR Real-time qPCR |
QC304F QC304R |
5'-CAAAATGCGTGAGTGCATCTC-3' 5'-GATTTAATACTCCTCTCCCTC-3' |
1363 bp |
| EsTUBAF EsTUBAR β-actin-F β-actin-R QEsTUBA-F QEsTUBA-R Qβ-actin-F Qβ-actin-R |
5'-TCACTGCCTCCCTGAGATTC-3' 5'-AATGGTCCTCTTGGTCTTGATG-3' 5'-CCGTGACCTCACAGACTACCT-3' 5'-CGGTGGTCGTGAAGGTGTAG-3' 5'-GATCGTGTCCTCCATCACTG-3' 5'-GTCACCAGAGGGAAGTGGAT-3' 5'-CTCCTGCTTGCTGATCCACATC-3' 5'-GCATCCACGAGACCACTTACA-3' |
311 bp 65 bp 114 bp |
Table 1: Primers used for gene cloning, semi-quantitative RT PCR and real-time quantitative PCR quantification (F mean forward primer and R mean reverse primer).
Expression profile of EsTUBA gene
Real-time quantitative PCR (RT-qPCR) was used to investigate the mRNA expression levels of EsTUBA in various tissues, including heart, gill, hepatopancreas, intestine and different larval physiological stages, as well as the temporal expression of EsTUBA transcripts in heart, gill, hepatopancreas, and intestine of crabs challenged with V. parahaemolyticus at 0, 6, 12, 24, 36 and 48 h. Meantime, semiquantitative reverse transcription PCR (RT-PCR) was used to investigate the EsTUBA expression pattern in different physiological stages.
Total RNA from various samples (collected tissue samples see Supplementary Table S1) was isolated using RNeasy Mini Kit (QIAGEN, Germany). The cDNA first-strand synthesis was carried out based on PrimeScript RT Reagent usage information (TaKaRa, China) using total RNA treated with DNase I (TaKaRa, China) as template. cDNA mix was diluted to 1:10 with Milli-Q water (Toyobo, Japan) and stored at −20ºC until used as a template for subsequent RT-qPCR and semiquantitative reverse transcription PCR. All primers (Table 1) used for RT-qPCR and RT-PCR were designed according to the corresponding sequences of E. sinensis. Gene-specific primers QEsTUBA-F and QEsTUBA-R for RT-qPCR were selected to amplify a 114bp product for EsTUBA. A constitutive expression gene, β-actin gene, was used as an internal control. Two β-actin primers Qβ-actin-F and Qβ-actin-R [27] were used to amplify a fragment of the Chinese mitten crab β-actin gene. Negative controls missing cDNA template were included in this experiment. The RT-qPCR assay was carried out using SYBR Premix Ex Taq™ II (TaKaRa, Japan) with a Rotor-Gene® Q (QIAGEN, Germany) real-Time PCR instrument. The real-time fluorescent quantitative PCR assay was carried out in triplicate on 96-well plate or 20 μl quantitative reaction volume per well containing 10 μl 2 × SYBR® Premix Ex Taq II (TaKaRa, Japan), 0.8 μl PCR forward primer (10 μM), 0.8 μl PCR reverse primer (10 μM), 2.0 μl cDNA temple, and 6.4 μl sterile deionized water. The thermal profile for SYBR Green RT-qPCR consisted of an initial step at 95ºC for 30 s, followed by 40 cycles of 95ºC for 5 s and 60ºC for 30 s. As the preliminary trial of the experiment had shown that β-actin gene had a steady expression in this experimental species, it was used as the housekeeping gene in all RT-qPCR assays [23,27]. RT-PCR primers (EsTUBAR and EsTUBAF, Table 1) were selected to produce a 311 bp amplicon. RT-PCR was performed in a final volume of 30 μl, and contained 15 μl of 2 × Taq PCR Master Mix (Lifefeng, China), 0.7 μl of 20 mM primer, 12.2 μl of sterile deionized water and 1.4 μl of first strand cDNA as template. PCR conditions were as follows: 25 cycles at 95ºC for 30 s, 54ºC for 30 s, and 72ºC for 50 s. Internal control PCR reactions for β-actin were performed in a separate tube, as described above with the exception of an alternative gene-specific primer pair (β-actin-R and β-actin-F, Table 1), which was designed based upon a cloned E. sinensis β-actin cDNA fragment (GenBank accession No. AY910691) to produce a 65 bp amplicon. Three biological replicates for each sample and three technical replicates were performed. RT-PCR products were size separated on an ethidium bromide stained 1.5% agarose gel, visualized under ultraviolet light, and images were captured with UVP DS7500 Gel imaging system (Upland, USA).
Then, the RT-qPCR data were analyzed with Rotor-Gene® Q Series Software v1.7.94 (QIAGEN, Germany). To maintain consistency, the baseline was set automatically by the software. The relative expression level of EsTUBA was calculated using the 2-ΔΔCt method [28]. The Ct for the target amplified EsTUBA, and the Ct for the internal control β-actin were determined for each sample. The Ct values for each set of three reactions were averaged for all subsequent calculations. Each data represents the average of three experiments. The results were subjected to one way analysis of variance (one-way ANOVA), followed by Tukey’s post hoc Duncan multiple range tests. All data were represented as mean ± standard error (S.E.) and p<0.05 was considered to be statistically significant.
