Isolation and Characterization of Filter Paper Degrading Bacteria from the Guts of Coptotermes formosanus
Received: 20-Apr-2018 / Accepted Date: 30-Apr-2018 / Published Date: 03-May-2018 DOI: 10.4172/2155-6199.1000440
Abstract
Background: The filter paper utilizing capabilities of Pseudomonas mendocina, Burkholderia pseudomallei, Chryseobacterium luteola, Klebsiella oxytoca and Klebsiella terrigena isolated from the gut of a local termite Coptotermes formosanus were analysed.
Design and methods: The isolates were inoculated into a buffered medium containing minerals and Whatman filter paper as sole source of carbon to observe the ability of these bacteria to digest solid substrate. The ability of the isolates to grow in this medium as well as to digest the filter paper was determined by visual observation after 30 days. Reducing sugar test and gravimetric analysis were also carried out at the end of 30 days.
Results: All bacteria cultures showed growth as the medium turned cloudy and the filter paper became macerated. The gravimetric analysis of the residual filter in the liquid medium at the end of 30 days incubation showed that Chryseobacterium luteola had the highest degradation rate of 95%, Pseudomonas mendocina had the degradation rate of 90%, whilst Burkholderia pseudomallei, Klebsiella oxytoca and Klebsiella terrigena had biodegradation rate of 75% each. Reducing sugar test and paper chromatography carried out for glucose production were positive showing their ability to convert cellulose to glucose.
Conclusion: The bacterial isolates showed a potential to convert cellulose into reducing sugars which could be readily used in many applications like feed stock for production of valuable organic compounds; for example in simultaneous saccharification and fermentation of cellulose into ethanol.
Keywords: Biodegradation; Chromatography; Amplification; Cloudy; Cellulose
Introduction
Termites are insects from the order Isoptera which in Greek isos means equal and pteron means wing [1]. They are usually called white ants. They are small to medium size with a dull white to light brown body [2] and characterized by their colonial behaviour. Termites are among the most important lignocelluloses-ingesting insects and possess a variety of symbiotic microorganisms in their hindguts, including bacteria, Archaea and Eukarya [2]. Termites can be classified into six families and fifteen subfamilies [3]. Higher termites make up about 85% of the known species of termites. Lower termites feed mainly on wood, utilizing the enzymes they make themselves, as well as those from bacteria, archaea, and protists in their guts to digest the wood [4]. The gut microbiota enables termites to efficiently hydrolyze cellulose. The cellulose activity of termite hindgut is attributed to cellulose-degrading bacteria. Termites are diverse in their feeding habits that lead to diverse microbiota. Many microorganisms have been reported with cellulolytic activities including many bacterial and fungal strains both aerobic and anaerobic. Chaetomium, Fusarium, Myrothecium, Trichoderma, Penicillium, Aspergillus and so forth are some of the reported fungal species responsible for cellulosic biomass hydrolysation. Cellulolytic bacterial species include Trichonympha, Clostridium, Actinomycetes, Bacteroides, Succinogenes, Ruminococcus albus, Methanobrevibacter ruminantium [5,6].
Materials and Methods
Sample collection
The termites used for the isolation and identification of cellulolytic microorganisms were collected from a termite hill at Oba, a town in Anambra State, Nigeria.
Isolation of filter paper degrading microorganisms
The termites collected were identified as Coptotermes formosanus (lower termites). The termites were taken out of their nests and placed in sterilized Petri dishes. Twenty worker termites were surface sterilized with 70% ethanol [7] and then washed in sterile distilled water and allowed to air-dry for 1 minute. Under sterile conditions, each termite was separated into its head and body. After removing heads with forceps, the bodies were dissected with a sharp blade with the aid of a magnifying Hand lens. The guts of the dissected termites were picked out using a sterile syringe and mixed in 20 ml of 0.85% NaCl.
