Characterization of Selected Maize Inbred Lines Adapted to Highland Agro-Ecologies of Ethiopia Using Morphological and Molecular Genetic Distances
Received: 11-Nov-2018 / Accepted Date: 05-Feb-2019 / Published Date: 15-Feb-2019 DOI: 10.4172/2329-8863.1000421
Abstract
Characterization of available genetic diversity is a crucial step in effective crop improvement which provide basis for the analysis of combining ability and exploitation of heterosis of inbred lines in hybrid breeding. Characterization involves evaluation of quantitative and qualitative attributes of a given genotypes in order to differentiate their usefulness, structure, genetic variability and relationships among them. The objectives of this study were to characterize elite maize inbred lines adapted to highland agro-ecologies and classify groups of similar inbred lines by means of cluster and principal component analysis based on morpho-agronomic and SNP markers data. A total of twenty-three maize inbred lines of highland breeding department at Ambo Plant Protection Research Center formed the core plant materials in the current experiment for 23 morpho-agronomic traits and SNP markers characterization. The evaluated lines were sown in alpha lattice design 12 × 2. It was planted in two replications at each locations (Ambo and Holetta agricultural research centers). The inbred-lines were highly significantly different for all traits evaluated (p ≤ 0.01). The PCA indicated that the first nine principal components (PCs) with eigen value greater than unity accounted for 83% of the entire diversity among 23 inbred lines for all traits. Cluster analysis performed on the basis morpho-agronomic traits using unweighted paired group method arithmetic averages (UPGMA) grouped 23 tested lines in to five distinct classes and an outlier genotype whereas cluster analysis based on molecular resulted from distance matrix of genetic categorized the entries into four main groups. Five inbred lines (L5, L8, L18, L12 and L7) with comparatively high yielder and other phenotypic characters were selected using the morpho-agronomic traits and SNP based genotyping for cultivar development and germplasm utilization.
Keywords: Elite; Genetic diversity; Genotype; Germplasm; Morphoagronomic; SNP marker
Introduction
Maize (Zea mays L. 2n=20) is a major crop of the world, categorized to the tribe Maydeae of the grass family poaceae. It has great worldwide significance as food, feed and as a source of industrial products [1]. Maize has a vital role in world financial system and market [2]. Apart from manufacturing of mixed feed, maize is a row material to produce corn starch, corn oil and corn syrups [3,4]. Globally, maize is considered as the first most-produced cereal followed by rice and wheat, but in terms of dietary intake, it is third after rice and wheat [5]. It is an important source of protein accounting for up to 60% of the daily human protein supply in sub-Saharan Africa [6]. Phenotypic and molecular characterization of elite highland maize inbred lines is the most vital to determine the existing genetic variability and the relationship among them for further utilization of germplasm in improved cultivar development. Characterization of available genetic diversity is a crucial step in effective crop improvement which provide the basis for the analysis of inbred lines combining ability and exploitation of heterosis. It involves evaluation of quantitative and qualitative attributes of given genotypes in order to differentiate them and determine their usefulness, structure, genetic variability and relationships among them [7,8].
The existing magnitude and nature of genetic variability among genotypes matters the preference of approaches of breeding for genetic improvement of a crop [9]. The probability that two randomly sampled alleles are different is genetic diversity [10]. The distance reflects definite amount of genetic difference present among the genotypes. These measures can be calculated by measuring morphological characteristics and/or using molecular markers. Even though, phenotypic evaluation has useful attribute for grouping inbred lines and populations, these phenotypic traits have limitations in distinguishing variation in highly related genotypes and elite breeding germplasm [11,12] due to genotype by environment interaction (GEI) Advances in molecular technology have produced a shift towards detecting individual difference using molecular marker such as SNP markers [13,14]. The nature and magnitude of genetic variability of every elite maize inbred lines is essential however limited number of highland maize inbred lines are characterized so far because of only certain researches have been conducted for the agro-ecology.
Concomitantly knowledge of magnitude of breeding material variation and associations among each new elite maize germplasm adapted to highland agro-ecology of Ethiopia is essentially significant to identify parents for further breeding in developing improved varieties. A number of previous research reports confirmed the existence of genetic variation among maize inbreds. A reasonable genetic variability was found among maize accessions adapted to highland agro-ecologies of Ethiopia [15,16]. Research reports also indicated large genetic differences among most lines of quality protein maize adapted to the highlands of Ethiopia [17]. On molecular characterization study, high genetic distance among most pairs of maize inbred lines were reported [18]. Maize inbred lines used in the present study were previously developed from Kenya foundation populations and advanced in the highland of Ethiopian maize breeding program at Ambo Plant Protection Research Center. However, they were not characterized to identify the usefulness and prospect how to use them in the breeding program. Therefore, the objectives of this study were to characterize elite maize inbred lines adapted to highland agro-ecologies and classify groups of similar inbred lines by means of cluster and principal component analysis based on morpho-agronomic and SNP markers data.
Materials and Methods
Plant material
A total of twenty-three maize inbred lines (Table 1) of highland breeding department at Ambo Plant Protection Research Center formed the core plant materials in the current experiment for 23 morpho-agronomic traits and SNP markers characterization.
