Research Article Open Access
Reduction of Selenium by Pseudomonas stutzeri Nt-I: Growth, Reduction and Kinetics
Wessels CE* and Chirwa EMN
University of Pretoria, Hatfield, Pretoria, South Africa
- *Corresponding Author:
- CE Wessels
University of Pretoria, Hatfield
Pretoria, South Africa
Tel: 27718700298
E-mail: Charlotte.elize@gmail.com
Received Date: March 19, 2017 Accepted Date: March 29, 2017 Published Date: March 30, 2017
Citation: Wessels CE, Chirwa EMN (2017) Reduction of Selenium by Pseudomonas stutzeri Nt-I: Growth, Reduction and Kinetics. J Bioremediat Biodegrad 8: 391. doi: 10.4172/2155-6199.1000391
Copyright: © 2017 Wessels CE, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Bioremediation of seleniferous water is gaining more momentum, especially when it comes to bacterial reduction of the selenium oxyanions. More and more bacterial strains that are able to reduce selenium are being isolated. These bacteria need to be studied further to determine whether they are suited for industrial application. In this study, the reduction of Se(VI) to Se(0) by Pseudomonas stutzeri NT-I was examined using batch experiments with the bacteria suspended in MSM. For the determination of the optimum conditions for the growth of the bacteria, the linearized rate during the exponential phase for different conditions were compared. A pH 7, temperature of 37°C, salinity of 20 g.L-1 NaCl and initial concentration of 5 mM selenate were found to be the best at promoting growth. To determine the optimum conditions for the reduction of selenium, the amount of Se (0) recovered from the plug after 16 hours of incubation was measured. A pH of 8, temperature of 37°C and salinity of 5 g.L-1 resulted in the most Se (0) recovered. The kinetics of the reduction of Se(VI) to Se (0) was found to follow the adapted Monod equation. An increase in the initial Se(VI) concentration positively affected the reduction rate indicating that substrate saturation had not yet been reached. One kmax could be fitted to each of the two reactions but not one Ks. It was found that Ks decreased with increasing initial selenate concentration. Visually it can be deduced that inhibition starts playing a role in the reduction of selenate at a concentration of 4 mM. Pseudomonas stutzeri NT-I is an exemplary selenium reducing agent and deserves more attention, not only for industrial application but also in the research world, for further understanding of the complex mechanism behind metal reduction in bacteria.
Keywords
Pseudomonas stutzeri NT-I; Selenium; Bioremediation; Aerobic bacterial growth; Aerobic bacterial reduction; Metal reduction; Reduction kinetics; Monod
Introduction
Selenium, first discovered in 1818, is a metalloid and chalcogen. It has multiple oxidation states, beginning with the most reduced state, namely selenate (SeO42-, Se(VI)), followed by selenite (SeO42-, Se(VI)) ,elemental selenium (Se0, Se(0)) and finally selenide (Se2-) Of the selenium released into the environment, 37.5-40.6% can be ascribed to anthropogenic activities. It is released from the earth’s crust by the mining of coal and various metals, oil production, use of agricultural products, as well as during the melting of non-ferrous metals [1]. Landfill ash disposal that generates toxic leachate poses a risk of groundwater contamination, which leads to the polluting of nearby water bodies [2].
Selenium is mostly present in water as selenate or selenite. According to different studies, acid mine drainage waters contain selenium at concentrations ranging between 2 × 10-4 and 6.2 × 10-3 mM of total selenium [3]. Waste water from oil refineries in the San Francisco Bay (USA) contains relatively low concentrations of selenium of about 50- 300 μg. L-1 [4]. The waste water from a selenium refinery plant in Japan contained an average of 30 mg. L-1 selenium, with the majority of the selenium present as selenite [4].
The upward trend in energy production from coal, along with the burning of fossil fuels contribute to this increase in selenium release. The high concentration of these selenium oxyanions poses a problem to the environment, since they are toxic and bio-accumulate readily. In one case it was found the concentration of selenium in the apex predators (birds and humans) in the area were 2 000 times higher than in the water [4]. Elemental selenium is not soluble in water and thus has a much lower bio-availability than the oxyanions. Elemental selenium’s toxicity is also much lower than that of the oxyanions [1].
Birth defects and reproductive problems among fish and waterfowl are examples of the effects of selenium poisoning. In the human body, selenium poisoning can have many adverse effects, including respiratory difficulty, gastrointestinal distress and liver damage. It also has a negative impact on the central nervous system [5-7]. In the US, the National Primary Drinking Water Standard is 50 ppb of total selenium, while the National Fresh Water Quality Standard is 5 ppb of total selenium [8]. For countries that don’t have a legislative framework for drinking water pollutants, the World Health Organization propose a value of 40 ppb [9].
Since the amount of selenium allowed in water bodies is regulated, industrial waste water is in need of effective selenium removal before it can be discharged into local water bodies. Different treatment technologies for the removal of selenium from water are available, including physical, chemical and biological methods. Biotic processes for the removal of selenium from water include the algal bacterial process; algal bioremediation; volatilisation; biological precipitation and wetlands [10].