Five milliliter of the suspension was inoculated into four different conical flasks (except the control), each containing seventy-five milliliter of Basal Salt Medium as described by Chakraborty et al. [8]. The medium contained in g/l, 2.2 K2HPO4, 1.5 KH2PO4, 1.3 (NH4)2SO4, 0.1 MgCl, 0.02 CaCl, 0.001 FeSO4.7H2O, five filter papers and the pH was adjusted to 7.2 (with 1M NaOH and HCl) and incubated at room temperature for 30 days. The medium was sterilized at 121°C for 15 minutes. The sterilized filter paper served as the main carbon source (cellulose) for the microorganisms. Cloudiness of the medium indicated growth and maceration of the filter paper indicated cellulolytic activity. After 30 days, the culture was plated out on nutrient agar and pure colonies were obtained by several subsequent sub-culturing and plating.
Identification and characterization of bacterial isolates
Colonial examination of the isolates was carried out to determine the type of shape, elevation and pigmentation pattern they exhibited. Microscopic examination including Gram staining and cellular morphological appearances were also carried out. API 20E and 20 NE kits were then used to carry out further identification tests on the isolates. The kit were prepared according to the manufacturer’s specification
Api 20E and 20NE identification
The API 20E (Biomérieux) strip contains 20 microtubules. The inoculums were prepared by culturing the organism on nutrient agar plate for 24 hours. The distinct colonies produced were then picked and transferred to 5 ml sterile normal saline to prepare a homogenous suspension. A sterile pipette was used to fill the tubules of CIT; VP and GEL positions were filled with the suspension. Mineral oil was used to overlay test ADH, LDC, ODC, H2S and URE to create anaerobiosis. The inoculated strip was placed in the incubation box into which sterile distilled water has been placed to create a humid condition during incubation. The strips were incubated at 30°C for 18-24 hours. Readings were taken after the 24 hours of incubation; other tests such as TDA, IND, VP and NIT were carried out by adding appropriate reagents into the tubules. Observations were recorded.
Filter paper degradation study
Each bacterial isolate was inoculated into a test tube containing nutrient broth, incubated overnight at room temperature and afterwards used as inoculum. The medium used for the cellulolytic activity study was as described by Chakraborty et al. [8]. The medium was sterilized at 121°C for 15 minutes in an autoclave. Seventy-five milliliter of the medium was poured into twenty-two conical flasks containing 0.4 g of sterilized filter paper as sole source of carbon (cellulose) for the microorganisms. Five milliliter of each isolate’s cell culture was washed and pipetted into each flask (except the control) and the incubated at room temperature for 30 days. Growth and cellulolytic activity was determined by observing the change in the medium as well as on the filter paper. Turbidity of the medium indicated growth and maceration of the filter paper indicated cellulolytic activity.
Gravimetric analysis
This analysis is by weight. All gravimetric analyses rely on final determination of weight as means of quantifying an analyte. Gravimetric analysis was used to determine the weight of residual filter paper present in the medium after 30 days of incubation and thus determined the degree of filter paper degradation. A standard profile was first obtained by determining the dry-weight of undegraded filter paper in the control experiment which was the same for all flasks before inoculation, incubation and degradation commenced. Media containing cellulolytic isolates and pieces of filter paper were filtered, washed and the residual filter paper pieces were dried to a constant weight at 60°C. Filter paper degradation by these cellulolytic microorganisms was determined as the differences between filter paper present at the beginning and the end products of the culture period. The treatment was repeated three times. The individual weight of residual filter paper pieces for each flask was determined. The weight of degraded filter paper for each flask was determined and converted to percentage.
Deteremination of reducing sugar with Fehling solution
Reducing sugar determination using Fehling Solution was done to determine the presence of reducing sugars in the filter paper/ cellulose degradation culture medium. The culture medium was centrifuged and 2 ml of the supernatant was added into a test tube. One ml of Fehling Solution was added to it and heated for 15 minutes. Formation of yellow to brick red precipitate showed the presence of reducing sugars such as glucose and fructose.
Paper chromatography
Paper chromatography is a technique that involves placing a dot or line of sample solution onto a strip of cellulose paper. The paper is placed in a jar containing a shallow layer of solvent and sealed. As the solvent rises through the paper, it meets the sample mixture, which starts to travel up the paper with the solvent.