Sequences analyses of EsTUBA gene
The open reading frame (ORF) of EsTUBA cDNA was determined using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/). Molecular mass and theoretical isoelectric point was predicted using Compute pI/Mw tool (http://www.expasy.org /tools/pi_tool.html). The EsTUBA amino acid sequence, plus other tubulin amino acid sequences retrieved from the National Center for Biotechnology Information (NCBI) protein database, was subjected to MAFFT version 7 (http://mafft.cbrc.jp/alignment/server/index.html) for multiple sequence alignment, respectively. Motif scan was respectively performed against databases of motifs (http://myhits.isb-sib.ch/cgi-bin/motif_scan) and by Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/). The putative signal peptide was predicted using SignalP 4.0 (http://www.cbs.dtu.dk/services/SignalP). The potential protein subcellular localization was predicted on the PRORT II (http://psort.ims.u-tokyo.ac.jp/form2.html). The secondary structure was predicted using SOPMA program (http://npsa-pbil.ibcp.fr/). Threedimensional domain structure of EsTUBA was predicted by SWISSMODEL Server (http://www.expasy.org/swissmod/SWISS-MODEL.html). Additional assessments of domain structures were performed on ProSA-Web (https://prosa.services.came.sbg.ac.at/prosa.php) and Verify3D Structure Evaluation Server (http://nihserver.mbi.ucla.edu/Verify_3D/). Transmembrane topology prediction was performed using TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and DAS-TMfilter server (http://mendel.imp.ac.at/sat/DAS/). The potential N-linked glycosylation sites were predicted according to the Asn-X-Ser/Thr rule (http://cbs.dtu.dk/services/NetNGlyc/). Proteinprotein interaction analysis was performed on STRING v.9.1 (http://string.embl.de/).
Multiple sequence alignment and phylogenetic analyses
The full-length cDNA sequence was subjected to homology analysis. Similarity search of amino acid sequences in database were conducted with BLAST programs [29] at the National Center for Biotechnology Information. The resulting sequences were verified and subjected to cluster analysis. Multiple sequences alignment was performed with the MAFFT version 7 Multiple Alignment program. The common names, species names of the taxa and the accession numbers of the amino acid sequences used to perform multiple sequence alignment and phylogenetic analyses are listed in ‘Supplementary Table S2’ online. The alignment was analyzed with the program Gblocks 0.91b [30], using default settings to select conservative regions of the putative amino acids. Model selection for the nucleotide dataset was performed with Modeltest version 3.7 [31]. The maximum likelihood (ML) amino acid phylogeny was reconstructed using PHYML [32]. Bayesian inferences (BI) of amino acid dataset was performed with MrBayes 3.0b4 [33] using the WAG+G model. Then unrooted phylogenetic trees were constructed with these programs, respectively.
Results
Identification and characterization of full-length cDNA encoding EsTUBA gene
The EsTUBA cDNA includes a 5'-terminal untranslated region (UTR) of 261 bp, a 78 bp 3'-terminal UTR and a 1,356 bp ORF encoding a 451 amino acid polypeptide with a predicted molecular mass of 49.95 kDa and theoretical isoelectric point of 4.89. Stop codon TAA, canonical polyadenylation signal (AATAAA located at nucleotide 1660-1665), and poly-(A) tail were found in the 3' end of EsTUBA cDNA and an important conserved structural domain (GGGTGSG) of EsTUBA was identified in the deduced amino acid sequence (Supplementary Figure S1). The C+G content in the sequence is 56.76%. Multiple alignments of EsTUBA with alpha-tubulin genes of other taxa showed highly conserved nature (Supplementary Figure S2). The BLAST results (Supplementary Table S2) of the deduced amino acid sequence of EsTUBA showed that the sequence obtained was, compared to other organisms, closest to American lobster (GenBank accession No. Q94572). EsTUBA shared significant homology at the protein (95–98%) level with the alphatubulin sequences reported in American lobster (98%), common water flea (96%), fruit fly (96%), mallard (96%), owl limpet (95%), etc. (Supplementary Table S2). The conservative characteristics and high similarity with known alpha-tubulin informed us that EsTUBA belonged to the α-tubulin gene family. A 1,695 bp nucleotide sequence, representing the full-length cDNA sequence of EsTUBA, was obtained by cluster analysis of the RACE-PCR amplification fragments and the EST sequences. The complete cDNA sequence was deposited in the GenBank database under the accession number KJ509188.
Phylogenetic analysis using alpha-tubulin amino acid sequences
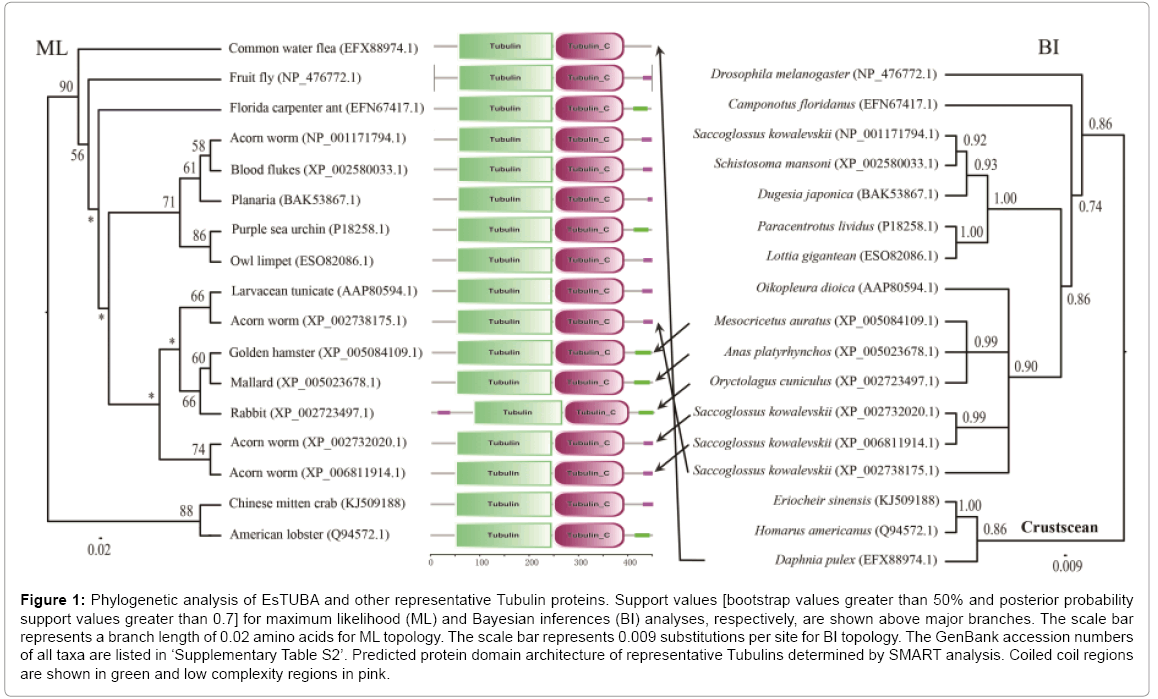
The phylogenetic analyses of the alpha-tubulin amino acid dataset generated similar tree topologies for BI and ML analyses. The results revealed that EsTUBA first clustered with crustacean tubulins, especially Homarus americanus tubulin (HaTUBA) and then with other tubulins (Figure 1).