Biological precipitation is the process where bacteria is used to reduce the selenium oxyanions to elemental selenium, which then precipitates out as agglomerated nanoparticles. A wide range of bacteria that reduce selenium oxyanions have been isolated. The advantages are high reduction efficiency, low production of residual solids due to slow growth rate, and eliminating exposure to wildlife and the environment. The disadvantages are the high cost associated with the addition of a carbon source, and the fact that the lowest residual soluble concentration achievable at this stage is 30 ppb, which means a polishing step will most probably be required [11].
Most of the identified selenate-reducing bacteria function under anaerobic conditions and, except for a few, can only reduce low concentrations of selenate. Several bacterial strains capable of reducing selenate under aerobic conditions have been isolated by different researchers. A few of them are Pseudomonas stutzeri [12]; Enterobacter cloacae SLD1a-1 [13]; Stenotrophomonas maltophilia [14], and Bacillus sp. STG-83 [15]. Two advantages of aerobic selenate reducing bacteria are firstly, that selenate reduction is not limited by the presence of oxygen and secondly, the aerobic conditions allow for a higher growth rate and thus an increase in cell production.
More than one Pseudomonas species that can metabolise selenate and selenite have been found. Examples are P. seleniipraecipitatus [16]; P. moraviensis stanleyae [17]; P. aeruginosa (SNT-SG1) [18]; P. stutzeri [12], and P. Stutzeri NT-I [19]. The genus Pseudomonas includes a wide variety of bacterial species. P. stutzeri is better known for denitrification under aerobic conditions, however an isolate of P. stutzeri has been found to reduce selenate/selenite aerobically [12]. This isolate has the same taxonomical features as P. Stutzeri NT-I, but these two strains differ in a few aspects. For example, the isolate could not reduce selenate at concentrations above 48.1 mM, nor could it reduce selenate anaerobically [19]. This study focuses specifically on Pseudomonas stutzeri NT-I. The bacteria was originally isolated from the drainage water of a selenium refinery plant in Hyogo, Japan by Masashi Kuroda and his team [19].
Materials and Methods
All chemicals used were of analytical grade and were obtained from Merck, Johannesburg, except for CaCl2, Na-EDTA, DAN, NaSeO4 and NaSeO3 which were obtained from Sigma Aldrich, Johannesburg.
Cultivation media: Tryptone Soy Broth and Agar (TSB, TSA) was used for the cultivation of the bacteria.
Reaction Media: Mineral Salt Medium (MSM) was used as the solution in which all the experiments took place. One litre of MSM was prepared according to the following recipe: 30 mM NH4Cl, 10 mM K2HPO4, 20 mM KH2PO4, 0.8 mM Na2SO4, and 0.2 mM MgSO4. For experiments where the MSM had to be at pH 6, 8 and 9, the ratio ofK2HPO4 to K2HPO4 was adjusted. 1 mL of a solution containing 50 μM CaCl2, 25 μM FeSO4, 0.1 μM ZnCl2, 0.2 μM CuCl2, 0.1 μM NaBr, 0.05 μM Na2MoO2, 0.1 μM MnCl2, 0.1 μM KI, 0.2 μM H3BO3, 0.1 μM CoCl2, and 0.1 μM NiCl2 was added to every 1 L of MSM.
Bacterial strain: The strain used in this study, Pseudomonas stutzeri NT-I, was furnished from the NITE Patent Microorganisms Depository (NMPD) in Chiba Ken, Japan. The strain ID is NITE BP- 685.
Experiments
Determination of exponential growth phase constant: The bacteria was cultivated by taking a loopful of bacterial cells from a TSA plate and inoculating it into 20 mL of glucose-MSM in a 100 mL serum bottle that was covered with cotton wool and foil and incubated for 24 hours at 37°C. Thereafter 5 mL was transferred to 100 mL of fresh glucose-MSM medium containing 1 mM selenate in a 250 mL Erlenmeyer flask. 3 mL samples were withdrawn with a syringe to measure the OD600 every 2 hours for at least 24 hours. These experiments were repeated at different temperatures, different pHs, different salinities and different initial concentrations. All experiments were conducted in triplicate. The specific growth rate was measured by the change in absorbance during the log phase of growth according to the following equation:
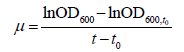
μ=Specific growth rate, h-1; t=time, h; OD600: Absorbance of cell suspension at 600 nm.