| Code | Pedigree of the inbred lines |
|---|---|
| L1 | TUXCML159 BC2F41-3-7-B-#-# |
| L2 | TUXCML159 BC2F36-2-2-B-#-# |
| L3 | TUXCML159 BC2F36-2-3-B-#-# |
| L4 | TUXCML159 BC2F18-5-7-B-#-# |
| L5 | SADVLACML176 BC2F6-8-3-B-#-# |
| L6 | SADVLACML176 BC2F6-8-6-B-#-# |
| L7 | SADVLACML176 BC2F6-8-11-B-#-# |
| L8 | SADVLACML176 BC2F56-1-1-B-#-# |
| L9 | SADVLACML176 BC2F51-1-9-B-#-# |
| L10 | SADVLACML154 BC2F7-1-1-B-#-# |
| L11 | SADVLA/CML154 BC2F54-4-1-B-# |
| L12 | P502 SRCML 384X176….98-2-1-2 BC2F6-2-4-B-#-# |
| L13 | P502 SRCML 384X176….98-2-1-2 BC2F4-1-3-B-#-# |
| L14 | P502 SRCML 384X176….98-2-1-2 BC2F30-1-6-B-#-# |
| L15 | P502 SRCML 384X176….98-2-1-2 BC2F30-2-2-B-#-# |
| L16 | P502 SRCML 384X176….135-2-2-2 BC2F30-4-2-B-#-# |
| L17 | P502 SRCML 384X176….135-2-2-2 BC2F30-4-4-B-#-# |
| L18 | P502 SRCML 384X176….135-2-2-2 BC2F30-4-9-B-#-# |
| L19 | P502 SRCML 384X176….135-2-2-2 BC2F21-3-4-B-#-# |
| L20 | P502 SRCML 384X176….135-2-2-2 BC2F20-1-4-B-#-# |
| L21 | P502 SRCML 384X176….135-2-2-2 BC2F2-1-1-B-#-# |
| L22 | CML144 |
| L23 | CML159 |
Table 1: Pedigree of inbred lines adapted to highland agro-ecologies of Ethiopia.
Study area description
This study was executed at two highland maize research testing sites: Ambo and Holetta, in the main season of 2014. Ambo is located at west of Addis Ababa at 8°59ꞌ N latitude and 37°51ꞌ E longitudes and an elevation of 2225 meters above sea level (m.a.s.l). It is in wet subhumid highland agro-ecological region of central Ethiopia. The area receives an average annual rainfall of 1115 mm. The rainy season spreads from April to October with maximum rainfall during June to August. The soil type of the experimental site is characterized by clay Vertisoil. Maximum and minimum temperatures of the centre are 26°C and 11°C respectively. Holetta is also located at West of Addis Ababa at 9°03ꞌ N latitude and 38°30ꞌ E longitude with an altitude of 2390 m.a.s.l. The average annual rainfall of the area is 1100 mm. The soil type of this site is characterized by Eutric Nitisol with minimum and maximum temperature is 6°C and 26°C respectively.
Experimental design and field management
The evaluated lines were sown in alpha lattice design 12 × 2. It was planted in two replications at each locations (Ambo and Holetta Agricultural Research Center). The trails were planted during the last week of May 2014 main season in both testing sites. Trials were hand planted at reliable moisture level of the soil to ensure good germination. Each entry was planted with two seeds per hill in a tworowed plot of 3.75 m long and 0.75 m with 0.25 m intra plant distance. After 35 days of germination each entry was thinned to a single seedling per hill to give a plant population of 53,333 ha-1.
Weed was controlled by the application of Primagram-gold 660SC Pre-emergence herbicide, at the rate of three liters per hectare. Fertilizers were applied at both locations, (DAP) was given 69 kg ha-1 of phosphate (P2O5) in the form of diammonium phosphate and onethird of 119 kg nitrogen (N) ha-1 in the form of urea as basal dressing at sowing time. The remaining nitrogen each one-third of 119 kg were side-dressed at 35 days after plant germination and before the stage of tasseling (female flowering time).
Measurements were taken on days to anthesis as the number of days from planting to 50% pollen shed. Anthesis silking interval was calculated as the difference between days to silking and anthesis. Leaf orientation was recorded at flowering. Silk, Stem and Tassel color was recorded on plot basis. Tassel size was recorded as small, medium and large while Tassel peduncle length and Tassel length was measured after milk stage. Leaf length was measured as the length of the leaf from ligule to apex. Leaf width was taken at mid-way along its length. Leaf area represented the area of the upper most ear leaf computed as maximum width × length × 0.75 in centimeter squares.
Number of primary branches on tassel were counted on plant basis. Ear diameter was measured at the mid-section along the ear length. Number of rows per ear was counted as the average number of kernel rows per ear. Number of kernels per row was counted as the average number of kernels per row. Plant height was measured as the distance from ground level to the first tassel branch while ear height was measured as the distance from ground level to the node bearing the uppermost ear.
Ear aspect was scored on 1-5 scale based on visual evaluation of harvested ears for general performance with regard to diseases and uniformity. Overall phenotypic appearance of the plant; where 1=excellent and 5=poor. Length of the ear from the base to tip; it was measured as the average length of 10 randomly sampled ears from each plot. Grain weight from all the ears of each experimental unit was measured and used to calculate grain yield, adjusted to 12.5% moisture content and expressed in ton ha-1.
DNA extraction and genotyping
Plants were raised on plastic trays at Biosciences for Eastern and Central Africa (BecA) hub in Nairobi, Kenya screen-house. A single leaf from each of 10 seedlings per sample were piled collectively, the tips trimmed off and around equal amount of leaf segment cut at once to make a bulk and transferred into 1.2 mL of narrow piece tubes that contained two 4-mm stainless steel grinding balls (SpexCetriPrep, USA).
Genomic DNA was extracted using CetylTrimethyl Ammonium Bromide (CTAB) method [19]. DNA concentration was measured using the Quant-iTTM PicoGreen® dsDNA assay kit (InvitrogenTM, Paisley, UK) and the Tecan Infinite F200 Pro Plate Reader (Grödig, Austria), and normalized to 50 ng/μL. The quality of the extracted DNA was checked by digesting 250 ng of the genomic DNA from 10 randomly selected samples with 3.6 units of ApeKI restriction enzyme (New England Biolabs, Boston, USA) at 75°C for three hours.
DNA samples were shipped to the Genomic Diversity facility at Cornell University for genotyping. DNA samples were genotyped using genotype by sequencing (GBS) [20]. GBS data was generated by the Genomic Diversity Facility, Cornell University using ApeKI as restriction enzyme and 96-plex multiplexing.