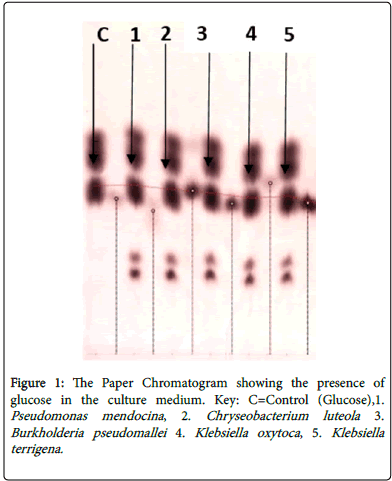
Paper chromatography was carried out using the supernatant from each of the bacterial isolate culture medium to determine the production of glucose from filter paper/ cellulose degradation by the bacterial isolates. Pure glucose was used as a control experiment. The spots on the Chromatograms were visualized by spraying them with ammonical silver nitrate and placing them in an oven for about 5-10 minutes (Figure 1). The reducing sugars appeared as brown spots and the Retardation factor of glucose on each Chromatogram was determined. Ammonia (1%) plus saturated phenol-water was used as the solvent for this procedure.
Results
Five bacterial species were isolated and identified using colonial morphology as shown in Table 1, biochemical tests and the use of API test kit. The organisms obtained were identified using API test kit 20E and 20NE as Bac-1, Pseudomonas mendocina , Bac-2, Burkholderia pseudomallei , Bac-3, Chryseobacterium luteola , Bac-4, Klebsiella oxytoca and Bac-5, Klebsiella terrrigena as shown in Tables 2 and 3.
| Characteristics | Bac-1 | Bac-2 | Bac-3 | Bac-4 | Bac-5 |
|---|---|---|---|---|---|
| Shape | Circular | Circular | Circular | Circular | Circular |
| Size | Small | Small | Small | Medium | Medium |
| Pigmentation | Cream | Cream | Cream | Cream mucoid | Cream |
| Elevation | Convex | Convex | Convex | Convex | Convex |
| Margin | Entire | Entire | Entire | Entire | Entire |
Table 1: Colonial Morphology of Bacteria on Nutrient Agar.
| Isolates | Bac-1 | Bac-2 | Bac-3 | |
|---|---|---|---|---|
| API | Cellular Morphology | Rods | Rods | Rods |
| Gram’s Reaction | - | - | - | |
| Catalase | + | + | + | |
| NO3 | + | + | + | |
| TRP | - | - | - | |
| GLU | + | + | - | |
| ADH | + | + | + | |
| URE | - | - | - | |
| ESC | - | - | - | |
| GEL | - | + | + | |
| PNG | - | - | + | |
| ARA | - | - | + | |
| MNE | - | + | + | |
| MAN | - | - | - | |
| NAG | - | + | + | |
| MAL | - | - | + | |
| GNT | + | + | + | |
| CAP | + | + | + | |
| ADI | - | - | - | |
| MLT | + | + | + | |
| CIT | + | + | + | |
| PAC | - | + | - | |
| OX | + | + | - | |
| Probably Organisms | Pseudomonas mendocina | Burkholderia pseudomallei | Chryseobacterium luteola | |
Table 2: Biochemical characteristics of isolates using API kit 20 NE.
| Isolates | Bac-4 | Bac-5 | |
| Cellular Morphology | Rods | Rods | |
| Gram’s Reaction | - | - | |
| Catalase | + | - | |
| API | ONPG | + | + |
| ADH | - | - | |
| LDC | - | + | |
| ODC | - | - | |
| CIT | + | + | |
| H2S | - | - | |
| URC | + | - | |
| TDA | - | - | |
| IND | + | - | |
| VP | + | + | |
| GEL | - | + | |
| GLU | + | + | |
| RHA | + | + | |
| MAN | + | + | |
| SOR | + | + | |
| MEL | + | + | |
| AMY | + | + | |
| ARA | + | + | |
| OX | - | - | |
| NO2 | + | + | |
| N2 | - | - | |
| MOB | - | - | |
| MCC | + | + | |
| OF – O | + | + | |
| OF –F | + | + | |
| Probably Organisms | oxytoca | Klebssiella | |
Table 3: Biochemical characteristics of isolates using API kit 20 E.
