Research Article Open Access
Molecular Docking Studies on Potent Adsorbed Receptor of Thrh Protein: A New Target for Biodegradation of Indigo Dye
Prabhavathi P1*, Rajendran R1, Karthik Sundaram S1, Dinesh Kumar S2,3, Santhanam P2, Premnath D4, Ponmari G4, Manikandan A1and Mi- Kyung Kim3
1PG and Research Department of Microbiology, PSG College of Arts and Science, Coimbatore, Tamil Nadu, India
2Department of Marine Science, School of Marine Sciences, Bharathidasan University, Tiruchirappalli, Tamil Nadu, India
3MCK Biotech Co. Ltd., Daegu R&D Fusion Center, Daegu, South Korea
4Department of Bioinformatics, Karunya University, Coimbatore, Tamil Nadu, India
- Corresponding Author:
- Prabhavathi P
PG and Research Department of Microbiology
PSG College of Arts and Science, Coimbatore
Tamil Nadu, India
Tel: +919787284784
E-mail: prabha_micro2007@yahoo.co.in
Received Date: May 12, 2016; Accepted Date: June 06, 2016; Published Date: June 13, 2016
Citation: Prabhavathi P, Rajendran R, Karthik Sundaram S, Dinesh Kumar S, Santhanam P, et al. (2016) Molecular Docking Studies on Potent Adsorbed Receptor of Protein: A New Target for Biodegradation of Indigo Dye. J Bioremed Biodeg 7:356. doi: 10.4172/2155-6199.1000356
Copyright: © 2016 Prabhavathi P, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Vat dyes are aromatic compounds widely used for denim textile industries, this result in a great wastewater problem from this industry due to recalcitrant nature of these dyes. The active protein (ThrH) was purified from Pseudomonas aeruginosa by DEAE-Sepharose A-50 column chromatography and this 3D crystal structure was reported recently. The present study aimed to demonstrate the binding energy between 3D crystal structures of indigo dye and ThrH. We have calculated the gliding score as well as gliding energy based on the hydrophobic interactions between targeted sites (amino acid and dye residue) and the main think is binding energy which was observed maximum level because of the presence of magnesium ions along with catalytic molecules located at the binding sites. The dye degraded mineralized compound was predicted by mass spectrum and infrared spectroscopy.
Keywords
Indigo domain; ThrH protein; Binding sites; Induced fit docking (IFD); Schrodinger program
Introduction
There are around 600 dyeing industries in and around Coimbatore district situated in the southern State of Tamil Nadu, India. In these dyeing industries about 30-60 L of water is consumed per kilogram of cloth dyed, and large quantities of effluents are released during processing. In textile industry, the dyeing and finishing process are very important which is widely employed in all kind dyeing process. Especially, vat dyes are extensively used for making denim cloth and they have two closely defined groups such as anthraquinonoid, heterocyclic quinines and indigoid containing two or more keto (=o) groups separated by a conjugated system of double bonds. Another important group inside the class of vat dyes is indigo. Intra and inter molecular hydrogen bonding are responsible for the dye’s extremely low solubility in water, diluted acids/ alkali. It is readily reduced by various reagents, such as sodium dithionite, hydroxyl acetone or by electrochemical methods. In an alkali medium the yellow brown sodium salt of leuco indigo is produced, which can be converted by acids to white indigo.
During textile dyeing and finishing processes, a large amount of wastewater is generated and discharged into environment directly without any treatment. This textile wastewater is scandalously known to contain strong color, a large amount of suspended solids and highly fluctuating pH. There are different physical and chemical methods are used to treat through adsorption, filtration, flocculation and oxidation [1,2] which are quite expensive and generate huge amount of wastes. Among being low cost and easy to everyone for wastewater treatment, the microbial system is familiar by their ability to treat and reduce color and suspended solid.
But vat dye requires to modify some complicated treatment technique using oxido-reduction process which has to initiate the oxidation and reduction mechanisms during dyeing. Basically the indigo dye is insoluble to water so from this point of view the industry people were doing these kinds of techniques. After this process the insoluble form is converted into soluble form (leuco) which has the ability to bind on denim fabric. But this process has some drawback because the soluble form is again re-oxidized to the original insoluble form. So from this point of view, the previous study revealed that to purify the cellulose enzyme which is used to bind indigo dye on denim fabric through washing process [3].
From this problem, the present research focuses on interaction and stability of indigo dye (ligand molecule) and ThrH protein which has responsible for oxido-reduction mechanisms. Initially, the molecular docking study was conducted under Schrodinger program (version 9.1). The results obtained from P. aeruginosa PA01 and this current scenario is expected to provide information regarding the hydrogen and hydrophobic interactions between the amino acid and indigo domain. The ThrH protein was purified from and which was downloaded from protein data bank. We conducted docking studies using Schrodinger’s glide molecule and got the good glide scores on the basis of ligands.