Figure 1: Phylogenetic analysis of EsTUBA and other representative Tubulin proteins. Support values [bootstrap values greater than 50% and posterior probability support values greater than 0.7] for maximum likelihood (ML) and Bayesian inferences (BI) analyses, respectively, are shown above major branches. The scale bar represents a branch length of 0.02 amino acids for ML topology. The scale bar represents 0.009 substitutions per site for BI topology. The GenBank accession numbers of all taxa are listed in ‘Supplementary Table S2’. Predicted protein domain architecture of representative Tubulins determined by SMART analysis. Coiled coil regions are shown in green and low complexity regions in pink.
Structural analyses of EsTUBA gene
No signal sequence was identified in the EsTUBA transcript using the SignalP software. Results from BLASTp in NCBI indicated that EsTUBA protein has clearly alpha-tubulin domain (Supplementary Figure S2-A). Many important sites, contain two N-glycosylation sites (128-131 and 380-383), one Amidation site (161-164), one Cell attachment sequence (320-322), one Tubulin subunit alpha, beta, gamma signature (142-148) and six Protein kinase C phosphorylation sites (38-40, 58-60, 94-96, 334-336, 337-339) etc., are located in GTPase domain and C-terminal domain (Supplementary Figures S1, S2, S3-A and S3-B). The predicted secondary structure by SOPMA indicated that EsTUBA had 42.79% α-helix, 30.60% random coil, 17.96% extended strand, and 8.65% β-turn. The three-dimensional structure analysis of EsTUBA revealed that twelve β-sheets are covered inside by fifteen α helices, two clearly distinguishable domains (GTPase domain and C-terminal domain) attached to each other by a relatively flexible loop (Figure 1 and Supplementary Figures. S2-A, S3-A and S3-B). Overall model quality that was −10.25 to Z-score and local model quality values were below zero generally. The averaged 3D-1D score is distributed generally from 0.14 to 0.96. Protein motif domains scanning from SMART diagram database indicated EsTUBA contains two domains (Tubulin, from 49 to 246 amino acid residue; Tubulin-C, from 248 to 393 amino acid residue) and coiled coil (from 415 to 443 amino acid residue), and it shares similar domain structural features with tubulin sequences reported in other animals (Figure 1). Transmembrane topology prediction indicated that EsTUBA is probably a nontransmembrane protein. The potential protein subcellular localization prediction indicated that EsTUBA is likely to locate in cytoplasm (52.2%), nucleus (17.4%), cytoskeleton (17.4%) and mitochondrion (4.3%), peroxisomal (8.7%). The prediction from SignalP program indicated that EsTUBA is non-secretory protein. The protein-protein interactions prediction from STRING indicated that EsTUBA possess similar protein-protein interaction model to tubulin from orangutan (Pongo pygmaeus). EsTUBA have close interactions with Tubulinfolding cofactor E, Tubulin-folding cofactor D, Tubulin-folding cofactor B, Von Hippel-Lindau-binding protein 1, and Tubulin-folding cofactor A, etc. (Supplementary Figures S3-C, S3-D, and S3-E).
