SSR Marker-based Genetic Diversity Analysis of Tidal and Flood Prone Areas in Rice (Oryza sativa L.)
Received: 24-Jul-2016 / Accepted Date: 04-Aug-2016 / Published Date: 11-Aug-2016 DOI: 10.4172/2155-952X.1000241
Abstract
One hundred and sixty rice varieties from the tidal and flood prone areas of south and south East Asian countries were analyzed. Samples sizes were: 50 varieties from Bangladesh (deepwater, tidal and flood prone rice and modern varieties), 14 varieties from India (flood prone rice), 16 varieties from Sri Lanka (flood prone rice), 7 varieties from Vietnam (tidal varieties), 69 varieties from Indonesia (tidal varieties) and 4 check varieties from IRRI. All 30 primer pairs created polymorphic bands among the 160 rice varieties from flood and tidal prone areas, which indicated that the microsatellites used were suitable for diversity analysis. A total of 337 alleles were detected with an average of 11 alleles per locus and the number of alleles per locus varied from 4 to 21. The highest PIC values were observed for the primer of RM474 (0.91), followed by RM5 (0.82), RM484 (0.81), RM214 (0.81), and RM19 (0.79). Cluster analysis divided the genotypes into four main clusters and six sub-clusters based on geographical origins and ecotypes. Microsatellite clustering (over 30 polymorphic loci) and submergence screening data indicated greater genetic diversity among 160 genotypes for molecular loci and for submergence tolerance. Tolerant genotypes in Cluster-1 are expected to have different tolerance genes. Finding relationship between tolerance and country of origin, highly tolerant varieties (FR13A and FR43B) were found from east India. Genetic diversity analysis among flood prone rice will be useful for identifying the varieties having maximum diversity with submergence tolerance and selected varieties will be useful for further studies.
Keywords: SSR, Genetic diversity, Tidal, Flood prone, Submergence, Rice (Oryza sativa L.)
250490Introduction
A linkage map is a chromosome map of a species that shows the position of its known genes and/or markers along each chromosomerelative to each other in terms of recombination frequency. The term molecular marker is taken to refer as markers for identifying variation at the level of DNA of individual organism. Genetic markers that are located in close proximity to genes (i.e. tightly linked) may be referred as gene ‘tags’ [1]. A DNA class of di-, tri-, tetra- or penta-nucleotide tandem repeats is described as microsatellites [2]. The use of Simple Sequence Repeats (SSRs) [3] or microsatellites have many advantages over RFLP and other PCR based markers, like RAPD, AFLP, CAPS and SCAR markers. They are distributed throughout the genome in many species [4]. They are characterized by great abundance [5], high variability [6], co-dominant inheritance, and locus-specificity. The PCR-based marker has become the main tool for genetic analysis. They are also used in genetic diversity analysis [7] and provide support for map-based cloning of genes, controlling trait of interest [8,9] and are useful in a molecular breeding programme [10,11]. PCR is a technique for amplifying DNA (or RNA) of any organism using two specific oligonucleotide primers, which flank the region of interest [12]. Gel electrophoresis techniques are used to separate the PCR product of different individuals and resultant polymorphisms can be observed directly.
Breeding to transfer tolerance to submergence into high yielding varieties has been ongoing for over three decades [13-15]. Introgression of Sub1 gene into popular high-yielding rice varieties of rainfed lowlands including Swarna (India), Samba Mahsuri (India), Sabitri (India), TDK1 (Laos), IR64 (IRRI), and BR11 (Bangladesh) have already been completed at IRRI. New submergence tolerance rice varieties with Sub1A gene might be able to resist floods that destroy vast tracts of paddy. Although FR13A has been successfully used as submergence tolerance source, additional sources are needed.Pyramiding several genes into the same background is the most effective breeding strategy, when multiple genes conferring a similar phenotype [16]. Rice diversity is crucial, because breeders are still striving to find additional sources of tolerance. Maintaining genetic diversity is important, in terms of responding to evolutionary and environmental forces [17]. Microsatellites provide a reliable means to measure intraspecific variation and genetic distance within populations [18,19]. The usefulness of markers for estimation of genetic diversity has been demonstrated in many crops, including barley [20], wheat [21,22], potato [23] and rice [24-26]. Due to their abundance, SSR markers are widely being used to determine the genetic structure and diversity patterns in different species [27,28]. Distinct climatic and ecological variations of flood prone areas - range from deepwater, swamp to rainfed, and with these wide ranges of zones being a contributing factor to greater varietal diversity of rice. The best known tolerant cultivar, FR13A was found from the submergence-prone area of Orissa, India [14]. Genetic diversity analysis among flood prone rice will be useful for identifying the varieties having maximum diversity with submergence tolerance and for further studies with selected genotypes. In view of the above mentioned introduction, the present studies were undertaken with the following major objectives: To evaluate genetic diversity of flood and tidal prone rice cultivars, using SSR markers.
Materials and Method
Plant materials
A total of 160 rice varieties (Table 1) were chosen from Genetic Resource Centre (GRC) of IRRI, representing traditional tall, deepwater, tidal and some modern varieties, grown in flood and tidal prone areas of south and southeast Asian countries (Bangladesh, India, Indonesia, Vietnam and Sri Lanka).