Data analysis
Data were collected on 23 morpho-agronomic traits according to maize descriptors of International Board for Plant Genetic Resource (IBPGR) and CIMMYT [21]. Abbreviations, list of traits used in the study and their descriptions are summarized in Tables 2 and 3. Phenotypic data were subjected to analysis using the PROC PRINCOMP of SAS version 9.1.3. [22] and MINTAB [23].
| Traits | Mean ± SE (m) | Range | CV (%) | F test |
|---|---|---|---|---|
| Grain yield | 2.4 ± 0.79 | 1.3-4.04 | 16.29 | ** |
| Anthesis date | 104.33 ± 1.7 | 100-111.5 | 15.8 | ** |
| Ear position | 0.41 ± 0.13 | 0.33-0.53 | 10.49 | ** |
| Ears per plant | 1.13 ± 0.03 | 0.61-1.61 | 14.6 | ** |
| Plant height | 124.8 ± 11.5 | 96.75-147.3 | 20.2 | ** |
| Ear height | 52.25 ± 6.15 | 33.39-65.76 | 7.65 | ** |
| Kernels rows per ear | 12.94 ± 0.84 | 10.66-15.66 | 11.6 | ** |
| Kerlels per row | 23.08 ± 0.81 | 18.16-27.66 | 10.6 | ** |
| Thousand kernel weight | 217.2 ± 0.84 | 142.5-314 | 13.11 | ** |
| Ear diameter | 36.45 ± 0.64 | 29.78-44.15 | 11.9 | ** |
| Leaf length | 66.63 ± 0.8 | 55.6-85.19 | 5.56 | ** |
| Leaf area | 482.47 ± 0.79 | 339.14-632.65 | 12.13 | ** |
| Number of leaves | 13.81 ± 0.87 | 11.52-16.22 | 5.27 | ** |
| Leaf width | 9.567 ± 0.83 | 8.04-11.38 | 7.32 | ** |
| Tassel length | 26.7 ± 0.89 | 20.8-35.5 | 5.92 | ** |
| Tassel peduncle length | 13.9 ± 0.85 | 8.53-18.6 | 12.4 | ** |
| Number of primary branch on tassel | 9.74 ± 0.89 | 5.0-20.0 | 21 | ** |
Table 2: Mean and standard error of the mean [SE (m)], range, coefficient of variation (CV) and F-test of 17 phenotypic characters tested at Ambo and Holetta.
| Inbred lines | L1 | L2 | L3 | L4 | L5 | L6 | L7 | L8 | L9 | L10 | L11 | L12 | L13 | L14 | L15 | L16 | L17 | L18 | L19 | L20 | L21 | L22 | L23 | mean of MD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L1 | -- | 3.24 | 4.29 | 2.94 | 7.63 | 7.51 | 5.66 | 5.02 | 4.97 | 1.03 | 4.55 | 3.97 | 5.08 | 3.15 | 4.5 | 2.86 | 2.88 | 2.58 | 3.41 | 4.19 | 3.04 | 9.59 | 4.69 | 4.40 |
| L2 | 0.3 | -- | 2.9 | 2.81 | 5.91 | 5.45 | 3.91 | 3.74 | 2.98 | 8.08 | 2.73 | 4.4 | 3.74 | 4.18 | 3.47 | 2.05 | 2.52 | 3.71 | 3.69 | 2.97 | 3.33 | 8.03 | 4.28 | 4.04 |
| L3 | 0.31 | 0.21 | -- | 3.44 | 4.26 | 3.55 | 2.94 | 2.32 | 3.11 | 6.42 | 2.54 | 3.68 | 4.96 | 4.85 | 2.1 | 2.36 | 2.21 | 5.52 | 3.93 | 3.97 | 3.37 | 5.55 | 3.97 | 3.75 |
| L4 | 0.31 | 0.31 | 0.31 | -- | 7.1 | 6.09 | 5.34 | 4.75 | 3.39 | 8.6 | 3.12 | 4.27 | 5.19 | 4.21 | 3.77 | 2.22 | 2.82 | 4.33 | 4.84 | 5.01 | 3.41 | 8.2 | 3.25 | 4.73 |
| L5 | 0.34 | 0.34 | 0.34 | 0.34 | -- | 3.59 | 2.79 | 3 | 5.19 | 4.99 | 5.6 | 5.64 | 7.98 | 7.46 | 5.19 | 6.03 | 5.54 | 8.8 | 5.78 | 5.6 | 5.74 | 4.58 | 7.61 | 5.62 |
| L6 | 0.34 | 0.34 | 0.34 | 0.34 | 0.22 | -- | 3.69 | 3.75 | 4.41 | 3.88 | 4.5 | 6.06 | 6.85 | 7.55 | 4.09 | 5.23 | 5.36 | 8.48 | 6.69 | 6.28 | 6.15 | 3.57 | 5.95 | 5.44 |
| L7 | 0.34 | 0.34 | 0.34 | 0.34 | 0.18 | 0.18 | -- | 1.42 | 3.6 | 6.46 | 4.22 | 3.9 | 6.12 | 5.11 | 3.42 | 3.94 | 3.85 | 6.59 | 4.11 | 4.07 | 3.75 | 6.15 | 6.13 | 4.55 |
| L8 | 0.34 | 0.34 | 0.34 | 0.34 | 0.2 | 0.17 | 0.18 | -- | 3.19 | 6.57 | 3.8 | 2.96 | 6.31 | 4.6 | 3.12 | 3.51 | 3.14 | 6.33 | 3.55 | 4.17 | 3.1 | 5.91 | 5.67 | 4.40 |
| L9 | 0.35 | 0.34 | 0.34 | 0.34 | 0.32 | 0.32 | 0.31 | 0.31 | -- | 6.71 | 2.5 | 3.84 | 6.11 | 4.71 | 3.94 | 3.24 | 3.66 | 6.16 | 4.79 | 5.12 | 3.71 | 6.99 | 5.17 | 4.76 |
| L10 | 0.34 | 0.34 | 0.34 | 0.34 | 0.31 | 0.29 | 0.3 | 0.29 | 0.33 | -- | 6.58 | 8.81 | 9.54 | 1.07 | 7.38 | 8.29 | 8.22 | 1.15 | 9.28 | 8.65 | 9.05 | 3.29 | 8.74 | 6.93 |
| L11 | 0.35 | 0.34 | 0.34 | 0.34 | 0.32 | 0.32 | 0.32 | 0.31 | 0.33 | 0.31 | -- | 4.5 | 4.91 | 5.3 | 3.8 | 3.1 | 3.61 | 5.78 | 4.53 | 4.55 | 4.42 | 6.7 | 4.79 | 4.67 |
| L12 | 0.34 | 0.34 | 0.34 | 0.34 | 0.35 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | -- | 7.19 | 2.82 | 4.08 | 3.69 | 3.32 | 5.78 | 3.44 | 5.35 | 2.16 | 7.93 | 5.99 | 4.70 |