The result of gravimetric analysis carried out on the residual filter paper from the filter paper degradation study culture medium by each isolate after 30 days of incubation is as follows: Chryseobacterium luteola gave the highest degradation value of 95% followed by Pseudomonas mendocina with the rate of 90%. Burkholderia pseudomallei, Klebsiella oxytoca and Klebsiella terrigena gave degradation rate of 75% each as shown in Table 4.
| Sample | Residual Filter paper (gl-1) | Biodegradation (%) |
|---|---|---|
| Control | 0.4 | 0 |
| Pseudomonas mendocina | 0.04 | 90 |
| Burkholderia pseudomallei | 0.1 | 75 |
| Chryseobacterium luteola | 0.02 | 95 |
| Klebsiella oxytoca | 0.1 | 75 |
| Klebsiella terrigena | 0.1 | 75 |
Table 4: Residual filter paper content in filter paper liquid culture medium at the end of a 30-day incubation period.
Reducing sugar determination test with Fehling Solution for each bacterial isolate culture medium was positive. Paper chromatography showed the production of glucose from the degradation of filter paper/ cellulose by the bacterial isolates. The Retardation factors were calculated and recorded as shown in Table 5.
| Isolate | Retention factor (Rf) |
| Control | 0.64 |
| P.mendocina, | 0.643 |
| B.pseudomallei | 0.642 |
| C.luteola | 0.643 |
| K. oxytoca | 0.639 |
| K. oxytoca | 0.634 |
Table 5: Retention factors (Rf) of glucose produced from cellulose degradation by bacterial isolates determined by paper chromatography.
Discussion
In this study, microorganisms capable of degrading filter paper were isolated from Coptotermes formosanus (lower termites). Termites are one of the planet’s most efficient bioreactors and may be capable of producing up to two litres of hydrogen from digesting a single piece of paper. Termites achieve this high degree of efficiency by exploiting the metabolic capabilities of about 200 different species of microorganisms that inhabit their hindguts. The complex lignocelluloses polymers within wood are broken down into simple sugars by fermenting bacteria in the termite’s gut, using enzymes that produce hydrogen as by-product. The potential for the production of biofuel from the fermentation of the simple sugars obtained from lignocelluloses decomposition in this way is tremendous. The world’s increasing need for an alternative source of energy has always urged scientists to turn to biofuel. Since many of the developed nations in the world have already started bioethanol production using cellulolytic bacteria obtained from many organisms including termites, it has become urgently necessary for our nation to exploit these organisms for biofuel production. In nature, cellulosic materials can be degraded by many bacteria. More than 50 species have been isolated. However, different strains possess different cellulose degradation capabilities [9].
Konig [10] had grouped bacteria from termites gut based on their lignocellulolytic activity into two, that is, hydrolytic and fermentative groups. In this study, all bacterial species were able to digest the filter paper as well used the products for growth. Previous studies by Borji et al. [11] also reported that Enterobacter and Acinetobacter species showed cellulolytic activity. Dugas et al. [12] had isolated and identified a strain of Chryseobacterium from the gut of the American cockroach which was fed with a high fibre diet. Ramin et al. [7] isolated and identified Chryseobacterium kwangyangense which belong to the family Flavobacteriaceae and Akpomie et al. [13] isolated and identified Klebsiella, Streptococcus, Celulomonas and Micrococcus . Pseudomonas was the most dominant group in the cellulolytic bacteria. Bacteria of the genus Pseudomonas can be found in many different environments including soil, water, plant and animal tissue, and these bacteria have the ability to metabolize a variety of diverse nutrient [14] but other Pseudomonas species also have been reported to degrade cellulose [15,16].
From this study, five species of cellulolytic bacteria were isolated from the gut of the lower termite, C. formosanus . They were identified as Pseudomonas mendocina, Chryseobacterium luteola, Burkholderia pseudomallei, Klebsiella oxytoca and Klebsiella terrigena . Gravimetric analysis showed that maximum and minimum rates of filter paper degradation were 95% and 75% respectively, estimated at thirtieth day of incubation. Chryseobacterium luteola and Pseudomonas mendocina exhibited biodegradative capabilities of 95% and 90% respectively.