Materials and Methods
Collection of sample
The Indigo dye containing denim effluent was collected from the KG fabrics, Perundhurai, Tamil Nadu, India. The effluent sample was collected in sterile plastic container and stored in cold condition until use.
Isolation and identification of adapted bacterial strain
P. aeruginosa was predominantly isolated from the indigo dye containing denim textile wastewater and this strain is capable of efficiently expressing the oxidoreductase (metal ion binding site: electrons are transferred from electron donors to electron acceptors) enzyme activity to decolorize the crude indigo dye particle via cometabolism. The P. aeruginosa strain was cultivated in nutrient broth, typically containing 0.1 g/L of glucose and urea. The isolate was examined for their gram staining and cell shape. Biochemical tests were performed according to Bergey’s manual of systemic bacteriology [4] and also done 16S rRNA gene sequencing based on PCR pattern.
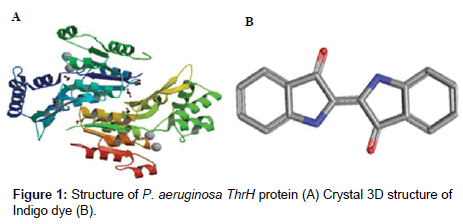
D Structure of indigo dye: The synthetic liquid Indigo dye (Indigotin) is 80% pure with chemical formula C12H8O2N2 and 3D molecular structure is shown in Figure 1A.
Purification of P. aeruginosa protein
P. aeruginosa broth culture was centrifuged at 10,000 rpm for 15 min at 4°C. The supernatant was separated from the pellet containing aliquot then the ammonium sulfate was added to the culture supernatant to get saturation level. After incubation (-20°C) the supernatant was again centrifuged at 5,000 rpm for 15 min at 4°C. Overnight dialysis was performed using dialysis bag for the precipitate against phosphate buffer. The pooled fractions were collected from DEAE Sepharose A-50 matrix column (1 × 25 cm). The column was previously equilibrated with 25 mM phosphate buffer (pH 8). The enzyme bound to the column was eluted with a linear gradient of 250 mM phosphate buffer (pH 8) at a flow rate was 15 ml/ h and 2 ml fractions were collected and checked for enzyme activity and further SDS- PAGE was performed using 12% polyacrylamide gel. The protein bands were visualized by staining coomassie brilliant blue and the molecular weight of the purified enzyme was determined by standard markers.
Protein estimation
Total amount of protein was measured according the standard method [5] used protein marker as a standard.
Study of indigo interaction with pooled bacterial enzyme factions
About 0.01% of Indigo dye was mixed with 0.1M phosphate buffer at pH 6 and added 0.5 ml of bacterial pooled fraction then the mixture was incubated at 47°C for 24 h. The liquid above the solution was removed and added equal volume of Sulphuric acid: Distilled water (1:1) and kept under water bath for 10 minutes for the dissolving of Indigo dye particles as well as the deactivation of enzyme. The absorbance of combined fractions of extracts was measured at 609 nm and then the decolorization pattern of indigo dye was calculated.
Decolorization and biodegradation analysis
The decolorization was quantitatively analyzed by UV-Visible spectrophotometer. During UV-Vis spectrum analysis, the absorbance spectrum in decolorized extract were recorded at range of 609 nm and were compared with result from control. Instead of the degradation was monitored by Fourier transform infrared spectroscopy (FTIR) and the identification of secondary metabolites were carried out by Liquid chromatography mass spectroscopy (LC-MS: Agilent Triple Quadra pole; Software: Mass Hunter Qualitative Work station; Infusion pump: Harward Pakd infusion pumps). After complete decolorization by Pseudomonas aeruginosa culture broth was centrifuged at 10,000 rpm for 20 min and equal volume of chloroform was used to extract secondary metabolites from visible supernatant then the extract was lyophilized to powder in lyophilizer, then the crystallized powder was dissolved in small volume of GC grade chloroform and used for further analysis. The biodegraded functional compounds were characterized by FTIR and after getting the results were compared with control indigo dye. The FTIR analysis was done in the mid IR region (400- 4000 cm-1).
MALDI-TOF
During peptide analysis collision induced dissociation and MS spectrum were performed through MS/ MS analysis then the spectrum was compared with NCBI nr databases. Finally the peak lists were generated on signal based filtering as well as de-isotoping parameters. The final peak list was searched and compared beside existing data bases such as Swiss prot and NCBI.
N-Terminal sequence analysis
The protein spots separated on the gel plate were transferred onto a membrane using a semi dry blotting apparatus (BIO-RAD Trans- Blot SD) at 2 mA/cm2 for 45 min and that membrane was stained with Coomassie Brilliant Blue and washed with 50% methanol. Coomassie stained protein spots were excised from the membrane and installed in the blot cartridge of an odel 491A protein sequencer (Perkin– Elmer, Foster City, CA, USA) for sequencing analysis. The obtained N-terminal sequence was used for protein identification by BLAST search of NCBI.
