Polymorphisms in Claudin 1 and Claudin 7 in Healthy Individuals in Sweden
Received: 08-Apr-2016 / Accepted Date: 28-Apr-2016 / Published Date: 02-May-2016 DOI: 10.4172/2476-2024.1000113
Abstract
Tight junction is the area between epithelial cells that controls the fluid between cells. It consists of three main groups of proteins, transmembrane proteins claudin and occludin, accessory proteins zonula occludence 1,2,3 and cytoskeletal protein Actin. Although their main function is to prevent leakage and diffusion of molecules across epithelial barriers they also contribute to the adherence of cells. In malignant tumors, tight junctions frequently show structural and functional abnormalities. Our aim in this study was to see the frequency of genotypes rs6174821 and rs9869263 in claudin 1, CLDN1 and rs4562, rs3744400 of claudin 7, CLDN7 among the healthy population in Sweden. 265 samples were collected from blood and plasma donors. DNA was extracted and mutational screening for CLDN 1 SNP rs6174821 and rs9869263 and CLDN 7 SNP rs4562 and rs3744400 was performed using pyrosequencing. The Hardy-Weinberg formula was used to calculate the genotype. The results show that there is a difference in the frequency of polymorphisms between men and women for several of the genotypes as well as differences compared to HapMap. In CLDN 1 SNP rs9869263 for men and women together the frequency for TT was 0.1959 for TC 0.385 and for CC 0.593 while the HapMap frequency for both men and women was for TT 0.035, for TC 0.230 and for CC 0.735. CLDN 7 SNP rs3744400 for men and women together genotype TT 1.931 and for TG 0.066, whereas GG were 0. When we compare to Hap map frequency for both men and women genotype TT 0.991, TG 0.009 and GG were 0. For men and women together the frequency for SNP rs4562 CC was 0.71, for TT 0.323 and for TC 0.97. The HapMap frequency for men and women was 0.292 for CC, 0.185 for TT and 0.523 for TC.
Keywords: Tight junction; Claudin; Colon cancer; Pyrosequencing
Introduction
Many cells in tissues are linked to one another and to the extra cellular matrix with the help of cell adhesion molecules. Tight junction play a critical role in maintaining the concentration differences of small molecules across epithelial cell sheets by sealing the plasma membranes of adjacent cells together in the apical surface and create a continuous impermeable or semi-permeable barrier to diffusion across the cell sheets [1]. Tight junction bar the movement of dissolved material from the lumen through the space between epithelial cells, there is no intercellular space where there is a tight junction. Tight junction consists of three main groups of proteins they are transmembrane proteins claudin and occludin, accessory proteins zonula occludence and cytoskeletal proteins Actin [1].
Claudins are the most important components of the tight junction [2]. The claudin family consists of 24 members and are small, 20-27 kDa, transmembrane proteins found in many organisms [3,4]. Their main function is to establish the paracellular barrier that controls the flow of molecules in the intercellular space between the cells of an epithelium.
Claudins span the cellular membrane 4 times, with the N-terminal end and C-terminal end both located in the cytoplasm. Recent studies have shown mutations in cell adhesion / tight junction genes claudin 1 exon 2 and claudin 7 exon 4 in patients diagnosed with colon cancer in both normal tissue and tumor tissue [4,5].
Genetic variations in claudin 1 have recently been found to be associated with susceptibility of Hepatitis C infection [6]. Also two SNPs in the intron region, rs 893051 and rs3774032 have been linked to asthma [7]. Hahn-Strömberg et al. found polymorphisms, rs9869263 and rs4562 in claudin 1 and 7 respectively in both the tumor tissue and the adjacent normal mucosa raising the question if people with these polymorphisms have a higher chance of developing colon cancer.
The role of claudins in cancer has not been clearly defined, however it is known that the claudins are tissue specific proteins that are up or down regulated depending on the disease [5,6]. Also, claudin-1 has been found to be dysregulated in colorectal carcinoma and identified as a probable target of beta catenin/Tcf signalling [8]. A study by Tokes et al, reported that loss of claudin-1 proteins in breast cancer cells is involved in the infiltration and metastasis of breast cancer [9]. In colorectal carcinoma claudin-7 has been found strongly overexpressed, and upregulation of claudin 7 has been described as an early event in gastric tumorgenesis [10]. The aim of this study was to investigate the frequency of polymorphisms in the claudin 1 and claudin 7 genes among healthy individuals in Sweden. Since these polymorphisms are present in coloncancer, these results will provide a better understanding of the relationship between cell adhesion genes and the development of colon cancer.