Batch experiments conducted with washed cell suspensions: For the reduction experiments the washed cell suspensions were prepared as follows: The bacteria was cultivated by taking a loopful of bacterial cells from a TSA plate and inoculating it into 20 mL of TSB medium in a 100 mL serum bottle capped with foil and cotton wool. The serum bottle was then placed in the incubator for 24 hours at 37°C. Subsequently 10 mL was transferred again to 100 mL of fresh TSB medium in a 250 mL Erlenmeyer flask and cultivated for a further 8 hours. The cells were harvested by centrifugation (6 000 rpm, 15 minutes, room temperature) and then washed with physiological saline (0.85% w/v NaCl in distilled water), whereafter it was resuspended in 50 mL of glucose-MSM (amended with 5 g.L-1 NaCl) with an OD600 far above 1.5, to ensure that further growth of the bacteria is marginal. The 50 mL of washed cell suspension was then distributed between three 100 mL serum bottles, each containing 15 mL of washed cell suspension in the end. An amount of 5 mL of the 4 mM selenate stock solution was added to the serum bottle to give a final concentration of 1 mM selenate in the adjusted washed cell suspension (final volume of 20 mL). The time at which the selenate stock solution was added was taken as t0. The extent of selenate reduction to elemental selenium by Pseudomonas stutzeri NT-I was quantified at different temperatures, pHs and salinities. The percentage of elemental selenium recovered after 16 hours was determined by taking the whole reaction volume and centrifuging it to separate the reaction volume into a plug containing the elemental selenium and bacterial cells, and the supernatant containing the unreacted selenium oxyanions.
For the kinetic batch experiments the bacteria was cultivated by taking a loopful of bacterial cells from a TSA plate and inoculating it into 20 mL of TSB medium in a 100 mL serum bottle capped with foil and cotton wool. Whereafter it was placed in the incubator for 24 hours at 37°C. Subsequently 10 mL was transferred again to 100 mL of fresh TSB in a 250 mL Erlenmeyer flask and cultivated for 8 hours. The cells were harvested by centrifugation (6 000 rpm, 15 minutes, room temperature) and then washed with physiological saline, following which it was resuspended in 30 mL of MSM amended with glucose, containing different amounts of Na2SeO4 (pH 8, glucose 10 g.L-1) with a high biomass (between 20 to 40 g.L-1 depending on experiment). The procedure followed for the baseline experiment (0.5 mM Selenate, 37°C, pH 8) was as follows: The 100 mL flask containing 30 mL of washed cell suspension was capped with cotton wool and foil whereafter it was incubated at 37°C with continuous shaking on a rotary shaker at 120 rpm. A 1 mL sample was taken at different time intervals and transferred to a 1.5 mL centrifuge tube. The reaction in the sample was stopped by centrifugation (6 000 rpm, 15 minutes, room temperature). The supernatant from the tube was decanted from the plug and diluted to a final volume of 25 mL with distilled water and filtered (Whatman 42, Sigma Aldrich, South Africa). 5 mL was used to analyse for selenite using the colorimetric method described below.
Analytical methods
Optical density: The wavelength at which the spectrophotometer measured the optical density (OD600) of the cell suspension was set at 600 nm. The blank used to zero the spectrophotometer was MSM containing no bacteria.
OD600 vs. dry biomass: Cells were cultivated following the same procedure as for the washed cell suspension, the only difference being that a litre of washed cell suspension was made and 4 samples of 200 mL were taken periodically. The OD600 of these samples were measured whereafter the sample was centrifuged. The plug, containing the cells, was resuspended in 10 mL saline and dried in an oven at 70°C for 5 days. After it was dried, the weight of the cells were measured.
Selenite: The measurement of selenite was carried out using the colorimetric method described in Standard methods for the examination of water and waste (APHA, 2012) (3500-Se C Colorimetric method, p 3-93). The absorbance at 480 nm was measured and the concentration was obtained using the linear calibration curve generated during the calibration of the instrument.
Elemental selenium in plug: The plug containing the elemental selenium and the cells was resuspended in 20 mL distilled water whereafter 10 mL concentrated HCl and 10 mL concentrated HNO3 were added, and the volume was made up to 50 mL by adding distilled water. With the addition of the acids, the characteristic colour of the elemental selenium disappeared, thereby confirming that the elemental selenium was oxidised back to the selenium oxyanions. After the volume was made up to 50 mL, 10 mL was taken and placed in a heating block at 100°C for 60 minutes. The next step was to make up the 10 mL sample to 50 mL using distilled water and then filtering the sample (Whatman 42), following which it was placed in a sample container and analysed using Inductively-Coupled Plasma-Optical Emission Spectrometry (ICP-OES) (ARCOS FHS12, Spectro Analytical Instruments, Kleve, Germany).
Results
Preliminary experiments
The OD600 of the bacteria at stationary phase was determined. This was done with no selenium present, since the discoloration due to the red elemental selenium forming when nearing the stationary phase interfered with the accurate determination of the complete growth curve. The OD600 at stationary phase was determined as 1.5. In terms of the relationship between optical density and dry biomass, it was found that at an optical density of 1, the weight of the dry biomass is 0.416 g. L-1.
