Malaria Parasite Pyrimidine Nucleotide Metabolism: A Promising Drug Target
Received: 13-Jan-2017 / Accepted Date: 02-May-2017 / Published Date: 05-May-2017
Abstract
Malaria is a major cause of morbidity and mortality in the tropical and subtropical endemic countries worldwide. This is largely due to the emergence and spread of resistance to most antimalarial drugs currently available, including the first-line treatment artemisinins. Thus, to fight this disease, there is an essential requirement to develop new antimalarial drugs for malaria chemotherapy. Plasmodium falciparum, the causative agent of the most lethal form of malaria in humans, cannot salvage preformed bases or nucleosides for pyrimidine synthesis and relies solely on pyrimidine nucleotides synthesized through the de novo biosynthetic pathway. In contrast, the human host cells have functionally operated both the salvage and de novo pathways. This mini review summarizes significant progress on understanding the pyrimidine nucleotide metabolism and the functional enzymes in the human parasite P. falciparum, which are different from the human host metabolic processes. Most recent information of the three-dimensional crystal structures and the catalytic mechanisms of the de novo pyrimidine enzymes: dihydroorotate dehydrogenase, orotate phosphoribosyltransferase, and orotidine 5'-monophosphate decarboxylase, as well as their inhibitors affecting these enzymatic activities are briefly reviewed in the context of their therapeutic potential against malaria.
Keywords: Malaria; Plasmodium falciparum; Pyrimidine nucleotide metabolism; Drug target; Drug development
66092Mini Review
Malaria, a major parasitic disease of humans, is caused by protozoa of the genus Plasmodium, classified in the phylum Apicomplexan. The disease afflicts 515 million clinical cases annually in 96 subtropical and tropical endemic countries. The death toll is reported at 1.3 million people each year, mostly young children in sub-Saharan Africa (90%) [1] of the five Plasmodium species that infect humans, including P. vivax, P. malariae, P. ovale, and P. knowlesi, P. falciparum is the most dangerous parasite and responsible for the majority of deaths. Based on the World Health Organization recommendation in 2001, artemisinin-based combination therapies are now used as the first-line drugs for treatment of P. falciparum malaria [2]. However by 2009, resistance to the drug treatment has been reported [3]. In addition to the lack of highly effective vector control and vaccines, the spread of drug-resistant malaria accompanied by a worldwide resurgence of the disease, it is necessary to develop quickly more effective novel antimalarial drugs, possessing a different mechanism of action, for malaria chemotherapy. This review highlights a better understanding of biochemical differences between the parasite P. falciparum and human metabolic processes, i.e., the pyrimidine nucleotide metabolic pathway, which may provide new candidate drug targets for intervention in the fight against this disease [4].
Nucleotide metabolism is one of the largest metabolic pathways, providing the building blocks for DNA and RNA synthesis in human cells. The nucleotides serve as key players in a wide range of cellular functions, including energy transduction, signal transduction, synthesis of biomolecules for carbohydrate and lipid metabolisms [5]. Purine and pyrimidine nucleotides are synthesized from de novo biosynthetic pathways or supplied via salvage pathways, where nucleobases, nucleosides and deoxynucleosides are recycled from nutrients or from degraded DNA and RNA. In human cells, both the de novo and salvage pathways are functioning at significant levels for the purine and pyrimidine nucleotide requirements, although the salvage pathways are more active than the de novo pathways [6]. This is true also for bacteria, plant and the free-living nematode Caenorhabditis elegans [7].