Molecular docking study
Receptors (Structure): From the data bases, the protein structure was predicted (Figure 1B) which was used in docking program (Schrodinger). The protein file (1RKU) was downloaded from PDB sum then the water molecules were removed (http://www.ebi.ac.uk/ thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl) application of Schrodinger and missing side chains were filled using prime options of protein preparation wizard. Followed by preprocess, analyze, optimization and minimization steps were performed to prepare the protein for docking.
Active site analysis: The Active sites and their residues obtained from PDB ligand explorer entry of ThrH protein of Pseudomonas aeruginosa PAO1 protein (PDB ID-1RKU) (http://www.schrodinger. com) and this entry contains five active site residues (Asp-78, ARG-81, ARG-102, GLY-105, PHE-106).
Ligand preparation: All the ligands in mol format from Pub Chem compound database are loaded individually in Schrodinger workspace and their geometries are cleaned was done by clean up geometry option in edit menu. These cleaned ligands in mol format are loaded in the Schrodinger workspace for ligand preparation. In this process Ligands Counter ions and excess hydrogen’s are removed using “desalt” option in task menu of Ligprep application in the Schrodinger (PDB sum) (PDB sum (http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/ pdbsum). In addition in presence of OPLS-2005 force filed ligfilter is used to produce the optimized ligand structure and also ionizer and generate tautomers options were used to generate possible states of every ligand. The output of this ligprep is in maestro format of every ligand is used for glide docking with receptor molecule.
Virtual screening: The ligand (indigo dye) was docked with ThrH gene of P. aeruginosa PA01 through glide docking program in Schrodinger then followed by the receptor grid was also generated. The shape and properties of the receptor are represented on a grid by several different sets of fields that provide progressively more accurate scoring of the ligands poses. Centroid of selected residues option is used to generate a grid. This option centers grids at the centroid of a set of active site residues which was calculated at five different active site residues and the size of x, y and z in receptor grid similar to 6.75, 30.87, 12.0 Å respectively. Grid generated receptor file is used for ligand docking. Ligand docking steps was as follows; In Schrodinger glide submenu of the ligand docking applications used for docking. Highthroughput virtual screening (HTVS) docking is intended for the rapid screening of very large numbers of ligands, Grid generated receptor molecule docked with ligands used to HTVS (High-throughput virtual screening), from this screening best ten lowest docked energy ligands were selected for SP (Standard-precision) docking which has used for the screening of ligands from large numbers. Ten ligands are selected from HTVS were individually docked to the receptor. Extra-precision (XP) docking and scoring is a more powerful and discriminating procedure, which takes longer to run than SP. The XP is designed to be used on ligand poses that have a high score using SP docking. First the database compounds run through SP docking, then take the top 10% to 30% of final poses and dock them using XP, so it gives the best drug candidates that interacted with the receptor to inhibit the process of the target.
Results and Discussion
Isolation and identification of adapted bacterial strain
Screening and characterization of ThrH protein (P. aeruginosa PA01) morphology and taxonomic feature was performed by Bergey’s manual of systemic bacteriology as well as MS spectrum. The amplicon of 16S rRNA gene was sequenced and aligned using BLAST and this isolate belongs to Pseudomonas sp. with the maximum relation to P. aeruginosa with 98% sequence similarities. This sequence was submitted in Genbank (Accession no: KC118965).
Study of Indigo dye interaction with adapted bacterial pooled enzyme fractions
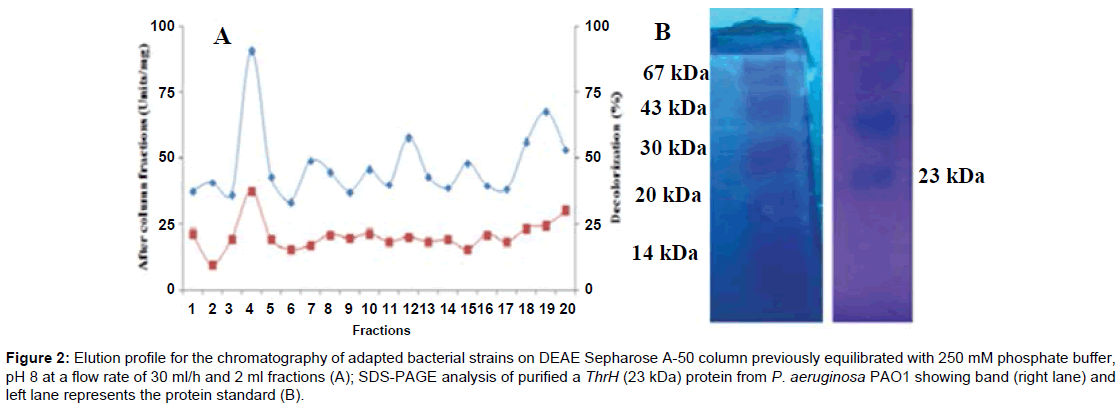
The Indigo dye decolorization was compared between the bacterial pooled fractions by means of absorbance with enzyme specificity. Already we have been taken for the fractions by using DEAE-Sepharose A-50 column chromatography from this various fractions of P. aeroginosa PA01 was able to study the indigo dye decolorization as well as the activity of enzyme, among these fractions only 4th fraction of PK18 isolate (P. aeruginosa) significantly influenced the enzyme action during the dye degradation and same is depicted in Figure 2. In case of the remaining fractions were not efficient for the dye decolorization. It was evident from the study that the 4th fraction of PK18 was used for the decolorization experiments. Although the mechanism of induction is still unknown (i.e., the indigo dye degradation pathway), some similar researches propose that microbial laccase enzymes may be responsible for partial decolorization of Indigo dye [6].