Growth experiments
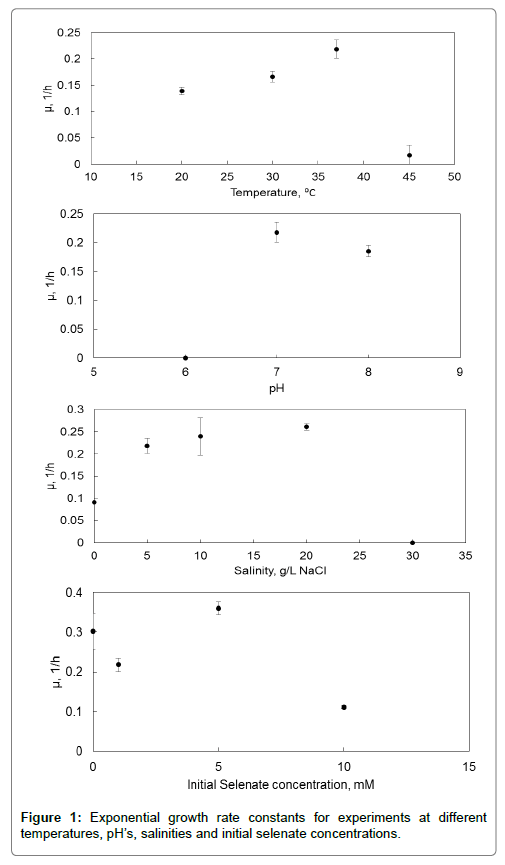
The exponential growth phase kinetic constants, μ (h-1), for the different experiments were compared. Before the results are given, it should be noted that the growth rates were determined at specific predetermined values and therefore the optimum values determined cannot be taken as the absolute optimum condition but only as the optimum for the values at which it was measured.
The optimum temperature for growth is 37°C with an exponential growth rate constant of 0.22 h-1. What can also be seen from the data is that there is a sharp decline in growth rate for temperatures above 37°C going as low as 0.02 h-1 whereas the relative difference in growth rate between 30 and 37°C is much less. In terms of pH, no growth was observed at a pH of 6; in fact the inoculum died soon after the experiment commenced. The optimum pH was found to be 7 with an exponential growth rate constant of 0.22 h-1. The decline in growth rate from pH 7 to 8 is not that significant, the implication being that the pH can be increased to a value between 7 and 8 without losing too much growth potential. For the growth rate evaluated at different salinities ranging from 0 to 20 g.L-1 NaCl, an optimum of 0.26 h-1 was determined at a salinity of 20 g.L-1 NaCl. For the experiments done with different initial selenate concentrations, optimum growth (0.36 h-1) was observed at a concentration of 5 mM selenate (715 mg.L-1), indicating that the selenate is useful for energy and growth. What is notable is that the growth rate for the experiment which contained no selenium is lower than the maximum growth rate.
Reduction rate experiments
In the study done by Kuroda et al. [19], it was determined that the bacteria could reduce selenate to selenite within 6 hours and the selenite can then be further reduced to elemental selenium in another 18 hours. This information was used to determine the length of the reduction experiments. However, for the experiments done as part of this study it was found that the reduction proceeded at a much faster rate. This may be due to a different carbon source being used (glucose instead of lactate) or the temperature of the experiments being different. In the paper referred to earlier, the temperature of the reduction reaction experiments is not given. Due to this inconsistency, the amount of elemental selenium recovered from the plug after 16 hours of incubation was taken as the criterion by which the optimum reduction conditions were determined. ICP-OES analysis confirmed that after 16 hours the reduction of both oxyanions were complete except for the experiment conducted at 30°C. Extra reduction experiments were performed, one without glucose, one with a heat-killed washed cell suspension and, lastly, one without any bacterial cells. No reduction was observed in these experiments. This means that the reaction is dependent on glucose as an electron donor and that the reduction occurs strictly due to microbial activity.
The experiment that was done at a temperature of 30°C was used to do a mass balance check. The supernatant was analysed on the ICPOES for total selenium and added to the amount of elemental selenium measured in the plug. After adding 79 mg.L-1 of selenium to the reaction mixture, at the end there was 19 mg.L-1 in the supernatant and 53.2 mg.L-1 in the plug. The difference between what was added and the amount at the end was 6.8 mg.L-1 and translates to a 91.4% recovery of selenium, which is within the acceptable range for mass balances done in microbial systems (Figure 1).
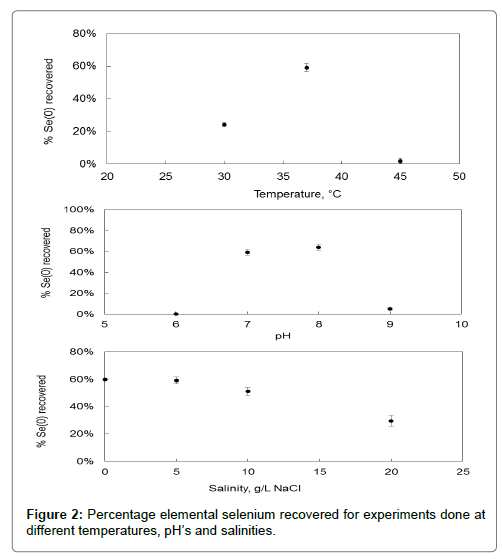
Figure 2 shows the impact temperature, pH, and salinity has on the amount of elemental selenium recovered after 16 hours of reaction time with a washed cell suspension of Pseudomonas stutzeri NT-I. The optimum temperature, 37°C, is the same as that of growth and is within the region of the temperature of industrial wastewater. The optimum pH for reduction of selenate is 8, which is different to the optimum pH of 7 for growth. When looking at Figure 2 it can be seen that from a salinity of 5 g.L-1 a further increase lowers the reduction capacity.