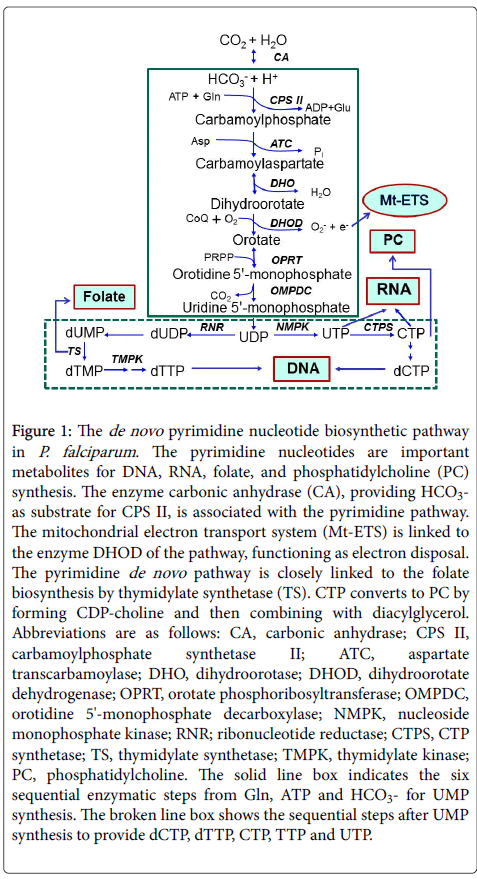
The de novo pyrimidine biosynthetic pathway is the most conserved metabolic pathway, and the six sequential enzymatic steps starting from bicarbonate ion (HCO3-), glutamine (Gln), and adenosine 5'- triphosphate (ATP), providing uridine 5'-monophosphate (UMP) (Figure 1, in solid line box), have remained intact throughout evolution, although the primary structures of the enzymes responsible for the de novo synthesis deviate significantly among prokaryotes, parasitic protozoa, fungi, animals, and mammals including humans [8,9]. Unlike human host cells, P. falciparum parasites have very limited ability to salvage preformed pyrimidine bases and nucleosides (e.g., uracil, uridine, thymidine, cytidine) from the host cell and extracellular environment, but rely mostly on nucleotide synthesized through the de novo pathway. All the enzymes required for de novo synthesis of UMP, the first pyrimidine nucleotide metabolite acting as the precursor for all pyrimidine nucleotides synthesis , including dCTP, dTTP, CTP, TTP, and UTP, were detected in cell extracts from all Plasmodium species so far examined [10]. The genes encoding each enzyme in all steps of the de novo pathway were identified in the parasite genome [11].
Figure 1: The de novo pyrimidine nucleotide biosynthetic pathway in P. falciparum. The pyrimidine nucleotides are important metabolites for DNA, RNA, folate, and phosphatidylcholine (PC) synthesis. The enzyme carbonic anhydrase (CA), providing HCO3- as substrate for CPS II, is associated with the pyrimidine pathway. The mitochondrial electron transport system (Mt-ETS) is linked to the enzyme DHOD of the pathway, functioning as electron disposal. The pyrimidine de novo pathway is closely linked to the folate biosynthesis by thymidylate synthetase (TS). CTP converts to PC by forming CDP-choline and then combining with diacylglycerol. Abbreviations are as follows: CA, carbonic anhydrase; CPS II, carbamoylphosphate synthetase II; ATC, aspartate transcarbamoylase; DHO, dihydroorotase; DHOD, dihydroorotate dehydrogenase; OPRT, orotate phosphoribosyltransferase; OMPDC, orotidine 5'-monophosphate decarboxylase; NMPK, nucleoside monophosphate kinase; RNR; ribonucleotide reductase; CTPS, CTP synthetase; TS, thymidylate synthetase; TMPK, thymidylate kinase; PC, phosphatidylcholine. The solid line box indicates the six sequential enzymatic steps from Gln, ATP and HCO3- for UMP synthesis. The broken line box shows the sequential steps after UMP synthesis to provide dCTP, dTTP, CTP, TTP and UTP.
Progress towards understanding structures, catalytic mechanisms and regulation of the mammalian and human enzymes for the de novo pyrimidine pathway has been significant in recent years [5,9]. Some key differences on the functioning organization of the enzymes, and their genomes including the six enzymes of the pathway from precursors HCO3-, Gln, and ATP to UMP synthesis warrants a closer look. The first three enzymes (carbamoyl phosphate synthetase, CPS II; aspartate transcarbamoylase, ATC; dihydroorotase, DHO) of the parasite were readily separated into three different molecular masses by analytical gel filtration chromatography [12], which is consistent with the presence of three discrete monofunctional enzymes. This is similar to that found in another species of protozoa, Crithidia fasciculata, and in many prokaryotic systems [12,13]. The characteristics are different from the humans, wherein the CPS II, ATC and DHO activities are carried on a 243-kDa multifunctional protein, namely CAD [14]. The malarial DHO enzyme has been purified from P. falciparum and its gene has been cloned, expressed and characterized in detail by our groups [15]. The DHO is a Zn2+ enzyme belonging to the amidohydrolase family, sharing characteristics of both mammalian type I and eubacterial type II DHO by overall amino acid sequence homology, structural characteristics, kinetic and inhibitor properties [15,16]. At present, the malarial CPS II and ATC enzymes are still poorly characterized and regulation of these pyrimidine enzymes is unknown, in contrast to the human CAD enzyme [9].