| L13 | 0.33 | 0.32 | 0.33 | 0.33 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.3 | -- | 6.38 | 4.65 | 4.14 | 4.91 | 4.06 | 5.73 | 4.08 | 6.18 | 9.34 | 4.86 | 5.43 |
| L14 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.33 | 0.34 | 0.29 | 0.28 | -- | 4.48 | 3.43 | 3.58 | 4.03 | 3.56 | 5.15 | 2.42 | 9.96 | 5.69 | 4.70 |
| L15 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.33 | 0.34 | 0.31 | 0.28 | 0.17 | -- | 2.21 | 2.45 | 5.23 | 4.44 | 4.3 | 3.6 | 6.38 | 3.2 | 3.98 |
| L16 | 0.34 | 0.33 | 0.33 | 0.33 | 0.34 | 0.34 | 0.34 | 0.33 | 0.34 | 0.34 | 0.34 | 0.28 | 0.29 | 0.3 | 0.29 | -- | 1.64 | 3.73 | 3.75 | 3.75 | 2.66 | 7.66 | 3.06 | 3.75 |
| L17 | 0.34 | 0.33 | 0.33 | 0.34 | 0.33 | 0.33 | 0.33 | 0.33 | 0.34 | 0.33 | 0.34 | 0.29 | 0.28 | 0.3 | 0.29 | 0.16 | -- | 4.06 | 3.22 | 3.51 | 2.38 | 7.29 | 3.49 | 3.99 |
| L18 | 0.33 | 0.33 | 0.33 | 0.34 | 0.34 | 0.34 | 0.34 | 0.33 | 0.33 | 0.34 | 0.34 | 0.3 | 0.27 | 0.29 | 0.28 | 0.18 | 0.17 | -- | 4.64 | 4.43 | 4.44 | 1.06 | 5.09 | 3.93 |
| L19 | 0.34 | 0.34 | 0.33 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.32 | 0.31 | 0.32 | 0.32 | 0.29 | 0.3 | 0.3 | -- | 2.94 | 3.34 | 8.67 | 6.2 | 5.29 |
| L20 | 0.34 | 0.33 | 0.34 | 0.34 | 0.33 | 0.33 | 0.33 | 0.34 | 0.34 | 0.34 | 0.34 | 0.29 | 0.27 | 0.29 | 0.29 | 0.28 | 0.28 | 0.28 | 0.31 | -- | 4.44 | 8.42 | 5.71 | 6.19 |
| L21 | 0.34 | 0.33 | 0.33 | 0.34 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.34 | 0.33 | 0.33 | 0.34 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | -- | 8.25 | 4.99 | 6.62 |
| L22 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.34 | 0.33 | 0.33 | 0.34 | 0.33 | 0.34 | 0.34 | 0.34 | 0.34 | 0.33 | 0.33 | -- | 7.77 | 4.85 |
| L23 | 0.27 | 0.3 | 0.28 | 0.32 | 0.33 | 0.32 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.32 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.32 | 0.29 | 0.32 | -- | 4.85 |
| mean of SNP | 0.33 | 0.3 | 0.33 | 0.34 | 0.31 | 0.31 | 0.32 | 0.33 | 0.34 | 0.33 | 0.34 | 0.31 | 0.3 | 0.3 | 0.31 | 0.27 | 0.3 | 0.32 | 0.3 | 0.3 | 0.31 | 0.3 | 0.3 | -- |
Table 3: Genetic distance matrix based on phenotypic traits (above diagonal) and SNP markers (below diagonal) for all pair-wise combinations of 23 inbred lines adapted to highland areas of Ethiopia.
During data analysis entries were considered as fixed factors whereas incomplete blocks within replicates and replications were as random factors. Multivariate analysis such as, principal component (PCA) and cluster analysis were performed. Assessment of the importance and contribution of the morpho-agronomic characters in PCA was done in explaining variation.
Mean data standardization was performed to make certain that all have equal weight in the analysis [24]. The standardized mean values (mean of each trait was subtracted from the data values and then divided by the standard deviation) were used to perform PCA. Euclidean distance matrices in both morpho-agronomic and molecular markers data were used in cluster analysis to determine phenotypic and genotypic interrelationship among the lines.
Correspondence between both the distance matrices was determined by the product moment correlation derived from mantel Z test using (NTSYS-pc version 2.1). Molecular distances in all each pair of inbred lines were calculated using genotyping by sequencing (GBS) analysis pipeline TASSEL version 4.0.28, software [25].
The filtered sequences were aligned to the maize reference genome B73RefGenv1 [26] using the Burrows-Wheeler alignment tool. This procedure provided a total 3, 825 polymorphic SNPs covering all the 10 chromosomes of the maize genome. SNP loci, having not less than 0.05 allele frequency and no missing value, were selected and used in the genetic diversity analysis of the inbred lines.
A dendrogram was constructed from the genetic distance matrix based on genetic dissimilarity using the unweighted pair group method with arithmetic average (UPGMA) and the resulting tree was visualized using MEGA version 6.0.