The temperature and pH at which the experiment was conducted is important since a change in temperature or pH leads to a change in the pKa value of an enzyme, which in turn impacts the enzyme’s ability to reduce the metal anion involved. Each enzyme has a pKa value specific to them. For E. Cloacae SLD1a-1 the optimum pH was found to be lower at 6.5 to 7 [20] whereas for T. selenatis the optimum pH was 8 [21]. Different selenium reducing bacteria operate best at various pH’s, which indicates that there are multiple reduction mechanisms involved depending on which bacteria is being used. The change in reduction capacity between pH 7 and pH 8 is not much. This makes the setting up of a reactor where growth and reduction occurs at the same time easier in that the pH only needs to be kept between 7 and 8 to still achieve a high reduction rate.
The decrease in reduction capacity at higher salinities may be due to the denaturing of the enzymes involved in the reduction pathway. This is the opposite of what is seen with the impact of salinity on growth. A compromise between reduction capacity and growth will have to be made when both are occurring in the same reactor. A salinity of 5 g.L-1 is suggested for such instances. In the other study already mentioned (Kuroda et al.), the optimum conditions for reduction of selenate by Pseudomonas stutzeri NT-I was 40°C, pH 8 and low salinity. This is once again in close agreement with the findings presented here.
Kinetic batch experiments
The bacteria took 2 hours to completely reduce 2 mM of selenate which translates to a reduction rate of 0.0313 mmol.(g.h)-1 When compared to other bacterial strains identified as selenium reducers, Pseudomonas stutzeri NT-I reduces the selenate to elemental selenium at a relatively high rate. E cloacae SLD1a-1 reduces selenate at a rate of 0.0184 mmol.(g.h)-1 [22]. Pseudomonas fluorescens only reduced 95% of 0.2 mM selenate after 45 hours [23] and Desulvibrio desulfuricans took 25-37 hours to reduce 86% of 1 mM of selenate [24] which is much slower than what was observed in this study.
Kinetic model
The bacteria took 2 hours to completely reduce 2 mM of selenate which translates to a reduction rate of 0.0313 mmol.(g.h)-1. When compared to other bacterial strains identified as selenium reducers, Pseudomonas stutzeri NT-I reduces the selenate to elemental selenium at a relatively high rate. E cloacae SLD1a-1 reduces selenate at a rate of 0.0184 mmol. (g.h)-1 [22]. Pseudomonas fluorescens only reduced 95% of 0.2 mM selenate after 45 hours [23] and Desulvibrio desulfuricans took 25-37 hours to reduce 86% of 1 mM of selenate [24] which is much slower than what was observed in this study.
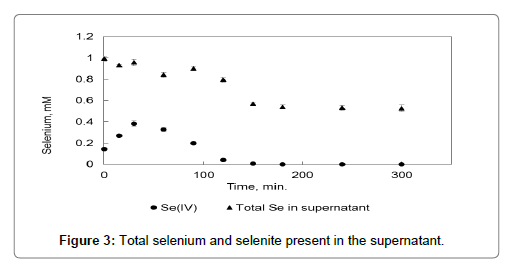
Total selenium in the supernatant was measured using ICP-OES. The motivation behind this was that the selenate concentration in the supernatant could be calculated then from the difference between the selenite concentration and the total selenium. However when the samples were analysed the amount of selenium in the sample did not go to zero as would be expected if only the selenium oxyanions were present and all the elemental selenium was present as nanoparticles that would be centrifuged out from the supernatant into the plug. This discrepancy can be explained by postulating that the elemental selenium is present in three states. Firstly, it is present as colloidal selenium within the cells, the colloidal selenium then form nanoparticle seeds which are transported to the outside of the cell where over time it ripens into nanoparticles that are big enough to centrifuge out with the cells forming the plug. The consequence of this is then that the total selenium measured in the supernatant is not only made up of the selenium oxyanions but also elemental selenium nanoparticle seeds that have been dispelled from the cells but have not yet evolved into nanoparticles heavy enough to centrifuge out. Figure 3 shows the concentration of selenium in the supernatant over time proving that the elemental selenium is spread between the plug and the supernatant making it very difficult to do a mass balance for the sake of the kinetic modelling. Therefore only the selenite concentration in the supernatant was used.