| S. No. | Variety | Country |
|---|---|---|
| 1 | Bashful | Bangladesh |
| 2 | Dholamota | Bangladesh |
| 3 | Kajal Sail | Bangladesh |
| 4 | LataMonaS | Bangladesh |
| 5 | LalChikon | Bangladesh |
| 6 | LohaSura | Bangladesh |
| 7 | Nona Sail | Bangladesh |
| 8 | SadaChikin | Bangladesh |
| 9 | Tupu Sail | Bangladesh |
| 10 | Rajasail | Bangladesh |
| 11 | Sadasail | Bangladesh |
| 12 | Rupsail | Bangladesh |
| 13 | Changai | Bangladesh |
| 14 | Sitabgog | Bangladesh |
| 15 | Kachra | Bangladesh |
| 16 | Chapail | Bangladesh |
| 17 | Mach Ranga | Bangladesh |
| 18 | Motorsail | Bangladesh |
| 19 | Chamara | Bangladesh |
| 20 | Kalamanik | Bangladesh |
| 21 | HabiganjAman 1 | Bangladesh |
| 22 | Rayada 25 | Bangladesh |
| 23 | BR25 | Bangladesh |
| 24 | HabiganjAman 7 | Bangladesh |
| 25 | Birpala | Bangladesh |
| 26 | Kumragoir | Bangladesh |
| 27 | Fulkari | Bangladesh |
| 28 | Badal | Bangladesh |
| 29 | BR11 | Bangladesh |
| 30 | BR3 | Bangladesh |
| 31 | BRRI Dhan28 | Bangladesh |
| 32 | BRRI Dhan29 | Bangladesh |
| 33 | BRRI Dhan31 | Bangladesh |
| 34 | Patnai 23 | Bangladesh |
| 35 | BRRI Dhan32 | Bangladesh |
| 36 | Gopalbhog | Bangladesh |
| 37 | KachaChikon | Bangladesh |
| 38 | Kutiagni | Bangladesh |
| 39 | Kharigojal | Bangladesh |
| 40 | LaxmiBilash | Bangladesh |
| 41 | MatiChak | Bangladesh |
| 42 | HoglaPata | Bangladesh |
| 43 | Kachamota | Bangladesh |
| 44 | ModhuMalati | Bangladesh |
| 45 | Nakpechi | Bangladesh |
| 46 | Swarna | India |
| 47 | Chakkol | Bangladesh |
| 48 | Chandmoni | India |
| 49 | Tilakkachari | India |
| 50 | Kalamocha | India |
| 51 | Bishpair | India |
| 52 | Khejurchari | India |
| 53 | CN 540 | India |
| 54 | FR43 B | India |
| 55 | DA27 | India |
| 56 | Sadamota | India |
| 57 | Modhukar | India |
| 58 | NC492 | India |
| 59 | TCA 4 | India |
| 60 | Thavalu 15314 | Sri Lanka |
| 61 | Thavalu 15325 | Sri Lanka |
| 62 | Kurkaruppan | Sri Lanka |
| 63 | PeriyaKaruppan | Sri Lanka |
| 64 | BurumaThavalu | Sri Lanka |
| 65 | Madabaru | Sri Lanka |
| 66 | Kottamalli | Sri Lanka |
| 67 | Lumbini | Sri Lanka |
| 68 | GodaHeenati | Sri Lanka |
| 69 | Kalukanda | Sri Lanka |
| 70 | Kannimurunga | Sri Lanka |
| 71 | Ratawee | Sri Lanka |
| 72 | Jamis Wee | Sri Lanka |
| 73 | Devarenddiri | Sri Lanka |
| 74 | Kaharamana | Sri Lanka |
| 75 | Giau Dumont | Vietnam |
| 76 | Ca Dung | Vietnam |
| 77 | Doc Phung | Vietnam |
| 78 | MongChim | Vietnam |
| 79 | Nang Thom | Vietnam |
| 80 | VeVang | Vietnam |
| 81 | Samo Ran | Vietnam |
| 82 | PungNgeom | Vietnam |
| 83 | T 442-57 | Philippines |
| 84 | LebMueNahng 111 | Thail |
| 85 | Bayar Kuning | Indonesia |
| 86 | Duku | Indonesia |
| 87 | Ampai | Indonesia |
| 88 | KetanSerai | Indonesia |
| 89 | TempokongPutih | Indonesia |
| 90 | Kapuas | Indonesia |
| 91 | UmbangInai | Indonesia |
| 92 | UmbangKencana | Indonesia |
| 93 | BijiNangka | Indonesia |
| 94 | UmbangPutih | Indonesia |
| 95 | Bilis | Indonesia |
| 96 | LakatanHirang | Indonesia |
| 97 | Lemo | Indonesia |
| 98 | Randah Padang | Indonesia |
| 99 | BalimanPutih, | Indonesia |
| 100 | RandahPalas | Indonesia |
| 101 | Buntok | Indonesia |
| 102 | Dange | Indonesia |
| 103 | GedabungKuning | Indonesia |
| 104 | Dayang | Indonesia |
| 105 | Jambai | Indonesia |
| 106 | KudaUnga | Indonesia |
| 107 | KetanBiajuk | Indonesia |
| 108 | KetanNyalin | Indonesia |
| 109 | Ketan Perak | Indonesia |
| 110 | KetanSiling | Indonesia |
| 111 | Ketumbar | Indonesia |
| 112 | Kretek Sir Putih | Indonesia |
| 113 | Kujam | Indonesia |
| 114 | Kumai-Kumai | Indonesia |
| 115 | LakatanJanbu | Indonesia |
| 116 | LayangPutich 1 | Indonesia |
| 117 | Siam | Indonesia |
| 118 | Padi Koran | Indonesia |
| 119 | Lima | Indonesia |
| 120 | SLM Temerin | Indonesia |
| 121 | KetanDelang | Indonesia |
| 122 | KetanCina | Indonesia |
| 123 | KetanToman | Indonesia |
| 124 | KetanSingkawang | Indonesia |
| 125 | KetanSamak | Indonesia |
| 126 | KetanGadul | Indonesia |
| 127 | Ungat | Indonesia |
| 128 | GedabungPutih | Indonesia |
| 129 | PadiKuntum | Indonesia |
| 130 | PadiHitamMelayu | Indonesia |
| 131 | PadiEwangJanggut | Indonesia |
| 132 | KretekSirendahMerah | Indonesia |
| 133 | TigaDara | Indonesia |
| 134 | Aceh-Aceh | Indonesia |
| 135 | JanggutBugisHitam | Indonesia |
| 136 | KuatikKundur | Indonesia |
| 137 | Padi Air | Indonesia |
| 138 | BurukBakul 2 | Indonesia |
| 139 | EwangRendah | Indonesia |
| 140 | Ewang Wangi | Indonesia |
| 141 | Ewang Wangi | Indonesia |
| 142 | Kuatik Jambi | Indonesia |
| 143 | KuatikMerah | Indonesia |
| 144 | KuatikSeraiRendah | Indonesia |
| 145 | MumbangKelapa | Indonesia |
| 146 | KuatikDubi | Indonesia |
| 147 | KuatikPutih | Indonesia |
| 148 | KuatikMerahTinggi | Indonesia |
| 149 | KuatikNibung 1 | Indonesia |
| 150 | KuatikNibung 2 | Indonesia |
| 151 | TCA48 | India |
| 152 | Betichikon | Bangladesh |
| 153 | Pajam | Bangladesh |
| 154 | Rayada 77210 | Bangladesh |
| 155 | DA21 | Bangladesh |
| 156 | Ghiganj | Bangladesh |
| 157 | FR13A | India |
| 158 | IR64 | IRRI |
| 159 | IR42(Check) | IRRI |
| 160 | IR40931(Check) | IRRI |
Table 1: List of varieties and their origin used in the diversity studies.