Figure 2:Elution profile for the chromatography of adapted bacterial strains on DEAE Sepharose A-50 column previously equilibrated 250 mM phosphate buffer, pH 8 at a flow rate of 30 ml/h and 2 ml fractions (A); SDS-PAGE analysis of purified a ThrH (23 kDa) protein from P. aeruginosa PAO1 showing band (right lane) and left lane represents the protein standard (B).
Purification of ThrH protein
The ThrH obtained from P. aeroginosa PA01 was purified by ammonium sulfate precipitation, Ion-exchange chromatography on DEAE- Sepharose A-50 column. The fractions of P. aeroginosa (PK18), which were adsorbed on DEAE- Separose and eluted with 250 mM phosphate buffer, showed the highest dye decolorization as well as enzyme activity, upon this chromatography only one fraction found to be a large adsorbed peak at 4th fraction of PK18 was enriched with ThrH activity with highest decolorization pattern. Subsequently, other fractions of PK18 were not effective for the dye decolorization due to the lack of insufficient activity of extracellular enzymes. The enzyme (4th fraction of PK18) was purified 16.4 fold from the crude extract with 55.4% yield. The purified ThrH exhibited a total activity of 613.4 U/ml (Table 1) than the other fractions. Previous work had indicated that a laccase from monkey head mushroom was attained 15-fold with 12% yield. Its chromatographic procedure involved ion-exchange chromatography on DEAE-cellulose, CM-cellulose, and Q-Sepharose and FPLC-gel filtration on Superdex 75 [7]. This protein was selected and in-gel digested with trypsin for MS analysis and then successfully analyzed and one protein was identified by sequence homology using the elucidated amino acid sequences.
| Fractions | Protein yield (mg) | Total Activity (U/ml) | Purification (Fold) | Recovery (%) |
|---|---|---|---|---|
| Crude Extract | 1563 | 950 | 1 | 100 |
| PK18- 1 | 137 | <13 | 2.34 | 23 |
| PK18- 2 | 41 | <5 | - | - |
| PK18- 3 | 67 | <6 | - | - |
| PK18- 4 | 458 | 613.4 | 16.4 | 55.4 |
| PK18- 5 | 112 | <12 | 1.02 | 13 |
| PK18- 6 | 96 | <9 | - | - |
| PK18- 7 | 68 | <6 | - | - |
| PK18- 8 | 83 | <8 | - | - |
| PK18- 9 | 34 | <3 | - | - |
| PK18- 10 | 48 | <4 | - | - |
| PK18- 11 | 59 | <5 | - | - |
| PK18- 12 | 79 | <7 | - | - |
| PK18- 13 | 95 | <9 | - | - |
| PK18- 14 | 102 | <11 | 1 | 10.3 |
| PK18- 15 | 125 | <12 | 1.21 | 18 |
| PK18- 16 | 69 | <6 | - | - |
| PK18- 17 | 97 | <9 | - | - |
| PK18- 18 | 60 | <6 | - | - |
| PK18- 19 | 75 | <7 | - | - |
| PK18- 20 | 83 | <8 | - | - |
Table 1: Yields and enzyme activity of aqueous extract and various chromatographic PK18 isolate fractions.