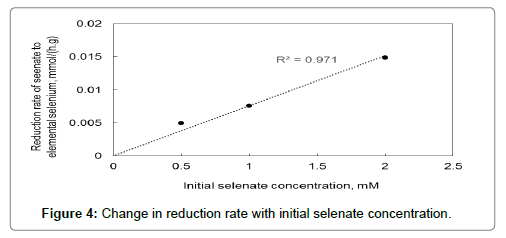
Figure 4 plots how the reduction rate changes with initial selenate concentration used. The reduction rate increases with initial selenate concentration. This trend where reaction rate increases linearly with increasing initial selenate/selenite concentration have been observed by other researchers as well and is most likely due to substrate saturation not being reached yet. Substrate saturation occurs when all the enzymes available to catalyse the reaction involved is saturated with the substrate.
Two kinetic models were considered when parameter estimation was performed, namely a first order rate equation and an adapted Monod equation. The first order rate equation did not fit the data but the Monod equation, adapted for metal reduction did. Differential equations were used in the software package AQUASIM [25] to estimate the parameters for the reduction of selenate to selenite and selenite to elemental selenium. AQUASIM estimates the parameter values by minimising the sum of squares of the weighted deviations between calculated results and measured values.
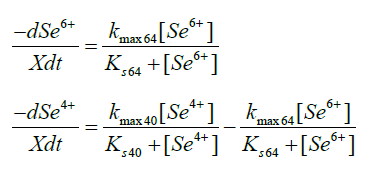
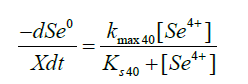
kmax64 Maximum reaction rate for the reduction of selenate to selenite, mol.(g.min)-1
kmax40 Maximum reaction rate for the reduction of selenite to elemental selenium, mol.(g.min)-1
Ks64 Substrate saturation coefficient for the reduction of selenate to selenite, mM
Ks40 Substrate saturation coefficient for the reduction of selenite to elemental selenium, mM
X Biomass concentration, g.L-1
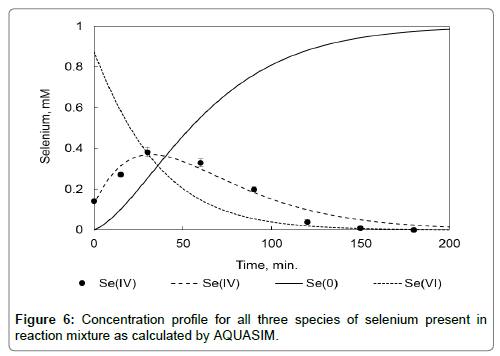
Since only selenite measurements could be used, the AQUASIM model was set up to fit the parameters of the equation for the reduction of selenite to the data points from three experiments where the initial concentration of selenate differed (0.5, 1 and 2 mM). Figure 5 shows the results of this fit and Figure 6 shows the predicted concentrations for all three species (selenate, selenite and elemental selenium) for the experiment with an initial concentration of 1 mM. From these fits it can be seen that the assumption that the change in biomass is negligible and that the biomass concentration is directly proportional to the reaction rate holds. Other observations that can be made are that there is no delay time for the reaction of selenate to selenite and that selenite reduction is faster than that of selenate. Ma et al. [22] as well as Lindblow-Kull et al. [26] also found that selenite was reduced faster than selenate.
The values of the fitted parameters are given in Table 1. The kmax values for E. Coli K12 was 0.00681 and 0.00076 mmol.(g.min)-1 for selenite and selenate reduction respectively [26]. The reduction rate for both oxyanions is slower than Pseudomonas stutzeri NT-I. The ks values are higher than that of other selenium reducing bacteria like Enterobacter cloacae SLD1-a1 which had Ks values of 3.1 and 0.72 mM for selenate and selenite reduction respectively [22]. One kmax value for each of the two reactions in all three of the experiments were fitted since the maximum reduction rate for a specific temperature and pH is constant. One Ks value for each of the three initial concentrations could not be fitted and was therefore optimised for each experiment.
| S No | Initial selenate concentration (mmol. L-1) | kmax64 (mmol.min-1. g-1) | kmax40 (mmol.min-1. g-1) | Ks64 (mmol. L-1) | Ks40 (mmol. L-1) | R2 |
|---|---|---|---|---|---|---|
| 1 | 0.5 | 0.0109 | 0.0533 | 12.6 | 58.7 | 0.993 |
| 2 | 1 | 0.0090 | 0.0575 | 7 | 49.8 | 0.964 |
| 3 | 2 | 0.0106 | 0.0547 | 2.9 | 18.8 | 0.997 |
| Average | 0.0102 | 0.0552 | ||||
Table 1: Parameters for the adapted Monod equation fitted to experiments 1-3.
The ks value for each of the reaction is also supposed to remain constant like the kmax. The fact that one ks value could not be fitted, but that the unique values fitted decreases almost linearly as initial selenate concentration is increased points to some form of increasing interference with the two reactions. When looking at the data from the experiment with an initial concentration of 4 mM (Figure 7) it can be seen that some form of inhibition is becoming apparent. One can only speculate what the cause is behind the inhibition based on what other researchers have found.