Recent studies have mainly attended on dihydroorotate dehydrogenase (DHOD), the fourth enzyme in the pathway, particularly as a target for antimalarial agents [16-18]. The P. falciparum DHOD has been characterized, and immunogold labelling localized DHOD in the inner membrane of mitochondrion [18]. The three-dimensional crystal structure of the parasite enzyme has been elucidated and compared to the human DHOD structure [19,20]. Crystal structures of human and parasite DHOD identifies completely different binding sites for the antiproliferating leflunomide inhibitor. The overall structure is α/β-barrel, similarly to that of other type II DHOD of eukaryotic origin. It contains flavin mononucleotide coenzyme, ubiquinone binding site and active site for dihydroorotate substrate. Furthermore, the de novo pathway is tightly coupled to the mitochondrial electron transport system (Mt-ETS) through the DHOD and its coenzyme Q (CoQ) (Figure 1). The MtETS are valuable targets in malaria chemotherapy [21].
In our laboratory, we have characterized in detail the functional, kinetic, and structural properties of orotate phosphoribosyltransferase (OPRT) and orotidine 5'-monophosphate decarboxylase (OMPDC), the fifth and sixth enzymes of the de novo pathway [22-24]. The OPRT and OMPDC enzymes were purified directly from in vitro culture of P. falciparum. The native enzymes obtained from the parasite are organized in an α2β2 heterotetrameric quaternary structure having two subunits each of OPRT and OMPDC [22]. We have also expressed both genes in E. coli [23,24]. Co-expression recombinant P. falciparum OPRT and OMPDC genes are also exhibited the in vitro α2β2 complex formation [25,26]. Most recently, the parasites’ low complexity region is involved in the protein-protein interaction during the α2β2 heterotetrameric formation of the malarial OPRT and OMPDC enzymes, [(OPRT)2(OMPDC)2], as identified by means of a unique insertion of low complexity amino acid sequence characterized by single amino acid repeat, which was not seen in their homologous enzymes from other organisms [27]. Furthermore, three-dimensional crystal structures of the parasite OPRT and OMPDC have been elucidated [28-32], and compared to the recently characterized human OPRT and OMPDC structures [5], which are carried on a 52-kDa bifunctional UMP synthase protein [8,9]. Thus, the inhibitors of OMPDC have been designed and chemically synthesized for therapeutic potential against malaria by using the structure-based drug-design approach of the parasite enzyme [33].
Surprisingly, there is little information on the sequential enzymatic steps after UMP synthesis before yielding dCTP, dTTP, CTP, TTP, and UTP, which are the building nucleotide blocks for nucleic acid synthesis in the parasite (Figure 1, broken line box). Genes are present in the parasite genome but few enzymes have been studied to date. Ribonucleotide reductase (RNR) of P. falciparum catalyzes the production of deoxyribonucleotides from ribonucleotides, which is tightly associated with the thioredoxin reductase enzyme [34]. P. falciparum TMP kinase (TMPK), catalyzing the synthesis of dTTP and TTP, is classified as type I enzyme by amino acid sequence but has high efficiency in phosphorylation of azido-dTMP and dGMP as well as E. coli type II TMPK, sharing characteristics of both types [35]. Additionally, CTP synthetase (CTPS) catalyzes the production of CTP from UTP, is the only known enzyme for cytosine nucleotide de novo synthesis in P. falciparum [36]. Typically, CTP reacts with phosphocholine to form CDP-choline, which can combines with diacylglycerol to form phosphatidylcholine (PC) and other phospholipids by operating Kennedy pathway.