Results
Across locations variance analysis in phenotypic traits
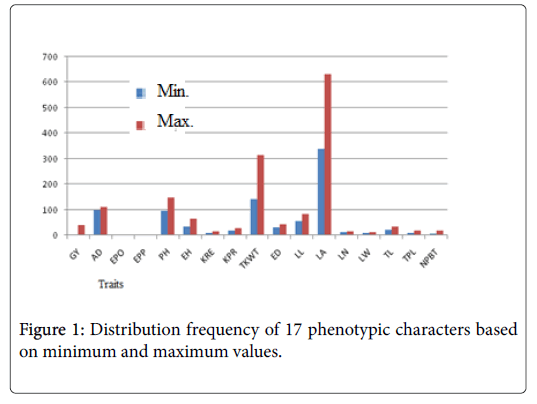
The overall variance analysis showed in the result of current work, Table 2 indicated that the 23 entries were highly significantly different for all traits evaluated (p ≤ 0.01). Substantial variability existed among the lines as was discovered by great variations between minimum and maximum values (Figure 1).
The minimum yield were recorded from L14 (1.13 ha-1) while L5 was the highest 4.04 t ha-1. The male flowering date lies between 100 (L7) to 112 days (L22). Inbred line L17 showed the highest kernel rows per ear (16) while L14 showed the lowest (10). Molecular distance matrix estimates based on SNP markers revealed as Euclidean distance between pair wise comparisons of all the 23 entries varied from 0.16 to 0.35 (Table 3) and the overall average genetic distance was 0.32.
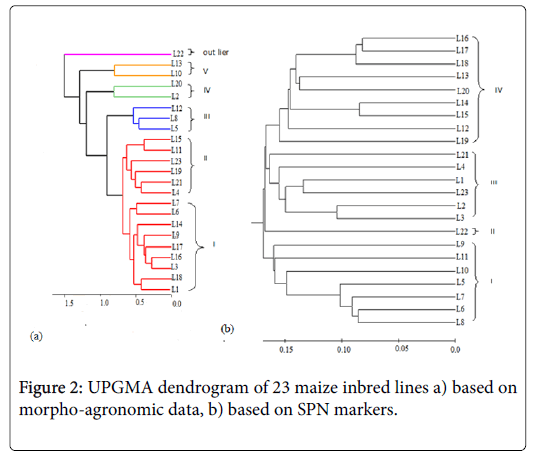
The highest value of genetic distance was recorded for L9 vs. L1, L11 vs. L1 and L12 vs. L5 (0.35), while L17 and L16 showed higher similarity with value 0.16. The UPGMA dendrogram resulted from distance matrix of genetic categorized the entries into four main groups (Figure 2). Cluster I consists of seven inbred lines and cluster II consists of one inbred line while cluster III consists of six inbred lines. Cluster IV consists of a greater number of lines, compared to the other groups.
The association analysis between molecular and morphological genetic distance and the correlation between them as tested by twoway mantel test was low (r=0.20) even though, their correlation result is low in magnitude, their association is positive.
Phenotypic cluster analysis
Cluster analysis performed on the basis of morpho-agronomic traits grouped 23 tested lines in to five distinct classes and an outlier genotype (Figure 2). The number of maize genotypes in a particular cluster ranged from two in cluster (IV and V) to nine in cluster (I).
Cluster I consisted of nine inbred lines characterized by having highest number of leaf, maximum anthesis date, highest ear height, highest thousand kernel weight, highest ear diameter and ear position. Cluster II consisted of six entries in which a traits (leaf orientation and silk color) showed the maximum while their stem color and ears per plant are the smallest. Genotypes in cluster III were designated by the highest leaf area, highest width of leaf, leaf length, high length of tassel, have more ears per plant and high grain yield. Inbred lines in cluster IV were represented by having highest tassel length, number of primary branch on tassel, plant height whereas their grain yield was the lowest. The unique features of genotypes in cluster V were greatest kernels rows per ear, more kernels per row on the contrary, plant aspects and ear aspects are poor (Table 4). Maize line L22 was observed as an outlier against the remaining lines for exhibiting either maximum or minimum value for the characters studied.
| Characters | C-I | C-II | C-III | C-IV | C-V |
|---|---|---|---|---|---|
| Leaf length | 62.22 | 70.12 | 73.67 | 59.58 | 57.58 |
| Leaf area | 432.85 | 517.42 | 578.00 | 365.2 | 362.06 |
| Leaf width | 9.18 | 9.87 | 10.28 | 8.04 | 8.37 |
| Leaf orientation | 1.79 | 2.78 | 2.63 | 2.25 | 2.25 |
| Stem colour | 1.50 | 1.43 | 1.44 | 1.75 | 2.75 |
| Silk colour | 3.42 | 3.55 | 2.69 | 3.50 | 2.50 |
| Number of leaves | 14.19 | 13.99 | 13.23 | 12.45 | 11.97 |
| Anthesis date | 105.33 | 104.25 | 104.50 | 102.00 | 101.00 |
| Tassel size | 3.38 | 3.07 | 2.79 | 3.40 | 3.05 |
| Tassel length | 25.07 | 27.17 | 31.18 | 25.9 | 27 |
| Tassel peduncle length | 13.02 | 14.54 | 13.47 | 18.10 | 14.35 |
| Number of primary branch on tassel | 9.96 | 9.19 | 6.37 | 13.05 | 6.50 |
| Plant aspect | 2.44 | 2.54 | 2.38 | 1.90 | 2.97 |
| Plant height | 121.94 | 127.08 | 127.7 | 129.3 | 96.75 |
| Ear height | 55.18 | 51.80 | 51.95 | 49.16 | 33.39 |
| Number of kernel rows per ear | 12.91 | 12.75 | 13.08 | 12.66 | 14.00 |
| Number of kernels per row | 23.32 | 22.62 | 24.96 | 20.08 | 26.42 |
| Thousand kernel weight | 231.92 | 214.35 | 220.4 | 181.5 | 142.50 |
| Ear diameter | 37.72 | 36.82 | 34.56 | 35.53 | 33.53 |
| Ear position | 0.46 | 0.40 | 0.40 | 0.38 | 0.34 |
| Ear per plant | 1.20 | 1.10 | 1.24 | 1.14 | 1.19 |
| Grain yield | 2.58 | 2.39 | 2.91 | 1.20 | 2.34 |
| Ear aspect | 2.82 | 2.99 | 2.77 | 2.76 | 3.26 |
Table 4: Clusters mean of the 23 morpho-agronomic traits of the 23 inbred lines grouped in five clusters.