Material And Methods
265 samples from blood/ plasma donors at Orebro University Hospital were collected randomly from both men and women after agreement, consisting of one EDTA tube of peripheral blood in connection with blood and plasma donation was collected and placed in –80°C immediately. The samples were anonymized and numbered from 1-265 and marked W (woman) or M (man). DNA was extracted from these blood/plasma samples and stored at -20°C. For mutational screening routine PCR and Pyrosequencing methods were used. PCR (polymerase chain reaction) and Pyrosequencing (Pyromark Q96 ID QIAGEN) was performed according to routine protocols at the Department of Laboratory Medicine, Orebro University Hospital, Orebro, Sweden. There were no exclusion criteria since these blood and plasma donors are subjected to high scrutiny. The Ethics Committee at EPN, Uppsala, Sweden approved the study.
DNA extraction
DNA extraction protocols have been implemented according to manufacturer’s protocol by (Viogene, GENTAUR Belgium BVBA). 1 ml sample was added into a 15 ml tube. 10 µl proteinas K was added as well as 1 ml EX buffer to the sample and incubated at 60°C for 20 min then at 7 for 20 min, 1050 µl of 99% ethanol was added to the tube after incubation.
Preheated 70°C 10 mM Tris-Hcl, pH 9.0 (2.5 ml/prep) was used for DNA elution. A genomic DNA midi column was placed in a 15 ml collection tube, the sample mixture was added to the column, and centrifuged at 3000 rpm for 3 min. The flow through was discarded and 2.5 ml of WS buffer was added and centrifuged at 3000 rpm for 3 min this was repeated twice.
The tube was then centrifuged at 4000 rpm for further 5 min to dry the column; the genomic DNA midi column was placed in a new 15 ml tube. The DNA eluted with 1ml of preheated 10 mM Tris-HCL, pH 9.0 was centrifuged at 3000 rpm for 10 min. DNA was stored at -20°C.
PCR (Polymerase Chain Reaction)
The PCR primers were designed using the primer 3 programme (http://www.embnet.sk/cgi-bin/primer3_www.cgi).
The protocol used to optimize the PCR amplification can be seen below. The PCR reactions were carried out in a thermocycler (Eppendorf gradient) starting with 95°C denaturation for 10 min then 50 cycles á 95°C denaturation for 1 min, 61°C annealing for 1 min 72°C extension for 1 min. The mastermix consisted of 3-5 ng of DNA in a buffer containing 2.0 mM MgCl2, 200-400 µM dNTP, and reverse and forward primer mix 0.1-0.5 µM (for primer sequences see Table 1) and Taq polymeras 0.5 units.
| Gene | Primer | Tm © | Product size (bp) |
|---|---|---|---|
| Claudin-1 | Forward 5'-CAGAAGATGAGGATGGCTGTCATT-3' Reverse *5'-AAGGGGGGCACAGCCTCTATTA-3' *biotin |
58 | 117 |
| Claudin-7 | Forward 5'-GTTTGGCCCTGCCATCTTTAT-3' Reverse *5'-ACACATACTCCTTGGAAGAGTTGG-3' *biotin |
60 | 155 |
Table 1: PCR primer sequences and conditions.
Pyrosequencing
Pyrosequencing was carried out using Qiagens Pyrosequencer PyroMark Q96 ID, Qiagen, USA. For each sample a mixed buffer containing 3 µl streptavidin sepharos solution, 12 µl Milliq H2O, 40 µl binding buffer and 25 µl av PCR products was used, the enzyme mix contained DNA-polymerase, apirax, luciferase and sulfurylase, while the substrate mix contained adenosine 5' phosphosulfate and luciferin. The software used to analyze the SNP postion was Pyromark ID1.0 Biotage, Sweden.