Leaf material collection and DNA extraction
Pre-germinated seeds were grown in plastic trays and trays were placed in the green house of IRRI. Young and healthy leaves (2-3 cm long) from 14 days old seedlings were harvested in 1.5 ml tubes and were preserved on ice, immediately. The samples were stored at -80ºC and total genomic DNA from the leaf samples was extracted following SDS based protocol.
The DNA pellet was air-dried for 4 h. Finally, dried DNA pellets were re-suspended in 100 μl of 1X TE buffer. The quality and quantity of DNA were measured by the NanoDrop (NanoDrop Technologies). Subsequently, the samples were diluted to 1:20 (DNA:water) with nanopure water maintaining a concentration around 20-30 ng/μl and stored in -20ºC. The diluted 4 μl of each DNA sample was used as template.
SSR marker genotyping
Thirty rice SSR primer pairs, well distributed on 12 chromosomes in Table 2 were chosen, based on previously used marker for genetic diversity analysis in rice by Thomson and his colleagues.
| SL | Markers | Chromosomes | Motifs | Annealing temp(ºC) |
|---|---|---|---|---|
| 1 | RM433 | 8 | (AG)13 | 55 |
| 2 | RM5 | 1 | (GA)14 | 55 |
| 3 | RM55 | 3 | (GA)17 | 55 |
| 4 | RM215 | 9 | (CT)16 | 55 |
| 5 | RM514 | 3 | (AC)12 | 55 |
| 6 | RM214 | 7 | (CT)14 | 55 |
| 7 | RM11 | 7 | (GA)17 | 55 |
| 8 | RM144 | 11 | (ATT)11 | 55 |
| 9 | RM171 | 10 | (GATG)5 | 55 |
| 10 | RM237 | 1 | (CT)18 | 55 |
| 11 | RM133 | 6 | (CT)8 | 55 |
| 12 | RM259 | 1 | (CT)17 | 55 |
| 13 | RM287 | 11 | (GA)21 | 55 |
| 14 | RM250 | 2 | (CT)17 | 55 |
| 15 | RM507 | 5 | (AAGA)7 | 55 |
| 16 | RM161 | 5 | (AG)20 | 61 |
| 17 | RM124 | 4 | (TC)10 | 67 |
| 18 | RM283 | 1 | (GA)18 | 55 |
| 19 | RM162 | 6 | (AC)20 | 61 |
| 20 | RM277 | 12 | (GA)11 | 55 |
| 21 | RM431 | 1 | (AG)16 | 55 |
| 22 | RM154 | 2 | (GA)21 | 61 |
| 23 | RM484 | 10 | (AT)9 | 55 |
| 24 | RM105 | 9 | (CCT)6 | 55 |
| 25 | RM536 | 11 | (CT)16 | 55 |
| 26 | RM125 | 7 | (GCT)8 | 55 |
| 27 | RM19 | 12 | (ATC)10 | 55 |
| 28 | RM541 | 6 | (TC)16 | 55 |
| 29 | RM413 | 5 | (AG)11 | 55 |
| 30 | RM474 | 10 | (AT)13 | 55 |
Table 2: List of 30 microsatellite markers and their motifs.
PCR reactions were conducted in a reaction volume of 20 μl, using 80-120 ng of template DNA (4 μl) with 16 μl of master mixture ( 8.5 μl of NP water, 2 μl of 10x TBE buffer, 2 μl of dNTP's (Appendix 4), 1 μl of each reverse and forward primers, 1 μl of DMSO and 0.5 μl of Taq polymerase). The PCR plates were placed in a G-storm thermal cycler machine for amplification of target DNA fragments and was programmed with condition of: initial denaturation at 94ºC for 5 min; 35 cycles of 45 s at 94ºC, annealing at 55-67ºC for 45 s, 1.5 min at 72ºC; and plus a finial extension step at 72ºC for 5 min. In the thermal cycler, annealing temperature was set up appropriate for each primer pairs (Table 2) to ensure successful amplification.
Phenotyping
The experimental procedures of IRRI were followed for submergence screening of rice seedlings in concrete water tank, as described by Pamplona and his colleagues. The experiment was laid out in a randomized complete design (RCB) with three replications. Before seeding, plastic trays were filled with soil, and fertilizer was applied @ 4 g ammonium sulphate/tray. Pre-germinated 25 seeds of each variety were placed in a row, keeping almost equal distance and covered with dry soil. Seedlings were grown in trays for 14 days and then, plant height of 5 plants for each entry was measured. The trays were transferred to concrete water tank and were submersed by raising water depth up to 75 cm and maintained for 5 days (Figure 1). Afterwards, water was again raised up to 100 cm to submerge the plants completely and maintained for 12 days. IR42 (susceptible check), was observed and tank was desubmerged, when 70% to 80% plants of susceptible check (IR42) become soft at shoot-root junction. After draining of water, the plants were allowed to recover for 7 days and the varieties were scored visually to categorize into 4 groups: tolerant (score 1-3), moderately tolerant (score 5), moderately susceptible (score 7) and highly susceptible (score 8-9), comparing with tolerant, IR40931 and susceptible check, IR42.