Expression patterns of EsTUBA gene in response to brachyurization development and immune challenge stress
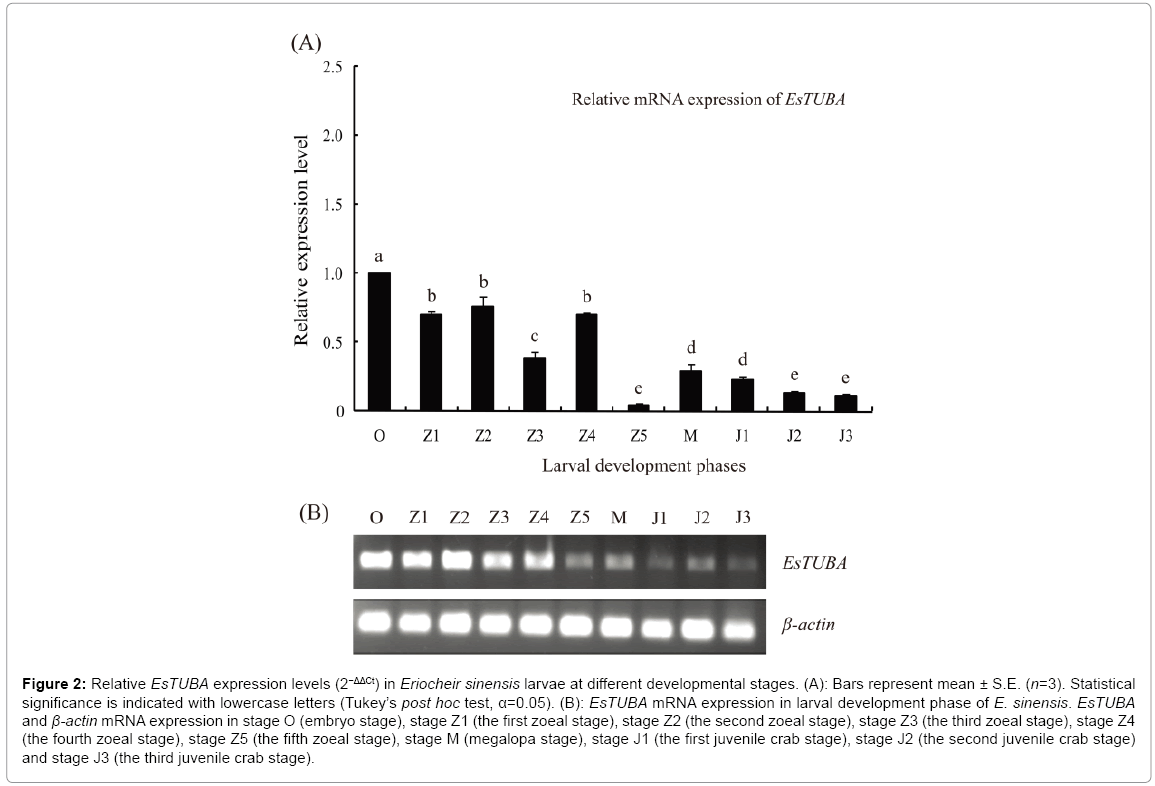
RT-PCR analyses showed that EsTUBA is largely distributed in stage O, stage Z1, stage Z2, stage Z3 and stage Z4 but significantly reduced in stage Z5, stage M, stage J1, stage J2 and stage J3. Internal control constitutive expression gene, β-actin, consistently expressed in same levels at different juvenile stages (Figure 2B). In addition, RT-qPCR analysis exhibited an expression pattern consistent with the former during brachyurization development. Expression levels were highest at stage O and lowest at stage Z5 (Figure 2A).
Figure 2: Relative EsTUBA expression levels (2−ΔΔCt) in Eriocheir sinensis larvae at different developmental stages. (A): Bars represent mean ± S.E. (n=3). Statistical significance is indicated with lowercase letters (Tukey’s post hoc test, α=0.05). (B): EsTUBA mRNA expression in larval development phase of E. sinensis. EsTUBA and β-actin mRNA expression in stage O (embryo stage), stage Z1 (the first zoeal stage), stage Z2 (the second zoeal stage), stage Z3 (the third zoeal stage), stage Z4 (the fourth zoeal stage), stage Z5 (the fifth zoeal stage), stage M (megalopa stage), stage J1 (the first juvenile crab stage), stage J2 (the second juvenile crab stage) and stage J3 (the third juvenile crab stage).
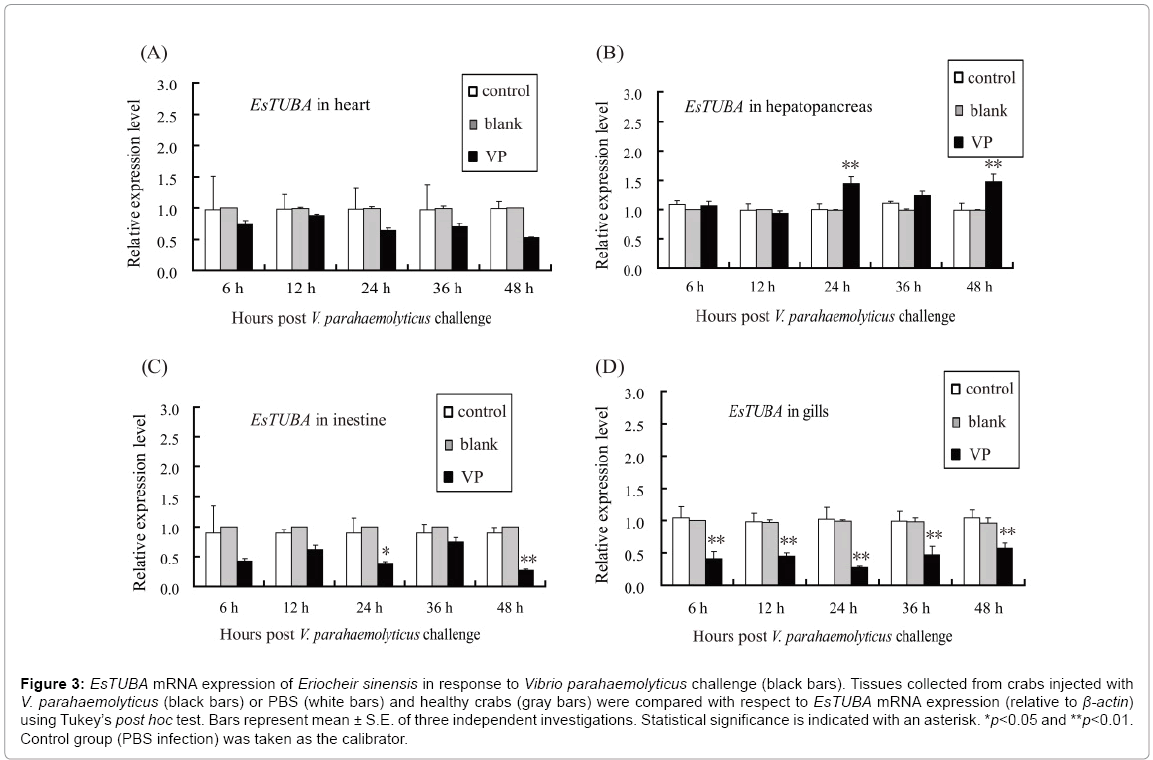
Moreover, RT-qPCR was used to analyze the temporal expression of EsTUBA in the heart, hepatopancreas, intestine, and gill when challenged with V. parahaemolyticus. The levels of EsTUBA mRNA expressed in hepatopancreas, intestine and gill, were significantly induced by V. parahaemolyticus (VP) at 6 h–48 h post-injection compared to controls (Figures 3A-3D, 4A-4F). The result showed that EsTUBA in heart was not significantly induced and only slightly upregulated or downregulated from 6 h to 48 h after V. parahaemolyticus challenge (Figure 3A). By contrast, EsTUBA in hepatopancreas was slightly upregulated from 24 h to 48 h post-injection, peaked at 48 h after challenge (Figure 3B). EsTUBA in intestine was rapidly down-regulated after VP challenge, slightly upregulated at 36 h and then reached the lowest level at 48 h post-injection (Figure 3C). In addition, EsTUBA in gill exhibited an expression pattern similar to it in intestine. EsTUBA was signif?icantly down-regulated after challenge, reach the lowest level at 24 h postinjection and then it was upregulated significantly (p<0.05) at 48 h postinjection (Figure 3D). Significant differences (p<0.05) were detected at nearly all time points between control and challenge groups, except for 6 h, 12 h, and 36 h after VP challenge in hepatopancreas, intestine and whole time after VP challenge in heart. Control reactions, in which PBS was used for induction or blank, yielded no significant variation in expression levels of EsTUBA (white and gray bars in Figure 3).