Determination of molecular mass and N-terminal sequence
From SDS-PAGE result, ThrH protein band was showed and confirmed at the range of ~23 kDa (molecular weight) matched with standard marker in SDS gel plate. The N- terminal sequence of that protein is shown in Table 2 which is exhibited and homology to published data bases of P. aeruginosa PA01. From the results, it was suggested that the ThrH from P. aeruginosa may be a novel protein and encoded by different genes. The purity of adapted bacterial enzymes and their molecular weight were determined by Sodium Dodecyl Sulphate Poly Acrylamide Gel Electrophoresis (SDS-PAGE) using 12% gel according to the standard method [8]. The low molecular weight 23 KDa was showed the purity of ThrH protein. The standard protein marker was also run parallel along with the bacterial protein sample.
| SNo | MW of peptides | Start - End | N- Terminal Sequences (Protein sequence) |
|---|---|---|---|
| 1. | 1972.0022 | 23 - 41 | R. VVMAPMTRNFSPGGVPNAK. V |
| 2. | 1545.8892 | 101 - 113 | R. IVPQLWHVGSVRR. L |
| 3. | 1760.8290 | 114 - 130 | R. LGVEPDASVPGYGPMEK. A (oxidation) |
| 4. | 869.4793 | 136 - 143 | K. VLVHGMSK. A |
| 5. | 1494.8558 | 339 - 351 | R. ALLVDPEWAVKVR. E |
MW: Molecular weight
Table 2: Matched peptides shown in P. aeruginosa.
Docking studies on Indigo domain binding site
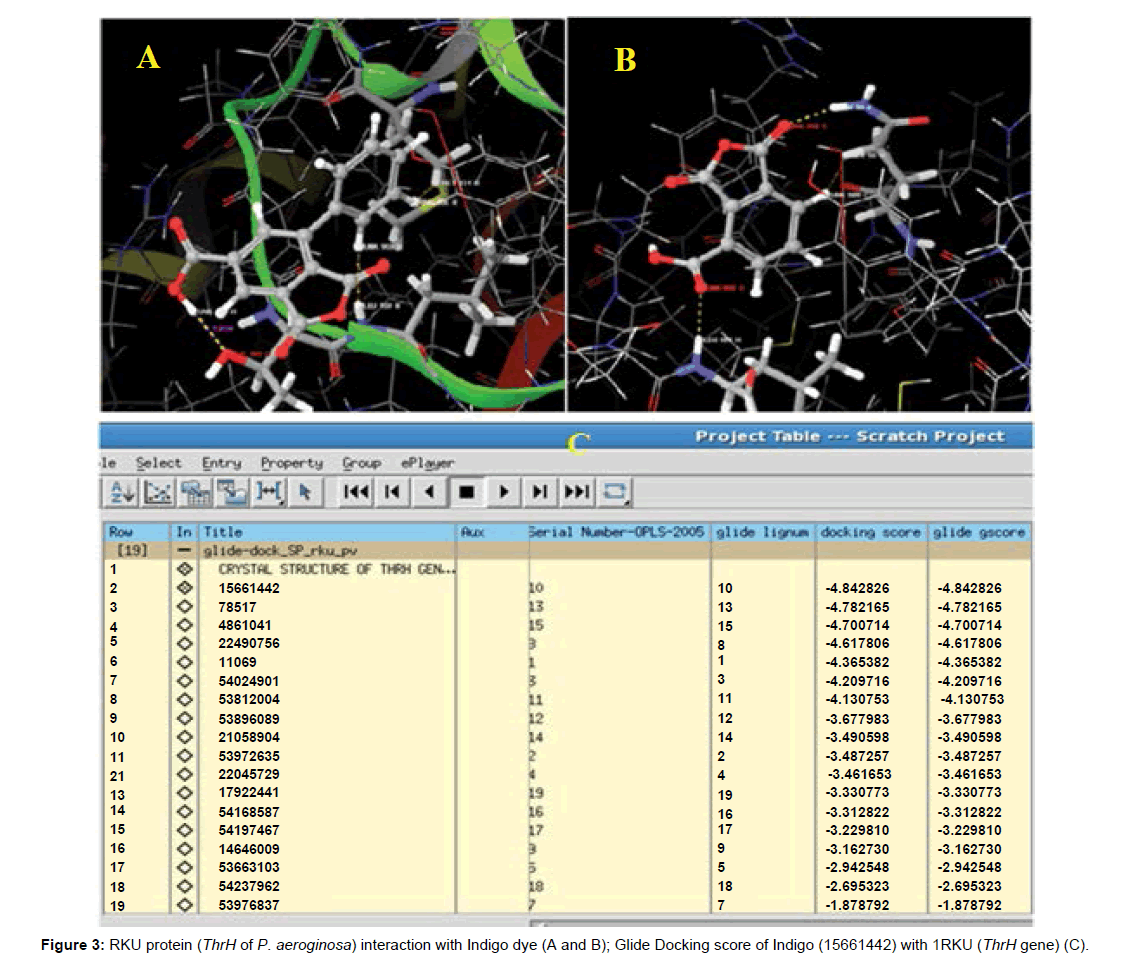
To study the molecular basis of interaction and affinity of binding of ThrH protein (Figure 1A) and its analogues ligands were docked into the active site of 3D structure of Indigo dye molecule. The ranking of ligands was based on the glide score was noted and the ligands accepted poses with the specific receptors. The difference in glide score on the basis of SP (Standard-Precision) and XP (Extra- Precision) docking study. The superposition of the docked ThrH in Indigo molecule revealed that the binding mode of this affinity was considered to be essentially similar in both docking methods. This confirms that the structural modification implemented in this study is significantly related to their activity. Also, this proved the reasonability and reliability of the docking results.