Score description
The varieties were scored visually based on standard evaluation system of IRRI (1996) and scale described by Gomosta.
| SL | Score description | Score | Survival% |
|---|---|---|---|
| 1 | Erect,dark green leaves, very little elongation | 1 | >95% |
| 2 | Erect, green leaves, little elongation | 3 | 80-95% |
| 3 | Droopy, pale green leaves, moderate elongation | 5 | 60-80% |
| 4 | Long, pale green leaves, elongated, few survived | 7 | 10-50% |
| 5 | Long, whitish leaves, elongated, completely dead | 9 | <10% |
Data analysis
Molecular weight of each band was measured to determine band size by using Alfa Imager version 5.5 software program. The presence of each informative band was measured, while its absence was scored as zero.
The polymorphic information content was based on the formula: PIC=1-Σ(Pi)2, where, 'Pi' is the frequency of the ith allele calculated for each microsatellite locus [29].
Power Marker software was used to calculate the average number of alleles, gene diversity and polymorphic information content (PIC) values. Binary data were used to compute genetic distance (GD) using C.S. Chord distance and un-rooted neighbor-joining tree was created based on C.S. Chord [30]. Phylogenetic reconstruction was based on the neighbor-joining method implemented in PowerMarker version 2.7 [31]. The genetic distances were interpreted based on theory of Nei [32] and Saitou and Nei [33]. Marker data was compared with submergence tolerance scores to select possible new source of tolerance from different clusters.
Results and Discussion
One hundred and sixty rice varieties from the tidal and flood prone areas of south and south East Asian countries were analyzed. Samples sizes were: 50 varieties from Bangladesh (deepwater, tidal and flood prone rice and modern varieties), 14 varieties from India (flood prone rice), 16 varieties from Sri Lanka (flood prone rice), 7 varieties from Vietnam (tidal varieties), 69 varieties from Indonesia (tidal varieties) and 4 check varieties from IRRI.
Genetic diversity in flood and tidal prone rice varieties
The number of alleles per locus, average number of alleles for all loci and PIC values were used for genetic diversity analysis. In this study, all 30 primer pairs created polymorphic bands among the 160 rice varieties from flood and tidal prone areas, which indicated that the microsatellites used were suitable for diversity analysis. A total of 337 alleles were detected with an average of 11 alleles per locus and the number of alleles per locus varied from 4 to 21 (Table 3). More alleles were found at each locus and the range in size of the alleles was higher, because of the larger population size and diversity of the samples. In accordance with previous reports, the number of alleles per locus was much larger than those reported in previous studies using different types of markers [34,35], RFLPs [26,36] and SSRs [25,26,37].The highest number of alleles (21) and the highest level of gene diversity (0.91) were detected at the locus RM474. Ni [38] also found significantly higher genetic diversity on chromosomes 6 and 7 of japonica cultivars compared with Indica at 111 microsatellite markers loci, through evaluating thirty-eight rice cultivars. In rice, molecular markers have been used to determine the genetic structure and pattern of diversity [24-26,37,39-41] and to identify rice accessions [42].
| Marker | Chr | Major allele | No of rare allele | No ofalleles | Size ranges (bp) |
Gene Diversity | PICvalue | Null alleles | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Size (bp) | Frequency (%) | |||||||||
| RM11 | 7 | 131 | 38 | 5 | 14 | 115-172 | 0.77 | 0.74 | 0 | |
| RM259 | 1 | 186 | 34 | 7 | 14 | 152-195 | 0.77 | 0.74 | 1 | |
| RM105 | 9 | 134 | 59 | 2 | 8 | 124-154 | 0.60 | 0.56 | 5 | |
| RM124 | 4 | 269 | 58 | 4 | 8 | 254-288 | 0.59 | 0.54 | 1 | |
| RM144 | 1 | 225 | 52 | 7 | 16 | 225-293 | 0.69 | 0.67 | 1 | |
| RM133 | 6 | 175 | 55 | 2 | 10 | 163-203 | 0.65 | 0.63 | 1 | |
| RM214 | 7 | 114 | 28 | 4 | 13 | 102-192 | 0.83 | 0.81 | 1 | |
| RM283 | 1 | 155 | 46 | 2 | 11 | 145-174 | 0.74 | 0.72 | 2 | |
| RM413 | 5 | 118 | 39 | 3 | 12 | 98-154 | 0.78 | 0.75 | 0 | |
| RM161 | 5 | 80 | 27 | 3 | 11 | 55-100 | 0.84 | 0.82 | 2 | |
| RM474 | 10 | 240 | 16 | 6 | 21 | 232-325 | 0.91 | 0.91 | 1 | |
| RM154 | 2 | 159 | 55 | 3 | 9 | 147-192 | 0.64 | 0.61 | 1 | |
| RM514 | 3 | 156 | 64 | 1 | 4 | 234-272 | 0.53 | 0.48 | 3 | |
| RM536 | 11 | 234 | 49 | 2 | 6 | 234-268 | 0.66 | 0.60 | 2 | |
| RM541 | 6 | 185 | 42 | 8 | 17 | 156-203 | 0.77 | 0.76 | 3 | |
| RM5 | 1 | 110 | 23 | 3 | 11 | 95-120 | 0.84 | 0.82 | 2 | |
| RM55 | 3 | 141 | 33 | 10 | 18 | 141-257 | 0.77 | 0.73 | 1 | |
| RM215 | 9 | 170 | 48 | 7 | 12 | 156-181 | 0.68 | 0.64 | 1 | |
| RM237 | 1 | 133 | 52 | 4 | 12 | 120-145 | 0.66 | 0.63 | 1 | |
| RM250 | 2 | 156 | 53 | 7 | 15 | 156-186 | 0.68 | 0.66 | 0 | |
| RM171 | 10 | 318 | 53 | 2 | 7 | 318-352 | 0.63 | 0.57 | 0 | |
| RM287 | 11 | 107 | 66 | 4 | 9 | 89-122 | 0.54 | 0.51 | 1 | |
| RM507 | 5 | 257 | 63 | 0 | 4 | 257-296 | 0.55 | 0.50 | 1 | |
| RM277 | 12 | 123 | 68 | 4 | 9 | 117-151 | 0.52 | 0.49 | 1 | |
| RM431 | 1 | 254 | 69 | 2 | 7 | 254-274 | 0.48 | 0.44 | 0 | |
| RM125 | 7 | 125 | 63 | 2 | 7 | 116-150 | 0.56 | 0.52 | 0 | |
| RM19 | 12 | 254 | 31 | 5 | 13 | 231-272 | 0.81 | 0.79 | 0 | |
| RM484 | 10 | 300 | 33 | 5 | 15 | 283-307 | 0.83 | 0.81 | 1 | |
| RM433 | 8 | 226 | 35 | 4 | 12 | 208-260 | 0.76 | 0.73 | 2 | |
| RM162 | 6 | 211 | 57 | 6 | 12 | 203-240 | 0.64 | 0.61 | 1 | |
| Mean | 47 | 4 | 11 | 0.69 | 0.66 | 1 | ||||
Table 3: Number of rare, null and major alleles, total number of alleles, amplification size range and PIC values for SSR loci assayed in 160 rice germplasm.