Figure 3: EsTUBA mRNA expression of Eriocheir sinensis in response to Vibrio parahaemolyticus challenge (black bars). Tissues collected from crabs injected with V. parahaemolyticus (black bars) or PBS (white bars) and healthy crabs (gray bars) were compared with respect to EsTUBA mRNA expression (relative to β-actin) using Tukey’s post hoc test. Bars represent mean ± S.E. of three independent investigations. Statistical significance is indicated with an asterisk. *p<0.05 and **p<0.01. Control group (PBS infection) was taken as the calibrator.
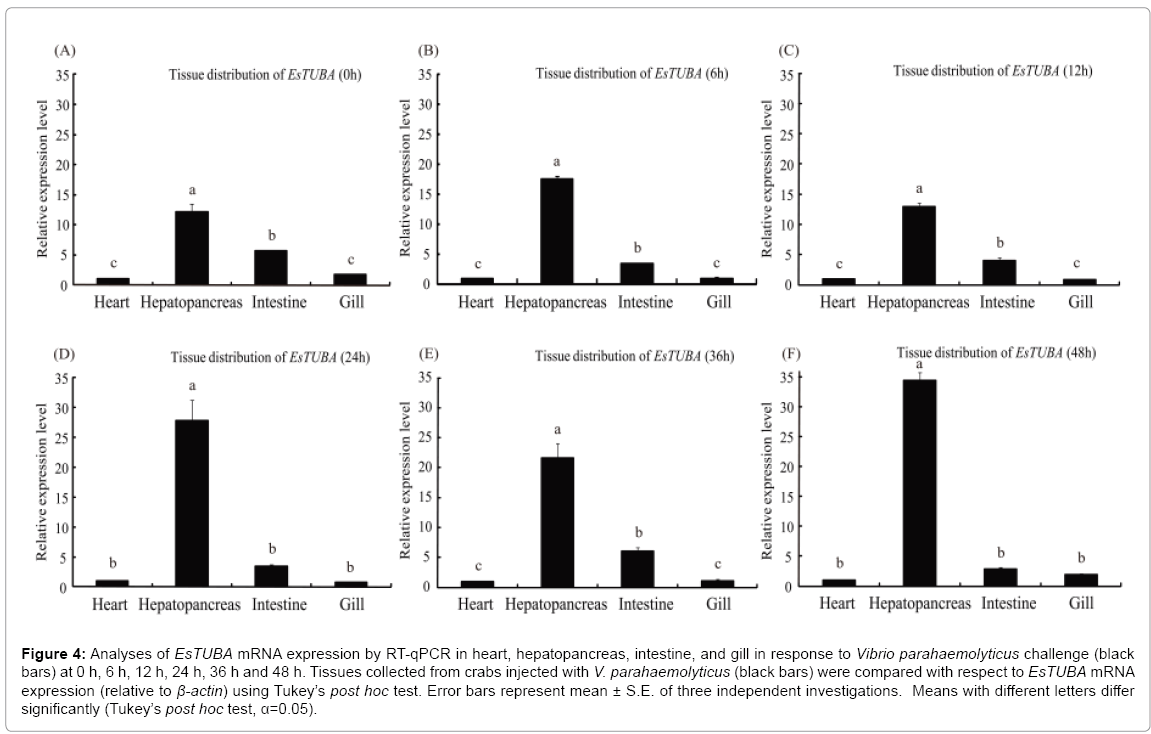
The mRNA transcript of EsTUBA could be detected in all the examined tissues with different expression levels under immune challenge stress. The obviously high levels of EsTUBA transcript were found in hepatopancreas, whereas only trace amounts were detectable in heart and gill at 0 h, 6 h, 12 h, 24 h post-injection (Figures 4A-4D), while EsTUBA expression level was even slightly lower in intestine than other at 24 h and 48 h post-injection (Figures 4D and 4F). RTqPCR results showed the highest EsTUBA expression was found in hepatopancreas at 48 h after VP challenge, which was significantly higher (~ 34-fold compared to that in heart, p<0.01) than in intestine, heart and gill (Figure 3F). The expression of EsTUBA in gill has reached the lowest level at 24 h post-injection (Figures 3D and 4D). The EsTUBA expression in different tissues maintains similar dynamic distribution at 12 h after challenge, whereas the EsTUBA mRNA transcripts in hepatopancreas showed slightly down-regulated (p>0.05) compared with in other tissues (Figures 4A-4C). The EsTUBA expression in hepatopancres was rapidly upregulated (~ 28.4-fold compared to that in heart, p<0.01) at 24 h after challenge, while the EsTUBA mRNA transcripts in other tissues maintain lower expression level (Figure 4D). The EsTUBA expression in intestine showed obviously upregulated after 36 h challenge compared with in other tissues (Figure 4E). The EsTUBA expression in hepatopancreas was rapidly upregulated (p<0.05) at 48 h after challenge, and the mRNA transcripts of EsTUBA in gill showed slightly upregulated while in intestine was slightly down-regulated (Figure 4F).