Principal component analysis
Agro-morphological variability was explained by a total of 23 components. The PCA indicated that the first nine principal components (PCs) with eigen value greater than unity accounted for 83% of the entire diversity among 23 inbred lines for all traits (Table 5). Under the first PC (17%) the major significant characters explaining the variation were weight of thousand kernel, height of plant, silk color and yield of grain. The 2nd PC, which contributed 13% of the entire variation, resulted predominantly from parameters like length of leaf, area of leaf, ear aspect and size of tassel.
| Variable | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | PC9 |
|---|---|---|---|---|---|---|---|---|---|
| Leaf length | -0.15 | -0.38 | 0.11 | 0.09 | 0 | -0.07 | 0.16 | 0.22 | 0.08 |
| Leaf area | -0.14 | -0.41 | -0.03 | 0.24 | 0.22 | -0.01 | 0.02 | 0.17 | 0.08 |
| Leaf width | -0.05 | -0.18 | -0.26 | 0.19 | 0.41 | 0.07 | -0.22 | 0.07 | 0 |
| Number of leaves | -0.14 | -0.12 | -0.22 | 0.11 | -0.42 | -0.21 | -0.04 | 0.01 | -0.02 |
| Tassel length | -0.07 | -0.22 | 0.16 | 0.25 | 0.21 | 0.2 | -0.28 | 0.23 | 0.09 |
| Tassel peduncle length | -0.02 | 0.05 | 0.51 | 0.06 | 0.07 | 0.05 | 0.06 | -0.2 | -0.12 |
| Number of primary branch on tassel | 0.08 | 0.08 | 0.2 | 0.4 | -0.19 | -0.24 | -0.03 | -0.15 | -0.19 |
| Number of kernel rows per ear | 0.18 | -0.18 | -0.25 | -0.16 | -0.27 | -0.3 | -0.13 | 0.22 | 0.08 |
| Number of kernels per row | 0.19 | -0.15 | 0.14 | -0.31 | 0.02 | -0.24 | 0.01 | 0 | 0.13 |
| Thousand kernel weight | -0.4 | 0.08 | -0.02 | 0.01 | -0.06 | 0.08 | 0.25 | -0.06 | 0.15 |
| Ear diameter | -0.4 | 0.08 | -0.02 | 0.01 | -0.06 | 0.08 | 0.25 | -0.06 | 0.15 |
| Grain yield | 0.27 | -0.23 | 0.1 | -0.2 | 0 | -0.19 | 0.04 | 0.19 | -0.31 |
| Anthesis date | -0.13 | 0.16 | -0.15 | -0.2 | 0.26 | 0.09 | 0.02 | -0.13 | -0.17 |
| Ear per plant | -0.08 | -0.03 | 0.03 | -0.08 | 0.38 | -0.4 | -0.04 | -0.28 | -0.31 |
| Root lodging | 0.18 | 0.2 | 0.24 | 0.16 | -0.11 | 0.1 | -0.21 | -0.04 | 0.39 |
| Plant height | -0.24 | -0.23 | 0.21 | 0.05 | -0.15 | -0.2 | -0.09 | -0.3 | 0.06 |
| Ear height | -0.21 | -0.15 | -0.04 | 0.22 | 0 | -0.26 | -0.14 | -0.36 | 0.28 |
| Ear aspect | -0.2 | 0.35 | 0.01 | 0.02 | 0.05 | -0.25 | -0.18 | 0.33 | 0.05 |
| Plant aspect | 0.19 | -0.04 | -0.12 | -0.05 | 0.11 | 0.09 | -0.3 | -0.3 | 0.05 |
| Stem color | 0.01 | 0.06 | -0.16 | 0.2 | 0.09 | 0.02 | 0.53 | -0.07 | 0.01 |
| Silk color | -0.3 | 0.06 | 0.05 | -0.12 | -0.09 | 0.19 | -0.37 | -0.03 | -0.19 |
| Leaf orientation | 0.11 | -0.01 | 0.3 | 0.26 | -0.01 | 0.05 | 0.1 | 0.32 | -0.28 |
| Kernel type | -0.36 | 0.11 | 0.06 | -0.12 | 0.03 | 0.13 | -0.18 | 0.07 | 0.05 |
| Tassel size | -0.18 | 0.35 | 0.05 | 0.08 | 0.01 | -0.33 | -0.14 | 0.26 | 0.06 |
| Eigenvalues | 4.67 | 3.36 | 3.24 | 2.79 | 2.11 | 1.94 | 1.73 | 1.58 | 1.08 |
| Individual% | 17 | 13 | 12 | 10 | 8 | 7 | 6 | 5.9 | 4 |
| Accumulated variation% | 17 | 30 | 42 | 52 | 60 | 67 | 74 | 79 | 83 |
Table 5: Eigenvectors, eigen values, individual and cumulative percentage of variation explained by the first nine principal components (PC) of 23 highland maize inbred lines.
The 3rd PC, which explained 12% of the whole variation, was governed by characters such as, tassel peduncle length and orientation of leaf. The most discriminating traits under the 4th PC were length of tassel, number of primary branch on tassel, kernels number per row and grain yield production which contributed for 10% of the entire diversity. Kernel row number per ear, grain yield, ears per plant, ear aspect, plant aspect and leaf orientation are important traits contributing to more than one principal components.