In human parasite P. falciparum, the de novo pyrimidine pathway is known to be linked with the de novo folate biosynthesis via thymidylate synthetase (TS) enzyme (Figure 1) [37]. The parasite TS is a part of the bifunctional dihydrofolate reductase-thymidylate synthase (DHFR-TS), a validated target for antifolate drugs used in malaria chemotherapy. The three-dimensional crystal structure of the parasite DHFR-TS enzyme and substrate channeling domains have been resolved [38]. It is well recognized that the de novo folate pathway is operating in the parasite, like in bacteria, whereas the human host is incapable of de novo synthesis [37,38].
Furthermore, functional and kinetic properties of carbonic anhydrase (CA) enzyme were studied in P. falciparum [39]. The parasite CA catalyzes the interconversion of HCO3- and CO2, having catalytic properties distinct from that of the human host CA isozyme I and II. However, low amino acid sequence identity in the primary structures and phylogenetic analyses of the P. falciparum CA is being tapped in preclinical phase for drug development [40-42]. Importantly, the CA supplies HCO3- as substrate for the first enzyme CPS II of the de novo pyrimidine biosynthetic pathway, linking the parasite CA to the pyrimidine pathway as shown in Figure 1 [42].
In conclusion, artemisinin-resistant parasites have already emerged and spread in Southeast Asian region by 2014 [43,44]. This phenomenon entails novel measures for malaria treatment and control. Fortunately, one triazolopyrimidine inhibitor, namely DSM265, targeting the parasite fourth enzyme DHOD of the de novo pyrimidine pathway would prove promising as it progresses to clinical phase I trials for drug development [45]. Moreover, as structure-based design of antimalarial drug development continues to be tapped three-dimensional crystal structure of the parasite enzyme, especially for the sixth enzyme OMPDC [30], the possibility of modulating potential toxicity through the pyrimidine pathway might have therapeutic potential against malaria [46].
Conflict of Interest
The author declares that there is no conflict of interest.
Acknowledgements
Our laboratory was supported by the UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (CHEMAL, TDR/WHO), the National Science and Technology Development Agency of Thailand (NSTDA Career Development Award), the Thailand Research Fund (TRF Basic Research), and Chulalongkorn University, Thailand.
References
- Hay SI, Okiro EA, Gething PW, Patil AP, Tatem AJ, et al. (2010)Estimating the global clinical burden of Plasmodium falciparum malaria in 2007. PloS Med 7: e1000290.
- White NJ (2008) Qinghaosu (artemisinin): the price of success. Science 320: 330-334.
- Dondorp AM, Nosten F, Yi P, Das D, Phyo A, et al. (2009) Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361: 455-467.
- Krungkrai SR, Krungkrai J (2016) Insights into the pyrimidine biosynthetic pathway of human malaria parasite Plasmodium falciparum as chemotherapeutic target. Asian Pac J Trop Med 9: 525-534.
- Welin M, Nordlund P (2010) Understanding specificity in metabolic pathways-structural biology of human nucleotide metabolism. Biochem Biophys Res Commun 396: 157-163.
- Weber G (1983) Biochemical strategy of cancer cells and the design of chemotherapy. Cancer Res 43: 3466-3492.
- Kim S, Park DH, Kim TH, Hwang M, Shim J (2009) Functional analysis of pyrimidine biosynthesis enzymes using the anticancer drug 5-fluorouracil in Caenorhabditis elegans. FEBS J 276: 4715-4726.
- Nara T, Hshimoto T, Aoki T (2000) Evolutionary implications of the mosaic pyrimidine biosynthetic pathway in eukaryotes. Gene 257: 209-222.
- Evans DR, Guy HI (2004) Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J BiolChem 279: 33035-33038.
- Gero AM, Brown GV, O’Sullivan WJ (1984) Pyrimidine de novo synthesis during the life cycle of the intraerythrocytic stage of Plasmodium falciparum. J Parasit 70: 536-541.
- Gardner MJ, Hall N, Fung E, White O, Berriman M, et al. (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498-511.
- Krungkrai J, Cerami A, Henderson GB (1990) Pyrimidine biosynthesis in parasitic protozoa: purification of a monofunctionaldihydroorotase from Plasmodium berghei and Crithidiafasciculata. Biochemistry 29: 6270-6275.
- O’Donovan GA, Neuhard J (1970) Pyrimidine metabolism in microorganisms. Bacteriol Rev 34: 278-343.
- Jones ME (1980) Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis. Annu Rev Biochem 49: 253-279.