Statistics
Statistical analyses were performed using PHASE version 2.1 and The Hardy-Weinberg formula was used to calculate the genotype.
Results
Our aim in this study was to see the frequency of polymorphisms in the genes of claudin 1 and claudin 7. For claudin 1 we were interested in SNP rs6174821 and rs9869263 and for claudin 7 SNP rs4562 and rs3744400 among the healthy population in Sweden. To analyze this, 265 blood and plasma donors were examined for polymorphisms in cldn 1 and cldn 7.
The Hardy-Weinberg formula was used to calculate the genotype. All were in Hardy-Weinberg equilibrium. The frequency results for CLDN 1 in SNP rs9869263 for female genotype TT were 0.027, 0,37 for TC and 0.607 for CC and for males genotype TT were 0.021, 0,4 for TC and 0.574 for CC. For men and women together the frequency for TT was 0.1959 for TC 0.385 and for CC 0.593 while the HapMap frequency for both men and women was for TT 0.035, for TC 0.230 and for CC 0.735.
The result of CLDN 7 frequency in SNP rs4562 for female genotype CC were 0.35, 0.143 for TT and 0.51 for genotype TC, and for male genotype CC were 0.36, for TT 0.18 and 0.51 for TC. In SNP rs3744400 the frequency result for female genotype TT were 0.97, for TC 0.04, 0.103 for CC and 0.027 for TG. For male genotype TT were 0.961, for TC 0.034, for CC 0.204 and for genotype TG 0.039. For men and female together the frequency for SNP rs4562 CC was 0.71, for TT 0.323 and for TC 0.97. The HapMap frequency for men and female was 0.292 for CC, 0.185 for TT and 0.523 for TC. SNP rs3744400 for male and female together genotype TT 1.931 and for TG 0.066, whereas GG were 0. When we compare to Hap map frequency for both men and women genotype TT 0.991, TG 0.009 and GG were 0.
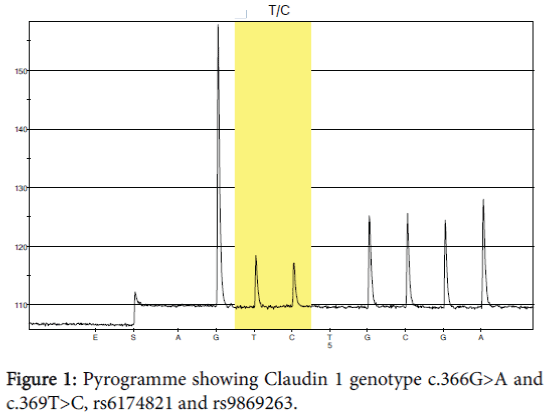
In CLDN 1 we studied exon 2 SNP c.366G>A rs6174821 but we could not find any mutations, in exon 2 c.369T>C rs9869263 shown in Table 2 and Figures 1 and 2 the mutation in this SNP is synonymous, in this case GGT=?GGC, both codes for (Glycin).
| Genotype | Female frequency | N | Male frequency | N |
|---|---|---|---|---|
| c.369T>C | 112 | 153 | ||
| TT | 0.027 | 3 | 0.033 | 5 |
| TC | 0.37 | 41 | 0.42 | 65 |
| CC | 0.607 | 68 | 0.54 | 83 |
Table 2: Frequency of claudin 1 genotype c.369T>C for men and women in the healthy population in Sweden.