Figure 4: Analyses of EsTUBA mRNA expression by RT-qPCR in heart, hepatopancreas, intestine, and gill in response to Vibrio parahaemolyticus challenge (black bars) at 0 h, 6 h, 12 h, 24 h, 36 h and 48 h. Tissues collected from crabs injected with V. parahaemolyticus (black bars) were compared with respect to EsTUBA mRNA expression (relative to β-actin) using Tukey’s post hoc test. Error bars represent mean ± S.E. of three independent investigations. Means with different letters differ significantly (Tukey’s post hoc test, α=0.05).
Discussion
An alpha-tubulin-like gene (EsTUBA) was identified from E. sinensis and the ORF of EsTUBA was 1,356 bp encoding a polypeptide of 451 amino acids. The deduced amino acid sequence of EsTUBA was found to share high levels of similarity with alpha-tubulins of other known organisms, especially with the one from American lobster H. americanus with 98% similarities. The major tubulin gene family is highly conserved from invertebrates to vertebrates as a result of structural and functional constraints on microtubule assembly [34]. The previous study on atomic model of the α/β tubulin dimmer indicated alpha-tubulin basically possess three functional domains contained an amino-terminal domain, an intermediate domain and a carboxy-terminal domain (which probably constitutes the binding surface for motor proteins) [2]. But EsTUBA possess a 145 amino acids carboxy-terminal domain and a 242 amino acids GTPase domain in length. Bioinformatics analyses indicated that EsTUBA has two N-glycosylation sites and six protein kinase C phosphorylation sites, which is consistent with the previous study and those functional sites may play an important role in post translational tubulin modification [2]. Study on European decapod crabs indicated tubulins appear only to be present in the cytoplasm [35]. In this study, the potential protein subcellular localization prediction indicated that EsTUBA is also located in cytoplasm. Multiple sequence alignments of tubulin sequences also revealed they are highly conserved among different animal tubulins. The BI and ML trees reconstructed based on amino acids sequences of EsTUBA and tubulins of other animals suggested that tubulin in E. sinensis first clustered with crustacean tubulins, especially Homarus americanus tubulin. Crustacean tubulins are more closely related to Drosophila melanogaster tubulin and Camponotus floridanus tubulin than other tubulins. One GTP nucleotide-binding site (GGGTGSG), located between amino acid positions 94–100. This site is necessary for the polymerization of TUA/β-tubulin [36] and was conserved in EsTUBA. The conservative characteristics and high similarity with known alpha-tubulins informed us that EsTUBA belonged to the tubulin gene family.
The RT-qPCR and RT-PCR results demonstrated that EsTUBA mRNA expression was widely expressed in all detected development stages, and it was significantly enhanced in stage O, stage Z1, stage Z2, stage Z3 and stage Z4, but significantly reduced in stage Z5, stage M, stage J1, stage J2 and stage J3. The results indicated EsTUBA plays a role during the brachyurization development of E. sinensis. As we know, the remarkable morphological features changes and brachyurization metamorphosis occurs veritably in the crab's cephalothorax and abdomen from stage Z5 to stage J1, in which the morphological features changes are most significant. The larvae of all crabs are shrimplike before brachyurization metamorphosis occurs in stage J1, with a depressed and broadened cephalothorax. During the brachyurization metamorphosis, two diametrically opposed processes can be distinguished: integration and differentiation. The segments, especially their appendages, are strongly differentiated in relation to those of other decapod Crustaceans. This differentiation is related with the maximum division of labor reached among the decapods. Some segments with reinforced functions, such as the thoracic ones, are intensified. But segments with diminished functions, such as the abdominal ones, are reduced. Some of the segments may be fused together, and the abdomen tucked away under the body, has lost all locomotory functions and has instead become specialized for reproduction. The brachyurization metamorphosis is accomplished basically from the megalops to the third stage J3 [37-40]. The larval development of the Chinese mitten crab undergoes five zoeal stages and one megalopa stage in estuarine and marine coastal waters [41,42], and starting megalopa stage and other several juvenile stages metamorphosis in freshwaters [43]. Desalination would impose osmotic stress challenge on crabs. Studies on rice and Arabidopsis indicated hyperosmotic stress induced α-tubulin phosphorylation [44]. Studies on land crab and lobster confirmed that tubulin localized in exoskeletal structures, it may serve both intracellular and extracellular functions in crustaceans [45]. In this study, we found that the EsTUBA mRNA expression f?luctuated with the morphological changes in the brachyurization metamorphosis process of E. sinensis. Expression levels are highest in stage O, while the lowest levels appeared in stage Z5. The significantly fluctuated expression profiles of EsTUBA implicate its roles in juvenile stages metamorphosis development of crabs. It is likely that this gene is required to support microtubule function during the entire course of development of E. sinensis, which is in agreement with the result found in M. alternatus [17]. Compared with those of stage Z5, the EsTUBA mRNA expression levels is significantly enhanced in stage M, and then the expression is gradually reduced from stage M to stage J3. The results indicated that the tubulin gene may be response to the osmotic stress challenge or involve in the shells exoskeleton structure formation.