- Krungkrai SR, Wutipraditkul N, Krungkrai J (2008) Dihydroorotase of human malarial parasite Plasmodium falciparum differs from host enzyme. BiochemBiophys Res Commun 366: 821-826.
- Krungkrai J, Krungkrai SR, Phakanont K (1992) Antimalarial activity of orotateanalogs that inhibit dihydroorotase and dihydroorotate dehydrogenase. Biochem Pharmacol 43: 1295-1301.
- Krungkrai J, Cerami A, Henderson GB (1991) Purification and characterization of dihydroorotate dehydrogenase from the rodent malaria parasite Plasmodium berghei.Biochemistry 30: 1934-1939.
- Krungkrai J (1995)Purification, characterization and localization of mitochondrial dihydroorotate dehydrogenase in Plasmodium falciparum, human malaria parasite. Biochim Biophys Acta 1243: 351-360.
- Hurt DE, Widom J, Clardy J (2006) Structure of Plasmodium falciparum dihydroorotate dehydrogenase with a bound inhibitor. ActaCrystallogr D BiolCrystallogr62: 312-323.
- Liu S, Neidhardt TH, Grossman T, Ocain T, Clardy J (2000) Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. Structure 8: 25-33.
- Rodrigues T, Lopes F, Moreira R (2010) Inhibitors of the mitochondrial electron transport chain and de novo pyrimidine biosynthesis as antimalarials: the present status. Curr Med Chem 17: 929-956.
- Krungkrai SR, Prapunwattana P, Horii T, Krungkrai J (2004) Orotatephosphoribosyltransferase and orotidine 5'-monophosphate decarboxylase exist as multienzyme complex in human malaria parasite Plasmodium falciparum. Biochem Biophys Res Commun 318: 1012-1018.
- Krungkrai SR, Aoki S, Palacpac NMQ, Sato D, Mitamura T, et al. (2004) Human malaria parasite orotatephosphoribosyltransferase: functional expression, characterization of kinetic reaction mechanism and inhibition profile. Mol Biochem Parasitol 134: 245-255.
- Krungkrai SR, DelFraino BJ, SmileyJA, Prapunwattana P, Mitamura M, et al. (2005) A novel enzyme complex of orotatephosphoribosyltransferase and orotidine 5'-monophosphate decarboxylase in human malaria parasite Plasmodium falciparum: physical association, kinetics and inhibition characterization. Biochemistry 44: 1643-1652.
- Kanchanaphum P, Krungkrai J (2009) Kinetic benefits and thermal stability of orotatephosphoribosyltransferase and orotidine 5'-monophosphate decarboxylase enzyme complex inhuman malaria parasite Plasmodium falciparum. BiochemBiophys Res Commun 390: 337-341.
- Kanchanaphum P, Krungkrai J (2010) Co-expression of human malaria parasite Plasmodium falciparum orotatephosphoribosyltransferase and orotidine 5'-monophosphate decarboxylase as enzyme complex in Escherichia coli: a novel strategy for drug development. Asian Biomed 4: 297-306.
- Imprasittichai W, Roytrakul S, Krungkrai SR, Krungkrai J (2014) A unique insertion of low complexity amino acid sequence underlines protein-protein interaction in human malaria parasite orotatephosphoribosyltransferase and orotidine 5'-monophosphate decarboxylase. Asian Pac J Trop Med 7: 184-192.
- Krungkrai SR, Kusakari Y, Tokuoka K, Inoue T, Adachi H, et al. (2006) Crystallization and preliminary crystallographic analysis of orotidine 5'-monophosphate decarboxylase from human malaria parasite Plasmodium falciparum. ActaCrystallogr Sect F StructBiolCrystCommun62: 542-545.
- Tokuoka K,Kusakari Y, Krungkrai SR, Matsumura H, Krungkrai J, et al. (2008) Structural basis for the decarboxylation of orotidine 5'-monophosphate (OMP) by Plasmodium falciparum OMP decarboxylase. J Biochem 143: 69-78.
- Takashima Y, Mizohata E, Krungkrai SR, Fukunishi Y, Kinoshita T, et al. (2012) The in silico screening and X-ray structure analysis of the inhibitor complex of Plasmodium falciparum OMP decarboxylase. J Biochem 152: 133-138.