Binding domains
The hydrogen atom of carbonyl group of ligand molecule has formed H-bond with the O atom of Thr108 at a distance of 1.86 Å and then H atom of Leu109 residue at a distance of 1.93Å (Figures 3A and 3B), respectively. The both residual distances were showed in Figures 4 and 5. The G-score of is - 4.84 and G-energy is -26.4 Kcal/mol (Figure 3C). From this study, the glide score as well as glide energy have attained maximum level which confirmed that the interaction of targeted ligand molecule and ThrH. The various final docked confirmation files were observed and retrieved which has evaluated from the number of H bonds were formed and their distance on active site and inhibitor. On the basis of this study reveals that the following amino acid in P. aeruginosa PA01 was involved in H bonding interactions with various inhibitors: MET101 (Methionine: non polar-sulphur or hydroxyl), LEU109 (Leucine: non polar-aliphatic), GLN98 (polar-acetic), THR108 (Threonine: polar-aromatic, hydroxyl or sulfur).
The active site of ThrH
The closest protein neighbor of ThrH has recently been well characterized structurally. Sequence and structure comparison of ThrH and PSP revealed a very similar active site configuration. The active site of ThrH is located at the junction of the two domains. Two Mg2+ ions are found in and near the active site. Their coordination exhibits a nearly ideal octahedral geometry. During substrate binding, some modifications in the residues but they have similar chemistry and catalytic activity of this particular protein. The above all the residues exhibited effective binding active site in ligand interaction which has used as an excellent carrier molecule as well as authenticate them as potential for remediation of textile wastes.
There are some domains like (polar residue; O2), Met-108 (hydroxyl or sulphur) and Leu-109 (non polar aliphatic) were observed and evaluated at the bottom as well as the right side of enzyme molecule. Similar domains formed by aromatic residues may be distinguished on the surface of ThrH amino acid. However, these residues may provide the initial binding and orientation of the enzymes on Indigo dye domain. Similar results have been reported about the hydrophobic cellulase enzyme activity which is used for most surface emulsifiers during bulk denim treatment process. This enzyme had attained the most binding activity on the surface of denim fabric which has used to remove the dye particles effectively [3]. The current research reveals that, the interaction and stability of these combinations have showed efficient scoring, energy and hydrogen bond. Hence, this work could be evaluated about the receptor-ligand interactions, which would help in designing of microbial proteins in indigo degradation. Hydrogen bonding plays an important role for the structure and function of biological molecules, especially for inhibition in a complex, which is a promising way of interfering with indigos role in pollution. Further the work can be evaluated experimentally to study the receptor-ligand interactions, which would help in designing of microbial proteins in indigo degradation.
Decolorization and biodegradation assay
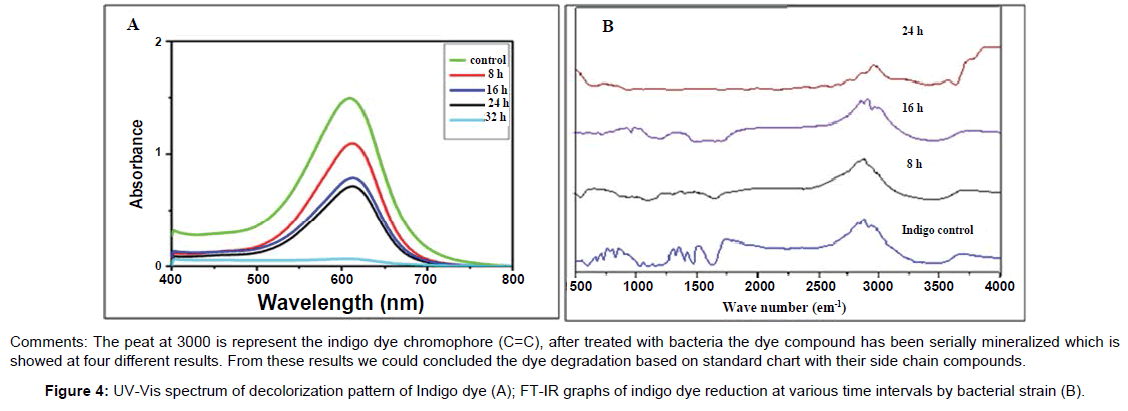
UV-Visible spectrophotometer: The decolorization of indigo dye was followed by UV-Vis analysis. From this result, the various spectrum was visualized at 610 nm after 48 hours incubation. The decreased intensity of visible peak of indigo dye which has not fully degraded after 24 hours incubation. After several hours incubation, showed peaks that could be evaluated the mineralized compounds (degraded compound) (Figure 4A). Furthermore the increased absorption in the visible spectra could be good evidence that polymerization reaction might have occurred. The breaking down of the dye into smaller fragments, including the breakage of the indigo pie bond can lead to a decrease in the absorbance of the visible spectra and into a colorless solution. However, according to the literature, from this reaction a product could simply lose the color due to a shift of the UV spectrum, rather than a direct degradation of the molecule into smaller fragments.