The transcriptional expression of EsTUBA showed a clear timedependent response in all detected tissues after the crabs were challenged with V. parahaemolyticus, but the level of mRNA expression differs between tissues. We have shown here the up-regulation of EsTUBA in hepatopancreas after V. parahaemolyticus challenge was consistent with previous studies in other genes such as the upregulation of Hsp70 in swimming crab challenged by V. alginolyticus [46] and the up-regulation of EsCTL in hepatopanceras of E. sinensis challenged by V. parahaemolyticus [21]. In addition, the previous study has shown the enhancement in the level of α-tubulin in influenza A virus-infected cells [47]. Hepatopancreas, gill and intestine are important organs involved in crustacean immunities, and play a crucial role in the immune response [48]. Whereas, the level of EsTUBA mRNA expression in heart, intestine and gill showed down-regulated after V. parahaemolyticus challenge, this is not consistent with previous studies. But RT-qPCR analyses revealed that the levels of EsTUBA mRNA expressed in immune associated tissues, such as hepatopancreas, gill and intestine, are significantly fluctuated and induced by the bacteria V. parahaemolyticus, while it is almost consistently expressed in heart at 0, 6, 12, 24, 36 and 48 h post-injection compared to control. These results suggest that EsTUBA may be involved in immune defense of crabs.
in summary, a α-tubulin gene designated as EsTUBA was isolated and identified from the Chinese mitten crab E. sinensis. According to the results of molecular characterization and expression patterns, it could be deduced that EsTUBA may play some roles in brachyurization metamorphosis development and innate immunity of crabs. However, further research is needed to clarify the more precise roles of EsTUBA in brachyurization metamorphosis and immune function of crabs
Acknowledgments
We are indebted to Dongxi Shen (Xinghua County, Jiangsu, China) and Bin Huang (Rudong County, Jiangsu, China) for providing the Chinese mitten crab materials. This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31000954), Program of Natural Science Research of Jiangsu Higher Education Institutions of China (Grant No. 13KJB180008) and Research Fund for the Doctoral Program of Higher Education of China (Grant No. 20103207120007), and the Priority Academic Program Development of Jiangsu Higher Education Institutions to PL.
Appendices Material
The online version of this article contains appendices material, which is available to authorized users.
References
- Nogales E, Whittaker M, Milligan RA, Downing KH (1999) High-resolution model of the microtubule. Cell 96: 79-88.
- Nogales E, Wolf SG, Downing KH (1998) Structure of the alpha beta tubulin dimer by electron crystallography. Nature 391: 199-203.
- Dutcher SK (2003) Long-lost relatives reappear: Identification of new members of the tubulin superfamily. CurrOpinMicrobiol 6: 634-640.
- Kaur R, Kaur G, Gill RK, Soni R, Bariwal J (2014) Recent developments in tubulin polymerization inhibitors: An overview. Eur J Med Chem 87: 89-124.
- Ludueña RF (1993) Are tubulin isotypes functionally significant. MolBiol Cell 4: 445-457.
- Sackett DL (2010) Evolution and coevolution of tubulin's carboxyterminal tails and mitochondria: Mitochondria: Structure, functions and dysfunctions. Nova Science Publishers 441-470.
- Varikmaa M, Bagur R, Kaambre T, Grichine A, Timohhina N, et al. (2014) Role of mitochondria–cytoskeleton interactions in respiration regulation and mitochondrial organization in striated muscles. BiochimBiophysActa 1837: 232-245.
- Tepp K, Mado K, Varikmaa M, Klepinin A, Timohhina N, et al. (2014) The role of tubulin in the mitochondrial metabolism and arrangement in muscle cells. J BioenergBiomembr 46: 421-434.
- Oakley RV, Wang YS, Ramakrishna W, Harding SA, Tsai CJ (2007) Differential expansion and expression of alpha- and beta-tubulin gene families in Populus. Plant Physiol 145: 961-973.
- Radchuk V, Sreenivasulu N, Blume Y, Weschke W (2008) Cloning and expression of the tubulin genes in barley. Cell BiolInt 32: 557-559.
- Yu Y, Li Y, Li L, Lin J, Zheng C, et al. (2009) Overexpression of PwTUA1, a pollen-specific tubulin gene, increases pollen tube elongation by altering the distribution of alpha-tubulin and promoting vesicle transport. J Exp Bot 60: 2737-2749.
- Howard J, Hyman AA (2003) Dynamics and mechanics of the microtubule plus end. Nature 422: 753-758.
- Roll-Mecak A (2015) Intrinsically disordered tubulin tails: Complex tuners of microtubule functions? Semin Cell DevBiol 37: 11-19.
- Bond JF, Fridovich-Keil JL, Pillus L, Mulligan RC, Solomon F (1986) A chicken-yeast chimeric beta-tubulin protein is incorporated into mouse microtubules in vivo. Cell 44: 461-468.
- Anthony RG, Hussey PJ (1999) Double mutation in Eleusineindica a-tubulin increases the resistance of transgenic maize calli to dinitroaniline and phosphorothioamidate herbicides. Plant J 18: 669-674.
- Anthony RG, Reichelt S, Hussey PJ (1999) Dinitroaniline herbicide-resistant transgenic tobacco plants generated by co-overexpression of a mutant alpha-tubulin and a beta-tubulin. Nat Biotechnol 17: 712-716.