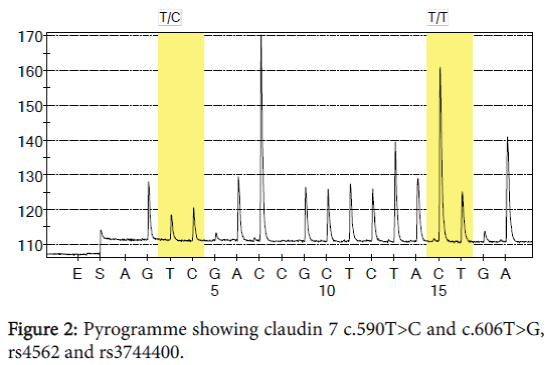
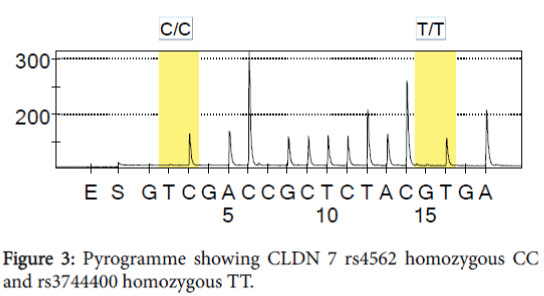
In CLDN 7 exon 4 both SNPs c.590T>C rs4562 and c.606T>G rs3744400, polymorphisms are shown in Table 3 and Figure 3. The mutation in rs4562 is missense, resulting in a codon that code for a different amino-acid. In rs3744400 the mutation is synonymous, CCT=?CCG and the final amino-acid is proline (Tables 4 and 5).
| Genotype frequency | HapMap Genotype frequency |
|||
|---|---|---|---|---|
| Genotype | Male and Female | N | Male and Female | N |
| c.369T>C | 265 | 113 | ||
| TT | 0.06 | 8 | 0.035 | 4 |
| TC | 0.79 | 106 | 0.23 | 26 |
| CC | 1.15 | 151 | 0.735 | 83 |
Table 3: Frequency of claudin 1 genotype c.369T>C for men and women together in the healthy population in Sweden compared to the HapMap frequency for the CEU population.
| CLDN 7 | Female | Male | ||
|---|---|---|---|---|
| Genotype | frequency | N | frequency | N |
| c.590T>C | 112 | 153 | ||
| CC | 0,35 | 39 | 0.36 | 55 |
| TT | 0,143 | 16 | 0.18 | 28 |
| TC | 0,51 | 57 | 0.46 | 70 |
| c.606T>G | 112 | 153 | ||
| TT | 0,97 | 109 | 0,961 | 147 |
| TG | 0,027 | 3 | 0,039 | 6 |
| GG | 0 | 0 | 0 | 0 |
Table 4: Frequency of claudin 7 genotype c.590T>C and c.606T>G for men and women in the healthy population in Sweden.
| CLDN7 | Genotypefrequency | HapMap Genotype frequency |
||
|---|---|---|---|---|
| Genotype | Male and Female | N | Male and Female |
N |
| c.590T>C | 265 | 65 | ||
| CC | 0,71 | 94 | 0.292 | 19 |
| TT | 0,323 | 44 | 0.185 | 12 |
| TC | 0,97 | 127 | 0.523 | 34 |
| c.606T>G | 265 | 113 | ||
| TT | 1.931 | 256 | 0.991 | 112 |
| TG | 0.066 | 9 | 0.009 | 1 |
| GG | 0 | 0 | 0 | 0 |
Table 5: Frequency of CLDN 7 genotype c.590T>C and c.606T>G for men and women together in the healthy population in Sweden compared to the HapMap frequency for the CEU population.
Discussion
Tight junction or zonula occludins, are the closely associated areas of two cells whose membranes join together forming a virtual impermeable barrier to against liquid and exist for example in cells in the intestine to prevent unwanted uptake in the body of the various components of spillage. The claudins are tissue specific and so the expression changes depending on the type of tissue studied [11].
Variants in CLDN 1 and CLDN 7 gene have recently been identified and correlated to several different diseases such as cancer, hepatitis C, atopic dermatitis and asthma [6]. These variations are of critical importance in the formation of tight junction and their function.
In this study we analyzed the frequency of polymorphisms in claudin 1 SNP rs6174821 and rs9869263 and in claudin 7 SNP rs4562 and rs3744400. Peripheral blood from 265 blood and plasma donors was used. Altogether 112 women and 153 men were analyzed for theses polymorphisms.