However, the previous literature studied; from this final end product could result that is simply losing the color due to a shift of the UV spectrum, rather than a direct degradation of the molecule into smaller fragments. This degradation action was initiated with the help of oxido-reductase enzyme on microorganisms which might be a simple shift in the UV spectrum. Balan and Monteiro [9] suggested that after oxidation reaction an extracellular enzyme disrupts the aromatic ring and on the results were obtained secondary metabolites by LC-MS.
Analysis of FT-IR spectrum: FT-IR spectra were obtained from samples treated showed the presence of several peaks at different time intervals. The crude indigo dye is subjected to FTIR and the graph obtained was compared with that of the samples treated at the end of 8 h, 16 h and 24 h (Figure 4B). The presence of peaks between the range of 1500-600 cm-1 represents the presence of the C=C stretch which is the chromophores of the indigo dye. The peak present at the range of 1300 cm-1 represents the O-H bond. The reduction in the colour of the indigo dye over a period of enzymatic action gives rise to the removal of C=C bond, and the presence of stretches at the range of 1500 cm-1 for C=O and peaks at the range of 2900 cm-1 represents the N-H bond. These functional groups suggest that the indigo chromophore was reduced and was converted to another ringed structure with the presence of peaks at the range of 1000 cm-1. The presence of peaks at the range of 1100 cm-1 suggests that the ringed structure was broken down by the enzymes though peaks at the range of 1000 cm-1 was still intact. This suggests that the four aromatic rings present in the structure of the indigo dye on reduction gives rise to a two ringed compound and there would be a further breakage in one of the ringed structure though the other ring was still intact (presence of peak at 1000 cm-1). The carboxylic acid and NH2 group was observed at the range of 1400-1500 cm-1 and 1400-1450 cm-1 respectively. The reduction of peaks at the end of treatment showed no peaks at the range of 1000 cm-1 which proved that the ringed structure is completely broken down and that the indigo dye was mineralized completely [10].
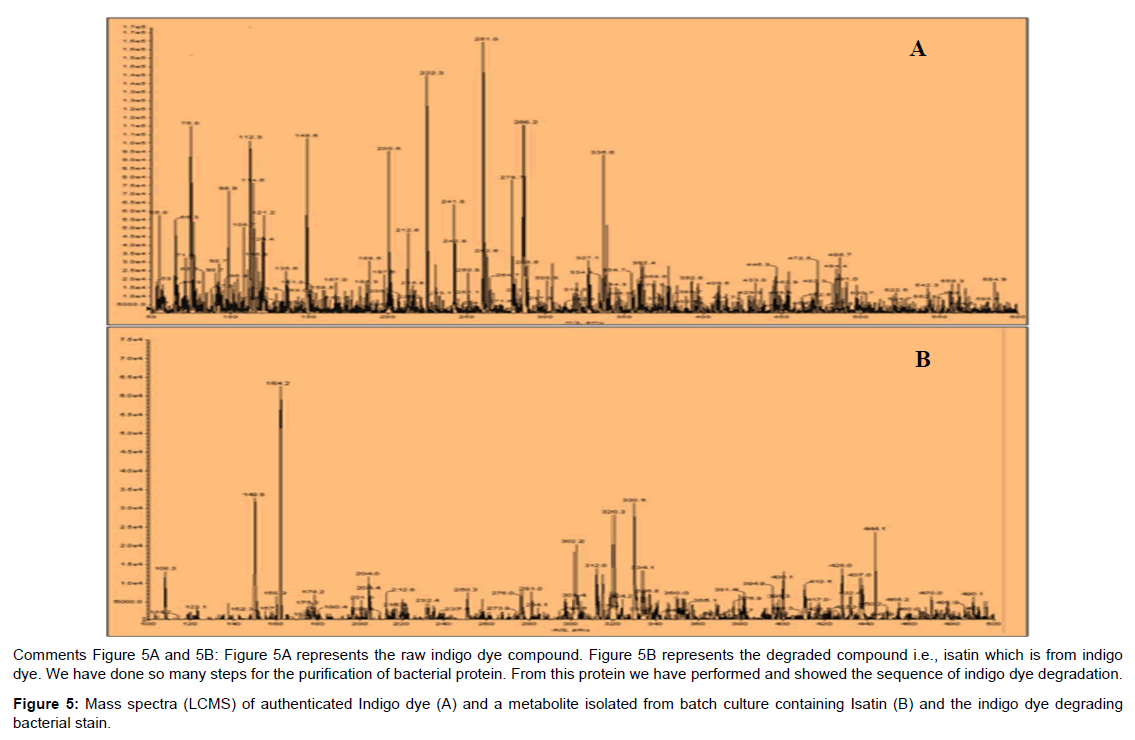
LCMS analysis: LCMS were carried out to investigate the secondary metabolites observed during degradation of indigo dye. In control dye sample (Figure 5) was collected at 0 h showed a major peak with the retention time of 0.76 min (261.9 m/z). after 8 h the sample was treated with similar study and got a one new peak while other peaks were disappeared at a time 1.21 min with m/z value of 261.9 m/z, which suggests that the compound whose molecules are composed of two identical, when the spectrum was taken for the same treated sample after 24 h and had greatest extent to note that the indigo dye peak was disappeared then additionally a new peak was formed (164 m/z) at the retention time of 1.75 min. at the end of 16 h incubation the isatin compound was formed. Since the electrons are necessary to reduce the oxygen molecule to water and finally to oxide indigo (aromatic ring- C=C- aromatic ring) dye to isatin (single aromatic ring) due to oxido reductase enzyme. The similar work was reported states that the degradation of indigo and bleaching on fabrics using purified laccases from T. hirsuta and S. rolfsii in combination with redox-mediators [6,11].