The PIC values ranged from 0.44 to 0.91 with an average of 0.66 (Table 3). The highest PIC values were observed for the primer of RM474 (0.91), followed by RM5 (0.82), RM484 (0.81), RM214 (0.81), and RM19 (0.79). In general, higher PIC values were observed for SSRs having higher numbers of alleles. Primer 474 had the highest PIC value (0.91) and the greatest number of alleles (21); therefore it detected the highest level of polymorphisms. Similar trend in PIC values for SSRs in barley were reported previously [43-46]. Primer RM431 (0.44) and RM514 (0.48) showed the lowest PIC value, indicating narrow genetic base at these loci. Thus, SSR primers have high resolving power for detecting polymorphism levels of rice cultivars (Figure 2).
Major allele is described as the allele with the highest frequency. Rare alleles are described as alleles with a frequency less than 5%.
Null and rare alleles
The null alleles can arise from point mutation(s) in one or both of the primer sites. In the case of null alleles, PCR amplification was repeated with failed samples to exclude failed PCR reaction. The lower number of null alleles was detected (average one/locus) with 30 primer pairs. The highest number of null alleles was scored at the locus RM105 (5) and 7 primers did not have any null allele, including RM507. Rare bands were amplified by 29 of the 30 primers. A total of 124 rare bands were found with an average number of 4 per locus (Table 3). The highest number of rare alleles was scored at the locus RM55 (10) followed by RM44 (7), RM259 (7), RM215 (7) and RM474 (6). Only 1 rare allele was detected at RM514 locus.
Clustering of cultivars based on SSR markers
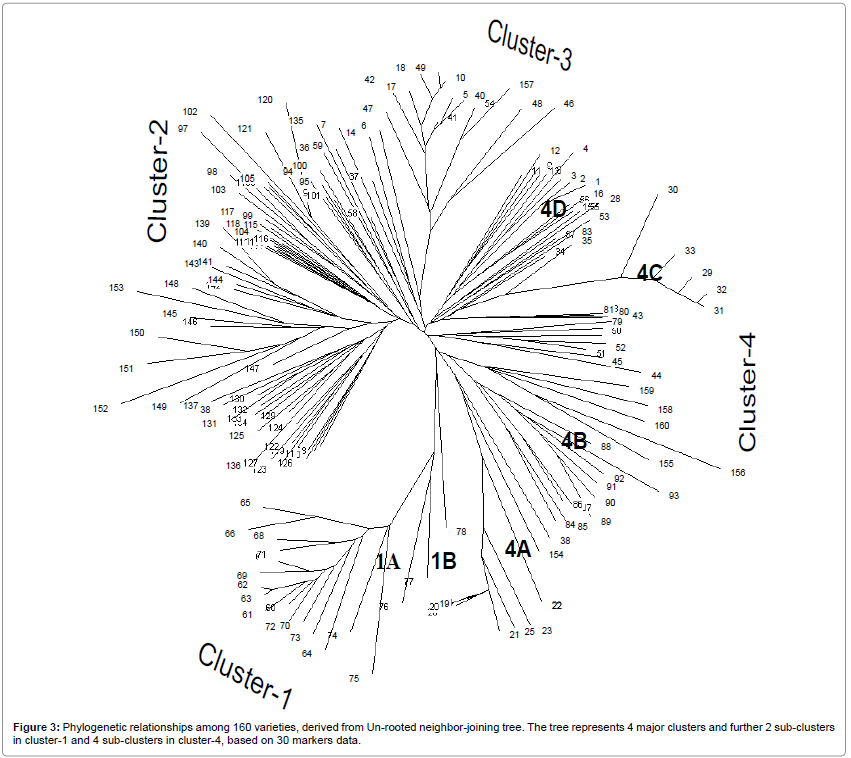
Un-rooted neighbor-joining tree based on C.S. Chord [30] showed 4 distinct major clusters (Figure 3), corresponding to the country of origins with additional sub-clusters under cluster-1 and cluster-4. In general, below the main cluster-1, genotypes from Sri Lanka and Vietnamese origin grouped separately in two sub-clusters: 1A- Sri Lanka, 1B- Vietnam. In fact, cluster-1 was mainly for the genotypes of Sri Lankan origin, only few varieties of Vietnam grouped loosely near cluster-1. It is likely that in cluster-1 all accessions from Sri Lanka, including accession 61 (Thavalu 153259), 62 (Kurkaruppan), 65 (Madabaru) and 66 (Kottamalli) were grouped. The cluster-2 did not have any sub-clusters, representing only the accessions from Indonesia. While those accessions derived from India grouped into Cluster-3 (49, Tilakkachari; 54, FR43B). Under the main cluster-4 (mainly Bangladesh origin) there were 4 sub-clusters: 4A consisted of tropical deepwater varieties (20, Kalamanik; 21, Habiganj Aman 1); 4B consisted of tidal varieties (43, Kachamota; 44, Modhu Malati and 45, Nakpechi); 4C consisted of MVs varieties of Bangladesh origin, and 4D consisted of tall flood prone rice varieties (4, Lata Mona; 11, Sadasail; 12, Rupsail) (Figure 3).