The largest differences were seen for CLDN 7 polymorphisms rs4562 genotype c.590T>C where 35% of the women were homozygote for CC, while among the men 36% were homozygote for CC. Also 14% of the women were homozygote for TT while only 18% of the men were homozygote for the same genotype. When comparing our results with results from the Hap Map project we could see that for rs4562 the frequency of the AA genotype was 18.5%, AG 52.3% and GG 29.2% in men and women combined. This was for the CEU population, there were no results registered from other populations. For rs3744400 our results were slightly lower than those of the European ancestry, the Indian Mexican, Masaya in Kenya, Italy and Nigerian ancestry who showed between 96% and 99.1% frequency of genotype TT, while the Japanese and Chinese population showed only 50%-57% positive for this genotype. For T/G the frequency among the Swedish population was 3.7% which is similar to the GIH population of 4.0% and TSI 2.9%, YRI 2.7% and the MEX 5.3% population compared to much higher frequencies in CHB 33.8%, CHD 39.4% and JPT 38.9%. The lowest frequency was in the CEU population with 0.9% TG. For genotype GG the frequency was 0% for all populations except CHB 8.8%, CHD 10.1% and JPT 8.8% which is slightly lower that our results 15.4%.
For rs9869263 our results for genotype TT 1.95% are similar to that of the GIH population with 2.0%, the TSI population with 2.9% and the MEX population with 1.7%. The other ancestries varied from 0.9% JPT-7.7% MKK. For the TC genotype our results for the Swedish population 38.5% were very similar to the other populations, which varied from CEU 23%-43.5% for the YRI population. For the CC genotype there was little discrepancy between our results 59.3% and other populations which varied from 49%YRI-74.3% GIH.
The results indicate that the largest difference in genotype between men and women were for the c.590T>C genotype.
For rs4562 there is an amino acid substitution (Val197Ala) while for rs9869263 there is no substitution (Gly123Gly). For rs3744400 there is no amino acid substitution (Pro202Pro).
According to Hahn-Stromberg et al. [5] in colon tumors, the frequency of CLDN 7 SNPs, rs4562 was 57% in both tumors and normal mucosa from the same individuals who showed a homozygous polymorphism TC (Val197Ala). Hahn-Stromberg et al. also found that polymorphisms in Claudin 1 and Claudin 7 were associated with tumor differentiation and tumor stage of colon carcinoma [12]. In the normal population the frequency was 61%. The remaining 43% showed no polymorphisms in this gene either in either the tumor or in the normal mucosa. In claudin 1, rs9869263 65% showed a homozygous AG (Gly123Gly) polymorphism and 31% showed a heterozygous AG polymorphism.
In the development of tumors a growing number of mutations in the genome can be linked to cell adhesion, a function which is of central importance to the cancer development and which ought to be an important component in the development of different invasions type of tumors.
A previous published study in the journal of clinical investigation by Dhawan et al. has shown that in metastatic colon cancer cells express the highest levels of claudin-1 and exhibited the highest rate of cellular mislocalization [13]. The decreased expression of claudin1 has been associated with loss of differentiation in prostatic carcinoma and recommended as an important marker for gastric carcinoma while CLDN 7 has been found to be decreased as invasive ductal carcinomas of breast. In recent research CLDN 7 has been shown to be strongly over expressed in colorectal carcinoma [8,14-17].
The purpose of this study was to see the frequency of polymorphisms in cell adhesion genes CLDN 1 and CLDN 7 among healthy men and women in Orebro County Sweden. The results show that there is a difference in some genotype expression between men and women while other genotypes show very little difference. Also the frequency of these SNPs in colon cancer tumor was very similar to those of the normal population. In the future it would be interesting to see if these cell adhesion genes play a central role in the development of colon cancer and how these polymorphisms affect the prognosis of the disease.
Acknowledgement
Special thanks to Shlear Askari and Sanja Farkas for skillful technical assistance. This study was supported by grants from the Varmland County Council, the Orebro University Hospital, and the research committee of Orebro County Council, the Orebro Medical Research Foundation and Lions Cancer Research Foundation for Medical research in Central Sweden.
Conflict of Interest
We declare that we have no conflict of interest.
References
- Weber CR, Nalle SC, Tretiakova M, Rubin DT, Turner JR (2008) Claudin-1 and claudin-2 expression is elevated in inflammatory bowel disease and may contribute to early neoplastic transformation. Lab Invest 88: 1110-1120.
- French AD, Fiori JL, Camilli TC, Leotlela PD, O'Connell MP, et al. (2009) PKC and PKA phosphorylation affect the subcellular localization of claudin-1 in melanoma cells. Int J Med Sci 6: 93-101.