Conclusions
From the above results, we could concluded that the UV, FTIR and LCMS of various spectral data confirmed that the decolorized Indigo dye contained isatin and anthranilic acid, which were obtained through Indigo dye pie bond (Aromatic ring-C=C- Aromatic ring) when used mixed bacterial strains. The molecular basis of interaction and affinity of ThrH amino acid were studied and its analogues ligands were also docked into the active site of 3D structure of indigo dye domain. The best ranging of ligands accepted on the basis of both Glide score (-4.84 Kcal/mol) and Glide energy (-26.4 Kcal/mol) by SP and XP docking methods and also got the good hydrogen bond (1.86 Å and 1.93 Å) interaction within the ligands molecules. It is clear that further studies should be performed in order to better understand the “ThrH” protein of Pseudomonas aeruginosa PA01 mediated indigo dye degradation mechanism of suitable substrates. Finally this method is most efficient and applicable innovative technique for degradation as well as denim washing because of high interaction and stability of ligand molecule (Indigo dye) and ThrH protein by docking score. In vivo experiments will be necessary to understand the complex pathways of dye degradation products in the natural environmental.
Acknowledgements
The author (PP and SD) acknowledges the University Grants Commission (Rajiv Gandhi National Fellowship) and Department of Biotechnology for their financial support and also thanks the Principal and Secretary of PSG College of Arts and Science, Coimbatore, Tamil Nadu for their support.
References
- Hou H, Zhou J, Wang J, Du C, Yan B (2004) Enhancement of laccase production by Pleurotusostreatus and its use for the decolourization of anthraquinone dye. Process Biochemistry 39: 1415-1419.
- Zollinger H (1987) Color chemistry: syntheses, properties and applications of organic dyes and pigments. Federal republic of Germany: Verlag GmbH & Co 8: 197-199.
- Gusakov AV,Sinitsyn AP, Markov AV,Skomarovsky AA,Sinitsyna OA,et al. (2000) Indigo-binding domains in cellulase molecules. Biocatalists Fundamentals and Applications 41: 77-80.
- Sneath PHA (1994) Endospore forming gram positive rods and cocci. In: Hensyl WM (ed).Bergey’s Manual of Systematic Bacteriology. Williams & Wilkins Co., Baltimore, pp: 1104-1139.
- Lowry OH,Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. Journal of Biological Chemistry 193: 265.
- Campos R,Kandelbauer A,Robra KH,Cavaco-Paulo A,Gubitz GM (2001) Indigo degradation with purified laccases from TrameteshirsutaSclerotiumrolfsii Journal of Biotechnology 89: 131-139.
- Wang HX, Ng TB (2004)A new laccasefromdried fruiting bodies of themonkey head mushroom Hericiumerinaceum. Biochemistry Biophysics and Research Communications 322: 17-21.
- Laemmli UK (1970) Cleavage structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685.
- Balan DSL,Monteiro RTR (2001) Decolorization of textile indigo dye by lignolytic fungi. Journal of Biotechnology 89: 141-145.
- Dhaouadi H,M’Henni F (2009) Vat dye sorption onto crude dehydrated sewage sludge. Journal of Hazardous Matrials 164: 448-458.
- Xu F (1996) Oxidation of phenols, anilines, and benzenethiols by fungal laccases: correlation between activity and redox potentials as well as halide inhibition. Biochemistry 35: 7608-7614.
--
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 12972
- [From(publication date):
July-2016 - Aug 20, 2025] - Breakdown by view type
- HTML page views : 11943
- PDF downloads : 1029