- Takashima Y, Mizohata E, Tokuoka K, Krungkrai SR, Kusakari Y, et al. (2012)Crystallization and preliminary crystallographic analysis of orotatephosphoribosyltransferase from human malaria parasite Plasmodium falciparum. ActaCrystallogr F StructBiolCommun68: 244-246.
- Kumar S, Krishnamoorthy K, Mudeppa DG, Rathod PK (2015) Structure of Plasmodium falciparum orotatephosphoribosyltransferase with autologous inhibitory protein-protein interactions. ActaCrystallogr F StructBiolCommun71: 600-608.
- Meza-Avina ME, Wei L, Buhendwa MG, Poduch E, Pai EF, et al. (2008) Inhibition of orotidine 5'-monophosphate decarboxylase and its therapeutic potential. Min Rev Med Chem 8: 239-247.
- Rubin H, Salem JS, Li LS, Yang FD, Mama S, et al. (1993) Cloning, sequence determination, and regulation of the ribonucleotidereductase subunits from Plasmodium falciparum: a target for antimalarial therapy. ProcNatlAcadSci USA 90: 9280-9284.
- Whittingham JL, Carrero-Lerida J, Brannigan JA, Ruiz-Perez LM, Silva AP, et al. (2010) Structural basis for the efficient phosphorylation of AZTMP and dGMP by Plasmodium falciparum type I thymidylate kinase. Biochem J 428: 499-509.
- Yuan P, Hendricks EF, Fernandez HR, O’Sullivan WJ, Stewart TS (2005) Functional expression of the gene encoding cytidine triphosphate synthetase from Plasmodium falciparum which contains two novel sequences that are potential antimalarial targets. MolBiochemParasitol 143: 200-208.
- Krungkrai J, Webster HK, Yuthavong Y (1990) Folate and cobalamin metabolism in Plasmodium falciparum. Parasitol Today 6: 388-391.
- Yuvaniyama J, Chitnumsub P, Kamchonwongpaisan S, Vanichtanankul J, Sirawaraporn W, et al. (2003) Insights into antifolate resistance from malarial DHFR-TS structures. Nat StructBiol 10: 357-365.
- Krungkrai SR, Suraveratum N, Rochanakij S, Krungkrai J (2001) Characterisation of carbonic anhydrase in Plasmodium falciparum. Int J Parasitol 31: 661-668.
- Krungkrai J, Scozzafava A, Reungprapavut S, Krungkrai SR, Rattanajak R,et al. (2005) Carbonic anhydrase inhibitors. Inhibition of Plasmodium falciparum carbonic anhydrase with aromatic sulfonamides: towards antimalarials with a novel mechanism of action? Bioorg Med Chem 13: 483-489.
- Krungkrai J, Supuran CT (2008) The alpha-carbonic anhydrase from the malarial parasite and its inhibition. Curr Pharm Design 14: 631-640.
- Krungkrai J, Krungkrai SR, Supuran CT (2008) Carbonic anhydrase inhibitors. Inhibition of Plasmodium falciparum carbonic anhydrase with aromatic/ heterocyclic sulfonamides: in vitro and in vivo studies. Bioorg Med ChemLett 18: 5466-5471.
- Ashley EA, Dhorda M, FairhurstRM, Amaratunga C, Lim P, et al. (2014) Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371: 411-423.
- Krungkrai J, Krungkrai SR (2016) Antimalarial qinghaosu/artemisinin: the therapy worthy of a Nobel Prize. Asian Pac J Trop Biomed 6: 371-375.
- Phillips MA, Lotharius J, Marsh K, White J, DayanA, et al. (2015) A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria. SciTransl Med 7: 296ra111.
- Flannery EL, Chatterjee AK, Winzeler EA (2013) Antimalarial drug discovery-approaches and progress towards new medicines. Nature Rev Micro 11: 849-862.
Citation: Krungkrai J (2017) Malaria Parasite Pyrimidine Nucleotide Metabolism: A Promising Drug Target. Arch Parasitol 1: 101.
Copyright: © 2017 Jerapan K. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Open Access Journals
Article Usage
- Total views: 6118
- [From(publication date): 0-2017 - Aug 18, 2025]
- Breakdown by view type
- HTML page views: 5069
- PDF downloads: 1049