- Darido C, Buchert M, Pannequin J, Bastide P, Zalzali H, et al. (2008) Defective claudin-7 regulation by Tcf-4 and Sox-9 disrupts the polarity and increases the tumorigenicity of colorectal cancer cells. Cancer Res 68: 4258-4268.
- Fritzsche FR, Oelrich B, Johannsen M, Kristiansen I, Moch H, et al. (2008) Claudin-1 protein expression is a prognostic marker of patient survival in renal cell carcinomas. Clin Cancer Res 14: 7035-7042.
- Victoria HS, Henrik E, Lennart B, Lennart F (2010) Claudin 1 and Claudin 7 Gene Polymorphisms and Protein Derangement are Unrelated to the Growth Pattern and Tumor Volume of Colon Carcinoma. Int J Biomed Sci 6: 96-102.
- Bekker V, O'Brien TR, Chanock S (2009) Population genetics and comparative genetics of CLDN1, a gene involved in hepatitis C virus entry. Hum Hered 67: 206-216.
- Moon KY, Lee PH, Kim BG, Park CS, Leikauf GD, et al. (2015) Claudin 5 in a murine model of allergic asthma: Its implication and response to steroid treatment. J Allergy ClinImmunol 136: 1694-1696.
- Mees ST, Mennigen R, Spieker T, Rijcken E, Senninger N, et al. (2009) Expression of tight and adherens junction proteins in ulcerative colitis associated colorectal carcinoma: upregulation of claudin-1, claudin-3, claudin-4, and beta-catenin. Int J Colorectal Dis 24: 361-368.
- Tokes AM, Kulka J, Paku S, Mathe M, Paska C, et al. (2005) The expression of five different claudins in invasive breast carcinomas: comparison of pT1pN1 and pT1pN0 tumors. Pathol Res Pract 201: 537-544.
- Johnson AH, Frierson HF, Zaika A, Powell SM, Roche J, et al. (2005) Expression of tight-junction protein claudin-7 is an early event in gastric tumorigenesis. Am J pathol 167: 577-584.
- Kaarteenaho R, Merikallio H, Lehtonen S, Harju T, Soini Y (2010) Divergent expression of claudin -1, -3, -4, -5 and -7 in developing human lung. Resp Res 11: 59.
- Hahn-Stromberg V, Askari S, Befekadu R, Matthiessen P, Karlsson S, et al. (2014) Polymorphisms in the CLDN1 and CLDN7 genes are related to differentiation and tumor stage in colon carcinoma. APMIS 122: 636-642.
- Dhawan P, Singh AB, Deane NG, No Y, Shiou SR, et al. (2005) Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest 115: 1765-1776.
- Seo KW, Kwon YK, Kim BH, Kim CI, Chang HS, et al. (2010) Correlation between Claudins Expression and Prognostic Factors in Prostate Cancer. Korean J Urol 51: 239-244.
- Resnick MB, Gavilanez M, Newton E, Konkin T, Bhattacharya B, et al. (2005) Claudin expression in gastric adenocarcinomas: a tissue microarray study with prognostic correlation. Hum Pathol 36: 886-892.
- Myal Y, Leygue E, Blanchard AA (2010) Claudin 1 in breast tumorigenesis: revelation of a possible novel "claudin high" subset of breast cancers. J Biomed Biotechnol 2010: 956897.
- Kinugasa T, Akagi Y, Yoshida T, Ryu Y, Shiratuchi I, et al. (2010) Increased claudin-1 protein expression contributes to tumorigenesis in ulcerative colitis-associated colorectal cancer. Anticancer Res 30: 3181-3186.
Citation: Befekadu R, Hahn-Stromberg V (2016) Polymorphisms in Claudin 1 and Claudin 7 in Healthy Individuals in Sweden. Diagn Pathol Open 1:113. DOI: 10.4172/2476-2024.1000113
Copyright: ©2016 Befekadu R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 10638
- [From(publication date): 5-2016 - Sep 19, 2024]
- Breakdown by view type
- HTML page views: 9958
- PDF downloads: 680