Interestingly, flood prone Indica varieties of Bangladesh and India were clustered in neighboring branches, while CN540 (53) of Indian
origin placed in Bangladeshi sub-cluster 4D and some Bangladeshi varieties (5, Lal Chikon; 10, Rajasail; 18,Motorsail) also placed in Indian Cluser-3. In fact, the flood prone genotypes of Bangladesh- Indian clusters, derived from same origin and same types of varieties were grouped into cluster-3 or sub-cluster-4D. Same types of varieties from different origin could be placed in the same cluster. Sub-cluster- 4B for tidal varieties of Bangladesh also included some accessions from Indonesian tidal areas (85, Bayar Kuning; 91, Umbang Inai). Therefore, it is concluded that accessions from Bangladesh are mostly close to Indian varieties, but different from Indonesian varieties. This study showed that microsatellite loci are useful for studying the genetic variation and grouping of flood prone Indica rice accessions. In a study, [28] analyzed 234 accessions of rice (Indica and Japonica) at 169 SSR marker loci and they detected 5 distinct groups of rice, corresponding to indica, aus, aromatic, temperate japonica, and tropical japonica rices.
It is interesting to note that 160 accessions clustered together in 4 main groups, indicating a geographical bias for genetic similarity. But diversity within varieties of Bangladesh-Indian origin was more pronounced than for other geographical areas. Each of the sub-clusters of Bangladesh seemed to represent different ecotypes (deepwater, tidal, and flood prone varieties) within a geographical area. This result indicated that major classification of varieties should rely on the different geographical areas and then sub-grouping of varieties should be done based on ecotypes. Bangladesh and East Indian agro-ecological conditions (distinct variations in water regimes) are most diverse, and these conditions probably contributed to great varietal diversity for evolving sub eco-cultural types. Selected varieties from different clusters could be used as a source of genetic variability for different traits and could be exploited directly in breeding. The present study contributes to the knowledge of the genetic structure and molecular characterization of the flood and tidal prone rice varieties of south and south Asian countries.
Predicting submergence tolerance
Cluster analysis divided the genotypes into four main clusters and six sub-clusters based on geographical origins and ecotypes. Further, submergence screening classified the varieties into 4 groups: tolerant (score 1-2), moderately tolerant (score 4-5) and moderately susceptible (score 6-7) and highly susceptible (score 9). Comparison of tolerance level in different clusters and sub- clusters by microsatellite markers are shown in Table 4. Cluster-1 comprises mainly the accessions of Sri Lanka, which belongs to the most of the tolerant varieties having higher survival % than other varieties. Only 2 cultivars from Sri Lanka were moderately susceptible. Singh [47] classified rainfed lowland rice genotypes, using cluster analysis and most of the genotypes belonging in their cluster-1 were submergence tolerance. In this study, cluster-2 mostly comprised of accessions from Indonesia with lower survival, indicating their narrow ranges of tolerance to submergence stress. Cluster-3 and cluser-4 included accessions of eastern India and Bangladesh, respectively. These 2 clusters comprised all types of varieties including, tolerant, moderately tolerant, moderately susceptible, as well susceptible types. Microsatellite clustering (over 30 polymorphic loci) and submergence screening data indicated greater genetic diversity among 160 genotypes for molecular loci and for submergence tolerance.
| Origins | Clusters | Sub-clusters | Ecotypes | No. of accessions | |||
|---|---|---|---|---|---|---|---|
| T | MT | MS | S | ||||
| Bangladesh | Cluster-4 | 4A | Deep-water | 0 | 0 | 2 | 10 |
| 4B | Tidal | 0 | 6 | 7 | 3 | ||
| 4C | MVs | 0 | 0 | 3 | 5 | ||
| 4D | Flood | 0 | 6 | 4 | 4 | ||
| India | Cluster-3 | - | Flood | 2 | 5 | 4 | 3 |
| Indonesia | Cluster-2 | - | Tidal | 0 | 2 | 41 | 26 |
| Sri Lanka | Cluster-1 | 1A | Flood | 8 | 6 | 2 | 0 |
| Vietnam | 1B | Tidal | 0 | 0 | 3 | 4 | |
| Total | 10 | 25 | 66 | 55 | |||
Table 4: Differentiation of 160 varieties based on tolerance level (1-9 scale) under main and sub-clusters after 12 days of submergence.
Genetic diversity analysis among flood prone rice will be useful for identifying the varieties having maximum diversity with submergence tolerance and selected varieties will be useful for further studies. Tolerant genotypes in Cluster-1 are expected to have different tolerance genes.
Genotypes were classified into 4 categories: T (tolerant, score 1-2); MT (moderately tolerant, score 4-5); MS (moderately susceptible, score 6-7); and S (susceptible, score 9) based on their performance under 12 days of submergence than those in Cluster-4. Crossing of cultivars from different clusters or from different sub-clusters could be useful for improvement of submergence tolerance in rice. For improvement of tolerance level of tidal varieties (Kajalsail and Lachikon) of Bangladesh, those varieties could be crossed with derivative lines of FR13A. Indonesian and Vietnamese varieties are lack of tolerance traits for their successful cultivation in flood prone and tidal areas. The tolerance level and other adaptive traits of Indonesian and Vietnamese modern varieties could be improved by crossing with the best tolerant cultivars found in Sri Lanka.
The possible origin of tolerant rice
Tolerance level of representative varieties from the four clusters is shown in Table 5. Different level of tolerance among the genotypes from different microsatellite clusters was found. Interestingly, most of the Sri Lankan varieties (Devarenddiri and Thavalu) were tolerant. All Indonesian cultivars (Siam) were susceptible. Finding relationship between tolerance and country of origin, highly tolerant varieties (FR13A and FR43B) were found from east India. Previous report also indicated that the best known tolerant cultivar, FR13A was found from the submergence-prone area of Orissa, India [14].
| Clusters/Origins | Sub-clusters | Eco-types | Representative varieties | Tolerance |
|---|---|---|---|---|
| Cluster-4 Bangladesh | 4A | Deep-water | Fulkari | S |
| Habj. A.7 | S | |||
| 4B | Tidal | Mach Ranga | MT | |
| Patnai23 | MS | |||
| 4C | MVs | BR11 | S | |
| BR31 | S | |||
| 4D | Flood | Motorsail | MT | |
| LalChikon | MT | |||
| Cluster-3 India | Flood | FR13A | T | |
| Tillakkachari | MT | |||
| Cluster-2 Indonesia | Tidal | Siam | S | |
| Padi Koran | S | |||
| Cluster-1 Sri Lanka | 1A | Flood | Devarenddiri | T |
| Madabaru | MT | |||
| Thavalu 15314 | T | |||
| Vietnam | 1B | Tidal | Giau Dumont | S |
| Doc Phung | S |
Table 5: Tolerance level of representative varieties under different cluster and sub-clusters.