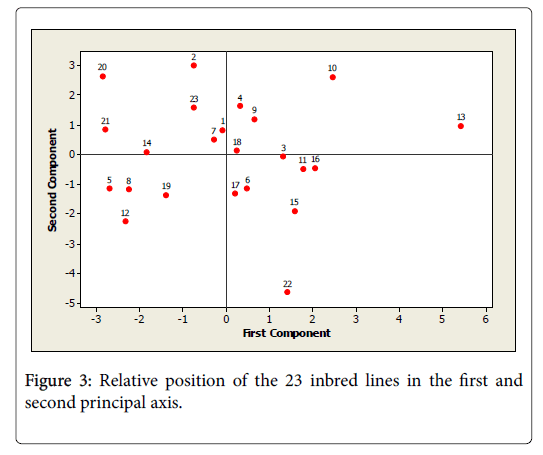
As it is clearly seen in Figure 3, inbred lines in this study were distributed in all the four quadrants of the principal component axis. Line L22, which remained solitary in the former clusters of Figure 2 was still plotted far apart from the other groups in Figure 3. The traits of genotype number 11, 16 and 13 were highly and positively contributed for the first principal component while that of inbred lines number 21, 5, and 8 were highly and negatively contributed to PCI. On the other hand, the traits of inbred lines number 2, 20, 10, 23, 13, 23, 21 and 4 contributed highly and positively to PCII whereas inbred lines 22, 12, 5, 8, 19 and 17 contributed highly but negatively to the same principal components.
Discussion
Characterization of genetic diversity and relationships among elite inbred lines within a given set of maize germplasm in the use of phenotypic and molecular markers is fundamentally useful in crop improvement for understanding how to use the assembled germplasm for further breeding, such as selecting parental lines [27], assigning heterotic groups [7,8] and creating a core set of germplasm [28].
Significant differences were found among the 23 inbred lines studied which indicates the existence of high level of variation for various characters which makes selection possible for improvement of grain yield and other agronomic traits. Both morpho-agronomic and molecular characterizations were effective in distinguishing the germplasm. A group inbred lines from the same group have a tendency of unnecessary contribution to crop genetic improvement therefore, genetically distinctive genotypes should be selected depending on their distance estimated from morpho-agronomic traits and/or marker information [18]. According to the current study, more variations were observed for the agronomical characteristics (LA, TKWT, PH, EH, KPR and GY) than were obtained for the morphological traits (Figure 1).
The broad range means of the inbred lines from morpho-agronomic data implies that there is great potential for the development of hybrids or OPVs using these materials. The wide range in leaf area (339.14-632.65) for instance, suggests the opportunity to develop a cultivar suitable for different purposes, like intercropping and animal feed. Similar results were indicated by earlier researchers on the area studied [29-32]. Morpho-agronomic data revealed the presence of significant genetic variability between the lines studied. In grouping of the inbred lines tested, the dendrogram also indicated the resolution power of morpho-agronomic characters. In most cases of this study lines clustered in the same group are related by their pedigree recordings. The best grain yielder inbred line in cluster III can be utilized in the formation of better yielding cultivars. Whereas, genotypes grouped in cluster II can be sought to breeding for reduced ear height and optimum variety in plant height. Previous reports by, Lucchin et al. [33] grouped 20 Italian flint maize landraces into clusters using agronomic characters and morphological traits. Similarly, 15 morphological characters were classified 62 traditional highland maize accessions in to three groups [15]. Diversity analysis of 45 maize inbred lines, [34] using morphological data grouped effectively closely related inbred lines which is in concurrence with pedigree information. On the other hand, reports consistently point out that the prevailing environmental conditions highly influenced morphological markers [15,34].
The broad range of variability observed between genotypes based on morpho-agronomic traits was furthermore substantiated by principal component analysis which showed that the general variation resulted possibly will not explained by a small number of eigenvectors. The percentage contribution of the first nine PCs to gross genetic variation obtained in the current study was 83%. Classification of entries was majorly due to grain yield and other agronomic characters including, plant height, ear height, ears per plant, thousand kernel weight, number of kernel row per ear, number of kernels per row and ear diameter. PCI represented variables reflecting grain yield and its contributor elements, whereas the supplementary components reflected morphological traits that contributed to yield. Morphoagronomic characters like, length of leaf, leaf numbers, anthesis date, grain yield, aspect of plant, kernels per row on ear, number of primary branch on tassel, width of leaf and stem color that had high values in the first nine components indicated their importance as maize descriptors and could be helpful for differentiation of maize genotypes. Overall, PCA enable to recognize the most crucial characters for classifying the variability within the genotypes. Previous reports Gissa and Kamara et al. [31,35] similarly used PCA to identify traits that explained for majority of the variation among different maize genotypes. The present study found, 83% of the entire variation which is higher than [15] who obtained 71.8%. According to their findings, traits like height of ear placement, kernels per row on ear, weight of thousand kernels, size of tassel, length of leaf are mainly contributing to the entire variability. The outcome of PCA was consistent with the results of cluster analysis, whereby the major differences between clusters were attributed to similar traits that contributed most to the first and second principal components.
The moderate molecular genetic distances exhibited by the inbred lines suggests reasonable genetic diversity, and hence enable a breeder to make crosses and maximize heterosis. The crosses involving parents from most divergent clusters are expected to manifest maximum heterosis and create variability in genetic architecture. Several research reports revealed grouping using the molecular marker UPGMA clustering methods follows pedigree data [32,36]. In this study too, almost all of SNP based clustering was the same with pedigree data and is in agreement with earlier studies [31,37,38]. According to the pedigree data of the current study, lines L1, L2, L3 and L4 showed a close relationship having a common parent TUXCML159 hence, grouped in the same cluster. Similarly, inbred lines L5, L6, L7, L8, L9, L10 and L11 were grouped in the same cluster and have line SADVLACML176 in common, just as lines L12, L13, L14, L15, L16, L17, L18, L19 and L20 were close due to the common parent P502 SRCML 384X176 in their pedigree. In some cases, the diversity analysis based on phenotypic and molecular markers resulted in a similar pattern of grouping of inbred lines however, the association of the two markers was low. Similar to the present result, earlier study [39] used morphological data and SSR markers in assorted maize inbred lines which found a correlation coefficient of 0.23 between the markers. A study on 36 genotypes of sorghum using molecular and morphological data also confirmed the absence of significant relationship between the markers [40]. The cause of low association between morphoagronomic and genetic marker is due to some reasons including distribution of markers in the genome, the magnitude of markers utilized and the nature of the evolutionary mechanism underlying the variation measured can affect the genetic distance estimates [41].