Adding a non-competitive inhibition term related to elemental selenium to the adapted Monod equation was considered due to the formation of insoluble seed-nanoparticles of elemental selenium within the cell. This was done with caution since the true Ks value is not known. Upon adding a product inhibition term the fit was not significantly improved. Other inhibition models were also tested for the sake of being thorough but none of them led to an improved fit.
The lower Ks values observed at a high reduction rate alludes to the fact that elemental selenium might be building up in the cell due to a slower transportation than reduction rate at high concentrations. In the case where the reduction rate is slower due to the lower initial selenate concentration, the elemental selenium seed-nanoparticles have time to be transported out of the cell and therefore interfere less with the reduction reaction. It has been shown in another study [27] that elemental selenium seed-nanoparticles can physically get in the way of different steps in the reaction chain, for example blocking the redox enzyme or interfering with the intermediate electron transfer sites. These seed-nanoparticles can also negatively influence the porosity of the cell membrane. Another possible reason for product inhibition may be that the elemental selenium seed-nanoparticles cause the cell viability to be lowered or physiological state altered. This possible explanation was not further investigated due to the scope of this study but do warrant further investigation. The interpretation of the data requires fundamental insight into the mechanisms involved, which include mass transfer of glucose (energy source for the detoxification reaction), the selenium oxyanions and the elemental selenium between the reaction sites.
The structure of the cell surface and membrane as well as the participation of different enzymes in the electron transport chain all add to the complexity of the detoxification mechanism. It has been suggested that unique redox enzymes are involved in the reduction of the valence state of metals and metalloids like zinc and selenium to avoid cellular damage and death [6]. A few research groups have studied this detoxification mechanism [14,28-30] and have found that in some cases the reduction occurs in the periplasm and in other cases in the cytoplasm. But they all require an energy source in the form of ATP since the reduction of selenate to elemental selenium is endogenic, the reduction reaction also requires NADH to act as the electron donor.
Conclusion
The growth and reduction studies show that Pseudomonas stutzeri NT-I is a suitable candidate for use in bioremediation of seleniferous industrial waste water. It is therefore recommended that further studies be conducted to determine the most effective reactor design for the facilitation of the reduction reactions by this bacterial strain. Augmenting current biological reactors with this bacteria to improve the reduction performance can also be investigated.
When considering the kinetics of the two consecutive reduction reactions, the Monod equation, adapted for metal reduction, is a good representation of the measured selenite concentration over time. The assumption that biomass is constant and directly proportional to the reaction rate holds, as would be expected for a biomass concentration exceeding 2 g.L-1. The three experiments shared a kmax value for each of the two reduction reactions. One Ks value for each of the two reactions in the three experiments could not be fitted though and was therefore optimised for each experiment. A linear trend, where Ks decreases with increasing initial selenate concentration, was observed. It is postulated that this is due to elemental selenium build-up which interferes with the reduction reaction.
Pseudomonas stutzeri NT-I is an exemplary selenium oxyanion reducing agent and deserves more attention, not only so that it can be used in industry for the bioremediation of industrial waste water, but also in the research world to further our understanding of the complex working of bacteria.
Acknowledgements
This project was funded by the University of Pretoria, South Africa.
References
- Lenz M, Lens PNL (2009) The essential toxin: the changing perception of selenium in environmental sciences. The Science of The Total Environment 407: 3620-3633.
- Lemly AD (2004) Aquatic selenium pollution is a global environmental safety issue. Ecotoxicology and Environmental Safety 59: 44-56.
- Lenz M, Van Hullebusch ED, Hommes G, Corvini PFX, Lens PNL (2008) Selenate removal in methanogenic and sulfate-reducing upflow anaerobic sludge bed reactors. Water Research 42: 2184-2194.
- Lawson S, Macy JM (1995) Bioremediation of selenite in oil refinery wastewater. Applied Microbiology and Biotechnology 43: 762-765.
- Satoshi MS, Hisamitsu T, Tsubasa K, Masaki M, Emi N, et al. (2012) Biotreatment of Selenium Refinery Wastewater using Pilot-scale granular sludge and swim-bed bioreactors augmented with a Selenium-reducing bacterium Psuedomonas Stutzeri NT-1. Japanese Journal of Water Treatment Biology 48: 63-71.
- Wu L (2004) Review of 15 years of research on ecotoxicology and remediation of land contaminated by agricultural drainage sediment rich in selenium. Ecotoxicology and Environmental Safety 57: 257-269.
- Kenward PA, Fowle DA, Yee N (2006) Microbial selenate sorption and reduction in nutrient limited systems. Environmental Science and Technology 40: 3782-3786.
- USEPA (1999) National Recommended Water Quality Criteria.
- World Health Orginization (2011) Guidelines for Drinking Water.
- Dunga RS, Frankenberger WT (1999) Microbial Transformations of Selenium and the Bioremediation of Seleniferous Environments. Bioremediation Journal 3: 171-188.
- Frankenberger Jnr WT, Amrhein C, Fan TWM, Flaschi D, Glater J, et al. (2004) Advanced Treatment Technologies in the Remediation of Seleniferous Drainage Waters and Sediments. Irrigation and Drainage Systems 18: 19-42.