Cluster-3 and cluster-4 consisted of accessions from flood prone areas of India and Bangladesh, where different types of tolerant and intolerant varieties (T, MT, MS and S) were found. Rice varieties of flood prone areas of east India-Bangladesh are consisting of: tall nonelongating [48], Rayada and Ashin [34], tidal swamp and deepwater rice varieties. Major flood-prone areas of Asia occur in Bangladesh (24%), and eastern India. Diverse hydrological conditions such as flash-flooding, stagnant water and tidal flooding probably contributed to great varietal diversity in East Indian and Bangladesh conditions. FR13A is a tolerant variety of Orissa, India and in the present study it was grouped in Cluser-3 of Indian origin. It is likely that hydrological conditions of east India favoured for the evolution of most tolerant variety, FR13A and FR43B. The evolution of plant populations is largely influenced by the stresses and thus susceptibility of plants to environmental extremes has driven the evolution of a wide range of stress tolerance mechanisms [49,50].
Tolerant FR13A (54) and FR43B (157) of Indian origin were grouped in cluster-3 and tolerant Thavalu-15314 from Sri Lanka was grouped in cluster-1. Thus, tolerant varieties of Indian and Sri Lankan origin are genetically dissimilar, except their submergence tolerance. The findings of Xu [51] suggested that grains from submergence tolerant plants have been transported over 1,000 km and subsequently introgressed into the local varieties in Sri Lanka. Location and close cultural relationship of India and Sri Lanka might able to utilize land routes for migration of tolerant varieties. Indonesia which is geographically far away and culturally isolated from east India did not allow the introgression of submergence tolerance gene into Indonesian varieties.
Rating of accessions as tolerant (T), moderately tolerant (MT), moderately susceptible (MS) and susceptible (S) was determined according 1-9 scale. The representative varieties were selected from different clusters and sub-clusters
Conclusion
One hundred and sixty rice varieties from the tidal and flood prone areas of south and south East Asian countries were genotyped using 30 SSR markers. The selected markers could be used in markerassisted selection program for the development of tolerant rice lines. The identified tolerant lines will be studied further to observe tolerance. FR13A and FR43B could be utilized to develop submergence tolerance rice lines/varieties with all desirable characters using marker-assisted breeding. The result indicated that the SSR markers are neutral and codominant and could be a powerful tool to assess the genetic variability of the cultivars. The information about the genetic diversity will be very useful for proper identification and selection of appropriate parents for breeding programs, including gene mapping, and ultimately for emphasizing the importance of marker-assisted selection (MAS) in aromatic rice improvement worldwide.
References
- Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK (2005) An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 142: 169-196.
- Litt, M, Lutty JA (1989) Ahyper variable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle action gene. Am J Hum Genet 44: 397-401.
- Hearne CM, Ghosh S, Todd JA (1992) Microsatellites for linkage analysis of genetic traits. Trends Genet 8: 288-294.
- Weber JL, May PE (1989) Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet 44: 388-396.
- Condit R, Hubbell SP (1991) Abundance and DNA sequence of two-base repeat regions in tropical tree genomes. Genome 34: 66-71.
- Schug MD, Hutter CM, Wetterstrand KA, Gaudette MS, Mackay TF, et al. (1998) The mutation rates of di-, tri- and tetranucleotide repeats in Drosophila melanogaster.MolBiolEvol 15: 1751-1760.
- Cho YG, Ishii T, Temnkh S, Chen X, Lipovich L, et al. (2000) Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.) TheorAppl Genet 100: 713-722.
- Xiao J, Li J, Grandillo S, Ahn SN, Yuan L, et al. (1998) Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryzarufipogon. Genetics 150: 899-909.
- Zou JH, Pan XB, Chen ZX, Xu JY, Lu JF, et al.(2000) Mapping quantitative trait loci controlling sheath blight resistance in two rice cultivars (Oryza sativa L.).TheorAppl Genet 101: 569–573.
- Mackill DJ, Nguyen HT, Zhang JX(1999) Use of molecular markers in plant improvement programs for rainfed lowland rice. Special issue: Adaptation of rainfed lowland rice. Field-Crops-Research. 64: 1-2,58,177-185.
- Zhang Z, Deng Y, Tan J, Hu S, Yu J, et al. (2007) A genome-wide microsatellite polymorphism database for the indica and japonica rice. DNA Res 14: 37-45.
- Gregorio GB (1997) Tagging salinity tolerance genes in rice using amplified fragment length polymorphism (AFLP). Ph.D. thesis.
- HilleRisLambers D, Vergara B (1982) Summary results of an international collaboration on screening methods for flood tolerance. In 'Proceedings of the 1981 International Deep-water Rice Workshop'. International Rice Research Institute: Los Baños, Philippines 347-353.
- Mackill DJ(1986)Rainfed lowland rice improvement in South and Southeast Asia: Results of a survey. In 'Progress in Rainfed Lowland Rice. International Rice Research Institute: Los Baños, Philippines 115-144.
- Singh RK, Dwivedi JL(1996) Rice improvement for rainfed lowland ecosystem: Breeding methods and practices in Eastern India. p. 50–77. In S. Fukai et al. (ed.) Breeding strategies for rainfedlowland rice in drought-prone environments. Proceedings of International Workshop, Ubon, Thailand. ACIAR Proceedings.
- Mackill DJ(2003)What molecular tools are available for selection for drought tolerance. In: Manual, Breeding rice for drought-prone environments 44: 55-57.
- Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity regulatory variation in mouse genes. Nat Genet 32: 432-437.
- Matsuoka Y, Mitchell SE, Kresovich S, Goodman M, Doebley J (2002) Microsatellites in Zea - variability, patterns of mutations and use for evolutionary studies. TheorAppl Genet 104: 436-450.
- Goldstein DB, Pollock DD (1997) Launching microsatellites: A review of mutation processes and methods of phylogenetic interference. J Hered 88: 335-342.
- Russell JR, Fuller JD, Macaulay M, Hatz BG, Jahoor A, et al. (1997) Direct comparison of levels of genetic variation among barley accessions detected by RFLPs, AFLPs, SSRs and RAPDs. TheorAppl Genet 95: 714-722.
- Pestsova E, Ganal MW, Röder MS (2000) Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome 43: 689-697.
- Huang XQ, Börner A, RöderMS, Ganal MW(2002) Assessing genetic diversity of wheat (Triticumaestivum L.) germplasm using microsatellite markers. TheorAppl Genet 105: 699-707.
- Primmer CR, Landry PA, Ranta E, Merila J, Piironen J, et al.(1996) Microsatellite analysis of relationships within cultivated potato (Solanumtuberosum). TheorAppl Genet 92: 1076-1084.
- Akagi H, Yokozeki Y, Inagaki A, Fujimura T (1997)Highly polymorphic microsatellites of rice consist of AT repeats, and a classification of closely related cultivars with these microsatellite loci. TheorAppl Genet 94: 61-67.
- Mackill DJ(1995) Classifying japonica rice cultivars with RAPD markers. Crop Science. 35: 889-894.
- Zhang Q, Maroof MA, Lu TY, Shen BZ (1992) Genetic diversity and differentiation of indica and japonica rice detected by RFLP analysis. TheorAppl Genet 83: 495-499.
- McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, et al. (2002) Development of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9: 199-207.
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169: 1631-1638.
- Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32: 314-331.
- Cavalli-Sforza LL, Edwards AW (1967) Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet 19: 233-257.
- Liu K, Muse S (2004) PowerMarker: New Genetic Data Analysis Software, Version 2.7
- Nei M (1972) Genetic distance between populations, American Naturalist106: 283-292.
- Saitou N, Nei M (1987) Theneighbour-joining method: A new method for reconstructing phylogenetic trees. MolBiolEvol 4: 406-425
- Glaszmann JC (1987) Isozymes and classification of Asian rice varieties. TheorAppl Genet 74: 21-30.
- Second G(1982) Origin of the genic diversity of cultivated rice (Oryza spp.): Study of the polymorphism scored at 40 isozyme loci. Jpn J Genet 57:25–57.
- Wang ZY, Tanksley SD (1989) Restriction fragment length polymorphism in Oryza sativa L. Genome 32:1113-1118.
- Yang GP, Maroof MA, Xu CG, Zhang Q, Biyashev RM (1994) Comparative analysis of microsatellite DNA polymorphism in landraces and cultivars of rice. Mol Gen Genet 245: 187-194.
- Ni J, Colowit MP, Mackill DJ(2002) Evaluation of genetic diversity in rice subspecies using microsatellite markers. Crop Science 42:601-607.
- Wu KS, Tanksley SD (1993) Abundance, polymorphism and genetic mapping of microsatellites in rice. Mol Gen Genet 241: 225-235.
- Ahn SN, Kwon SJ, Yang CI, Hong HC, Kim YK, et al. (2000) Diversity analysis of Korea bread rice cultivars. In International plant and animal genome VIII conference, January 9th-12th, Abstract. P. 499.
- Giarrocco LE, Marassi MA, Salernoa GL (2007) Assessment of the genetic diversity in argentine rice cultivars with SSR markers. Crop Sci 47:853-858.
- Virk PS, Ford-Lloyd BV, Jackson MT, Newbury HJ (1995) Use of RAPD for the study of diversity within plant germplasm collections. Heredity (Edinb) 74: 170-179.
- Pillen K, Binder A, Kreuzkam B, Ramsay L, Waugh R, et al. (2000) Mapping new EMBL-derived barley microsatellites and their use to differentiate German barley cultivars. TheorAppl Genet 101: 652-660.
- Saghai-Maroof MA, Biyashev RM, Yang GP, Zhang Q, Allard RW (1994) Extraordinarily polymorphic microsatellite DNA in barley: Species diversity, chromosomal locations and population dynamics U.S.A. ProcNatlAcadSci 91: 5466-5470.
- Dávila JA, Loarce Y, Ramsay L, Waugh R, Ferrer E (1999) Comparison of RAMP and SSR markers for the study of wild barley genetic diversity.Hereditas 131: 5-13.
- Struss D, PlieskeJJ(1998)The use of microsatellite markers for detection of genetic diversity in barley populations. TheorAppl Genet 97: 308-315.
- Singh S, Vijaykumar CHM, Sarkarung S, Singh RK, Singh ON, et al.(2002) Molecular diversity of rainfed lowland rice genotypes with varying sensitivity to drought and delayed planting. Rice Genetics Newsletters 19: 18.
- Catling D(1992) Rice in deep water. Growth and development. International Rice Research Institute P 121-169.
- Mahalingam R, Gomez-Buitrago A, Eckardt N, Shah N, Guevara-Garcia A, et al. (2003) Characterizing the stress/defense transcriptome of Arabidopsis. Genome Biol 4: R20.
- Chen WJ, Zhu T (2004) Networks of transcription factors with roles in environmental stress response. Trends Plant Sci 9: 591-596.
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, et al.(2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708.
Citation: Masuduzzaman ASM, Haque M, Ahmed MME, Mohapatra CK (2016) SSR Marker-based Genetic Diversity Analysis of Tidal and Flood Prone Areas in Rice (Oryza sativa L.). J Biotechnol Biomater 6:241. DOI: 10.4172/2155-952X.1000241
Copyright: © 2016 Masuduzzaman ASM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 14493
- [From(publication date): 9-2016 - Aug 23, 2025]
- Breakdown by view type
- HTML page views: 13147
- PDF downloads: 1346