- Song L, Liu XX, Zhang YA, Zhang QW, Zhao ZW (2008) The cloning and expression of alpha-tubulin in Monochamusalternatus. Insect MolBiol 17: 495-504.
- Ramírez CA, Requena JM, Puerta CJ (2013) Alpha tubulin genes from Leishmaniabraziliensis: Genomic organization, gene structure and insights on their expression. BMC Genomics 14: 454.
- Vukojevic K, Janjic T, Saraga-Babic M (2014) Developmental patterns of Ki-67, Oct-4 and α-tubulin proteins expression in the human spinal cord. ActaHistochem 116: 619-626.
- Chen DW, Zhang M, Shrestha S (2007) Compositional characteristics and nutritional quality of Chinese mitten crab (Eriocheirsinensis). Food Chemistry 103: 1343-1349.
- Huang Y, Huang X, Wang Z, Tan JM, Hui KM, et al. (2014) Function of two novel single-CRD containing C-type lectins in innate immunity from Eriocheirsinensis. Fish Shellfish Immunol 37: 313-321.
- Sun M, Jiang K, Zhang F, Zhang D, Shen A, et al. (2012) Effects of various salinities on Na+-K+-ATPase, Hsp70 and Hsp90 expression profiles in juvenile mitten crabs, Eriocheirsinensis. Genetics and Molecular Research 11: 978-986.
- Li P, Zha J, Zhang Z, Huang H, Sun H, et al. (2009) Molecular cloning, mRNA expression, and characterization of HSP90 gene from Chinese mitten crab Eriocheir japonica sinensis. Comp BiochemPhysiol B BiochemMolBiol 153: 229-235.
- Kong P, Wang L, Zhang H, Zhou Z, Qiu L, et al. (2010) Two novel secreted ferritins involved in immune defense of Chinese mitten crab Eriocheirsinensis. Fish Shellfish Immunol 28: 604-612.
- Lubega GW, Byarugaba DK, Prichard RK (2002) Immunization with a tubulin-rich preparation from Trypanosomabrucei confers broad protection against African trypanosomosis. Expparasitol 102: 9-22.
- Li P, Zha J, Sun H, Song D, Zhou K (2011) Identification of differentially expressed genes during the brachyurization of the Chinese mitten crab Eriocheir japonica sinensis. Biochem Genet 49: 645-655.
- Yu AQ, Jin XK, Guo XN, Li S, Wu MH, et al. (2013) Two novel Toll genes (EsToll1 and EsToll2) from Eriocheirsinensis are differentially induced by lipopolysaccharide, peptidoglycan and zymosan. Fish Shellfish Immunol 35: 1282-1292.
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) Method. Methods 25: 402-408.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402.
- Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. MolBiolEvol 17: 540-552.
- Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817-818.
- Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52: 696-704.
- Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754-755.
- Hutchens JA, Hoyle HD, Turner FR, Raff EC (1997) Structurally similar Drosophila alpha-tubulins are functionally distinct in vivo. MolBiol Cell 8: 481-500.
- Tudge CC, Justine JL (1994) The cytoskeletal proteins actin and tubulin in the spermatozoa of four decapod crabs (Crustacea, Decapoda). ActaZoologica (Stockholm) 75: 277-285.
- Kirschner M, Mitchison T (1986) Beyond self-assembly: From microtubules to morphogenesis. Cell 45: 329-342.
- Števcic Z (1971) The main features of brachyuran evolution. Systematic Biology 20: 331-340.
- Warner GF (1977) The biology of crabs. Van Nostrand Reinhold Company, New York.
- Du NS (1993) Carcinology. Beijing, Science Press.
- Guinot D, Bouchard JM (1998) Evolution of the abdominal holding systems of brachyuran crabs (Crustacea, Decapoda, Brachyuran). Zoosystema 20: 613-694.
- Kim CH, Hwang SG (1995) The complete larval development of the mitten crab Eriocheirsinensis H. Milne Edwards, 1853 (Decapoda, Brachyura, Grapsidae) reared in the laboratory and a key to the known zoeae of the Varuninae. Crustaceana 68: 793-812.
- Montú M, Anger K, Bakker C (1996) Larval development of the Chinese mitten crab Eriocheirsinensis H. Milne Edwards (Decapoda: Grapsidae) reared in the laboratory. HelgoländerMeeresuntersuchungen 50: 223-252.
- Liang XQ, Yen SL, Cheng TC, Kou TT (1974) The larval development of Eriocheirsinensis H. Milne Edwards. ActaZoologicaSinica 20: 61-68.
- Ban Y, Kobayashi Y, Hara T, Hamada T, Hashimoto T, et al. (2013) α-tubulin is rapidly phosphorylated in response to hyperosmotic stress in rice and Arabidopsis. Plant Cell Physiol 54: 848-858.
- Mykles DL, Haire MF, Skinner DM (2000) Immunocytochemical localization of actin and tubulin in the integument of land crab (Gecarcinuslateralis) and lobster (Homarusamericanus). J ExpZool 286: 329-342.
- Cui Z, Liu Y, Luan W, Li Q, Wu D, et al. (2010) Molecular cloning and characterization of a heat shock protein 70 gene in swimming crab (Portunustrituberculatus). Fish Shellfish Immunol 28: 56-64.
- Husain M, Harrod KS (2011) Enhanced acetylation of alpha-tubulin in influenza A virus infected epithelial cells. FEBS Lett 585: 128-132.
- Ji PF, Yao CL, Wang ZY (2009) Immune response and gene expression in shrimp (Litopenaeusvannamei) hemocytes and hepatopancreas against some pathogen-associated molecular patterns. Fish Shellfish Immunol 27: 563-570.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 15536
- [From(publication date):
September-2015 - Sep 02, 2025] - Breakdown by view type
- HTML page views : 10825
- PDF downloads : 4711