- Lortie L, Gould WD, Rajan S, McCready RGL, Cheng KJ (1992) Reduction of selenate and selenite to elemental selenium by a Pseudomonas stutzeri isolate. Applied and Environmental Microbiology 58: 4042-4044.
- Losi ME, Frankenberger WT (1997) Reduction of Selenium Oxyanions by Enterobacter cloacae SLD1a-1: Isolation and Growth of the Bacterium and Its Expulsion of Selenium Particles. Applied and Environmental Microbiology 63: 3079-3084.
- Dungan RS, Yates SR, Frankenberger WT (2003) Transformations of selenate and selenite by Stenotrophomonas maltophilia isolated from a seleniferous agricultural drainage pond sediment. Environmental Microbiology 5: 287-295.
- Saudi MR, Ghazvini PTM, Khajeh K, Gharavi S (2009) Bioprocessing of seleno-oxyanions and tellurite in a novel Bacillus sp. strain STG-83: a solution to removal of toxic oxyanions in presence of nitrate. Journal of Hazardous Materials 165: 71-77.
- Hunter WJ, Manter DK (2011) Pseudomonas seleniipraecipitatus sp. Nov.: A selenite reducing proteobacteria isolated from soil. Current Microbiology 62: 565-569.
- Staicu LC, Ackerson CJ, Cornelis P, Ye L, Berendsen RL, et al. (2015) Pseudomonas moraviensis subsp. stanleyae, a bacterial endophyte of hyperaccumulator Stanleya pinnata, is capable of efficient selenite reduction to elemental selenium under aerobic conditions. Journal of Applied Microbiology 119: 400-410.
- Gupta S, Prakash R, Prakash NT, Pearce C, Pattrick R, et al. (2010) Selenium Mobilization by Pseudomonas aeruginosa (SNT-SG1) Isolated from Seleniferous Soils from India. Geomicrobiology Journal 27: 35-42.
- Kuroda M, Notaguchi E, Sato A, Yoshioka M, Hasegawa A, et al. (2011) Characterization of Pseudomonas stutzeri NT-I Capable of Removing Soluble Selenium from the Aqueous Phase under Aerobic Conditions. Journal of Bioscience and Bioengineering 112: 259-264.
- Losi ME, Frankenberger WT (1997) Reduction of selenium oxyanions by Enterobacter cloacae SLDa-1: reduction of selenate to selenite. Environmental Toxicology and Chemistry 16: 1851-1858.
- Macy JM, Rech S, Auling G, Dorsch M, Stackebrandt E, et al. (1993) Thauera selenatis gen. nov., sp. nov., a Member of the Beta Subclass of Proteobacteria with a Novel Type of Anaerobic Respiration. International Journal of Systematic Bacteriology 43: 135-142.
- Ma J, Kobayashi DY, Yee N (2007) Chemical kinetic and molecular genetic study of selenium oxyanion reduction by Enterobacter cloacae SLD1a-1. Environmental Science and Technology 41: 7795-7801.
- Belzile N, Wu GJ, Chen YW, Appanna VD (2006) Detoxification of Selenite and Mercury by Reduction and Mutual Protection in the Assimilation of Both Elements by Pseudomonas fluorescens. Science of The Total Environment 367: 704-714.
- Tucker MD, Barton LL, Thomson BM (1998) Reduction of Cr, Mo, Se and U by Desulfovibrio desulfuricans immobilized in polyacrylamide gels. Journal of Industrial Microbiology and Biotechnology 20: 13-19.
- Reichert P (1994) AQUASIM - A tool for simulation and data analysis of aquatic systems. Water Science and Technology 30: 21-30.
- Lindblow-Kull C, Kull F, Shrift A (1985) Single transporter for sulfate, selenate, and selenite in Escherichia coli K-12. Journal of Bacteriology 163: 1267-1269.
- Liu C, Gorby YA, Zachara JM, Fredrickson JK, Brown CF (2002) Reduction kinetics of Fe(III), Co(III), U(VI), Cr(VI), and Tc(VII) in cultures of dissimilatory metal-reducing bacteria. Biotechnology and Bioengineering 80: 637-649.
- Etezad SM, Khajeh K, Soudi M, Ghazvini PTM, Dabirmanesh B (2009) Evidence on the presence of two distinct enzymes responsible for the reduction of selenate and tellurite in Bacillus sp. STG-83. Enzyme and Microbial Technology 45: 1-6.
- Heße F, Radu FA, Thullner M, Attinger S (2009) Upscaling of the advection-diffusion-reaction equation with Monod reaction. Advances in Water Resources 32: 1336-1351.
- Kessi J (2006) Enzymic systems proposed to be involved in the dissimilatory reduction of selenite in the purple non-sulfur bacteria Rhodospirillum rubrum and Rhodobacter capsulatus. Microbiology 152: 731-743.
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 4040
- [From(publication date):
May-2017 - Aug 31, 2025] - Breakdown by view type
- HTML page views : 3035
- PDF downloads : 1005