Conclusion
The bacterial isolates showed a potential to convert cellulose into reducing sugars which could be readily used in many applications like feed stock for production of valuable organic compounds; for example in simultaneous saccharification and fermentation of cellulose into ethanol. This study demonstrates that C. formosanus habour a dense and diverse community of cellulolytic bacteria in the hindgut and that the bacteria in the hindgut have an important role in the degradation of the roots and other organic matter consumed by this termite. These bacteria are able to produce enzymes useful for biofuel production.
References
- Thorne BL, Carpenter JM (1992) Phylogeny of the Dictyoptera. J Syst Entomol 17: 253-268.
- Varma A, Krishna BK, Paul J, Saxena S, Konig H (1994) Lignocellulose degradation by microorganisms from termite hills and termite guts: A survey on the present state of art. J FEMS Microbiol Rev 15: 9-28
- Lee KE, Wood TG (1971) Termites and soil. Acad Press, New York, Pp: 1-100.
- Matsui T, Tokuda G, Shinzato N (2009) Termites as Functional Gene Resources. Recent Pat Biotechnol 3: 10-18.
- Milala MA, Shugaba A, Gidado A, Ene AC, Wafar JA (2005) Studies on the use of agricultural wastes for cellulose enzyme production by A. niger. J Agric Biol Sci 1: 325-328.
- Schwarz WH (2001) The cellulosome and cellulose degradation by anaerobic bacteria. Appl Microbial Biotechnol 56: 634-649.
- Ramin M, Alimon AR, Sijam K, Abdullah N (2008) Filter paper degradation by bacteria isolated from local termite gut. Res J Microbiol 3: 565-568.
- Chakraborty N, Sarkar GM, Lahiri SC (2000) Cellulose degrading capabilities of cellulolytic bacteria isolated from the intestinal fluids of the silver crickets. Environment 20: 9-11.
- Yucai L, Xiaofen W, Ning L, Xiaojuan W, Masaharu I, et al. (2011) Characterization of the effective cellulose degrading strain CTL-6. J Environ Sci 23: 649-655.
- Konig H (2006) Bacillus species in the intestine of termites and other soil invertebrates. J Appl Microbiol 101: 620-627.
- Borji M, Rahimi S, Ghorbani G, Vand YJ, Fazaeli H (2003) Isolation and identification of some bacteria from termites’ guts capable in degrading straw lignin and polysaccharides. J Fac Vet Med Univ 58: 249-256.
- Dugas JE, Zurek L, Paster BJ, Keddie BA (2001) Isolation and characterization of a Chryseobacterium strain from the gut of the American cockroach, Periplanatamericana. Arch Microbiol 175: 259-262.
- Akpomie OO, Ubogun E, Ubogun M (2013) Determination of the cellulolytic activities of microorganisms isolated from poultry litter for sawdust degradation. J Environ Wat Resour 2: 062-066.
- Kodama K, Kimura K, Komagata K (1985) Two new species of Pseudomonas: P. oryzahabitans isolated from rice paddy and clinical specimens and P. luteola isolated from clinical specimens. Int J Syst Evol Microbiol 35: 467-474.
- Sindhu SS, Dadarwal KR (2001) Chitinolytic and cellulolytic. Antagonistic to fungal pathogens enhances nodulation by Mesorhizobium sp. Cicer in chickpea. Microbial Res 156: 353-358.
- Millward-Sadler SJ, Davidson K, Hazlewood GP, Black GW (1995) Novel-cellulose binding domains, NodB homologues and conserved molecular architecture in xylanases from aerobic soil bacteria Pseudomonas fluorescens subsp. Cellulose and Cellvibromixtus. Biochem J 312: 39-48.
Citation: Egwuatu TF, Appeh OG (2018) Isolation and Characterization of Filter Paper Degrading Bacteria from the Guts of Coptotermes formosanus. J Bioremediat Biodegrad 9: 440. DOI: 10.4172/2155-6199.1000440
Copyright: © 2018 Egwuatu TF, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 7089
- [From(publication date): 0-2018 - Dec 18, 2025]
- Breakdown by view type
- HTML page views: 5968
- PDF downloads: 1121