Conclusion
The existence of genetic variability and relationship within each all pairs of lines, obviously represents the uniqueness of the majority of the inbred lines used in the current work. The information resulted from the present experiment is significant in selecting best parent for improved variety development. In general, the two genetic distance measurements are associated positively. SNP markers grouped the genotypes into four using UPGMA clustering algorithm which is in accordance with their pedigree information. The output of the current work point out the strength of SNP markers for diversity analysis and clustering related inbred lines together more effectively than morphoagronomic characters. The resulted information is useful for better understanding of the genetic relationships and efficient use of the identified inbred lines in the breeding programs for the formation of improved hybrid maize varieties adapted to highland agro-ecological zone of Ethiopia.
Acknowledgements
The authors would like to thank the Ethiopian Institute of Agricultural Research (EIAR) for giving the opportunity and International Maize and Wheat Improvement Center (CIMMYT) for granting the scholarship of MSc for the first author, through the Nutritious Maize for Ethiopia (NuME) project. The author acknowledges Kassa Semagn (PhD) and Dagne Wegary (PhD) for supporting molecular work.
References
- White PJ, Johnson LA (2003) Corn: Chemistry and technology, American Association of Cereal Chemists. Inc Minnesota, USA pp: 71-74.
- Vasal SK (2000) The quality protein maize story. Food and Nutrition Bulletin 21: 445-450.
- Forrest AT (2004) Background of US hybrid corn II: Breeding, climate, and food. Crop Sci 44: 370-380.
- Selvi DT, Srimathi P, Senthil N, Ganesan KN (2013) Distinctness, uniformity and stability (DUS) characterization on phenological traits and assessing the diversity of inbreds in maize (Zea mays L.). AJAR 8: 6086-6092.
- Musila RN, Diallo AO, Makumbi D, Njoroge K (2010) Combining ability of early-maturing quality protein maize inbred lines adapted to Eastern Africa. Field Crops Research 119: 231-237.
- Flint-Garcia SA, Buckler ES, Tiffin P, Ersoz E, Springer NM (2009) Heterosis is prevalent for multiple traits in diverse maize germplasm. PloS One 4: e7433.
- Lu Y, Yan J, Guimaraes CT, Taba S, Hao Z, et al. (2009) Molecular characterization of global maize breeding germplasm based on genome-wide single nucleotide polymorphisms. Theoretical and Applied Genetics 120: 93-115.
- Hallauer AR, Miranda JB (1988) Quantitative genetics in maize breeding. Iowa State University Press, Iowa, Ames, USA.
- Hallauer AR, Carena MJ, Miranda Filho JD (2010) Quantitative genetics in maize breeding. Springer Science & Business Media.
- Goodman MM, Brown WL (1988) Races of corn. In: Corn and corn improvement. ASA, CSSA, and SSSA. Madison, WI.
- Bernardo R (1992) Relationship between single-cross performance and molecular marker heterozygosity. Theoretical and Applied Genetics 83: 628-634.
- Hamblin MT, Warburton ML, Buckler ES (2007) Empirical comparison of simple sequence repeats and single nucleotide polymorphisms in assessment of maize diversity and relatedness. PloS One 2: e1367.
- Jones ES, Sullivan H, Bhattramakki D, Smith JSC (2007) A comparison of simple sequence repeat and single nucleotide polymorphism marker technologies for the genotypic analysis of maize (Zea mays L.). Theoretical and Applied Genetics 115: 361-371.
- Beyene Y, Botha AM, Myburg AA (2006) Genetic diversity in traditional Ethiopian highland maize accessions assessed by AFLP markers and morphological traits. Biodiversity & Conservation 15: 2655-2671.
- Ababulgu D, Shimelis H, Laing M, Amelework B (2018) Phenotypic characterization of elite quality protein maize (QPM) inbred lines adapted to tropical highlands and the association studies using SSR markers. Aust J Crop Sci 12: 22.
- Abakemal D, Hussein S, Derera J, Semagn K (2015) Genetic purity and patterns of relationships among tropical highland adapted quality protein and normal maize inbred lines using microsatellite markers. Euphytica 204: 49-61.
- Semagn K, Magorokosho C, Vivek BS, Makumbi D, Beyene Y, et al. (2012) Molecular characterization of diverse CIMMYT maize inbred lines from eastern and southern Africa using single nucleotide polymorphic markers. BMC Genomics 13: 113.
- Semagn K (2014) Leaf tissue sampling and DNA extraction protocols. In molecular plant taxonomy, Humana Press, Totowa, NJ pp: 53-67.
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, et al. (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PloS One 6: e19379.
- IBPGR C (1991) Descriptors for maize. International Maize and Wheat Improvement Center, México City. International Board for Plant Genetic Resources.
- SAS Institute (2004) The SAS system for windows. Version 9.1.3. SAS Inst., Cary, NC.
- MINTAB (2003) Minitab statistical software. Version 14.13.00. MINITAB Inc.
- Manly BF, Alberto JAN (2016) Multivariate statistical methods: A primer. Chapman and Hall/CRC.
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, et al. (2007) TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633-2635.
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, et al. (2009) The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112-1115.
- Barrett BA, Kidwell KK (1998) AFLP-based genetic diversity assessment among wheat cultivars from the Pacific Northwest. Crop Science 38: 1261-1271.
- Mohammadi SA, Prasanna BM (2003) Analysis of genetic diversity in crop plants-salient statistical tools and considerations. Crop Science 43: 1235-1248.
- Hadji TH (2004) Combining ability analysis for yield and yield-related traits in quality protein maize (QPM) inbred lines. MSc Thesis, School of Graduate Studies, Alemaya University, Ethiopia.
- Akande SR, Lamidi GO (2006) Performance of quality protein maize varieties and disease reaction in the derived-savanna agro-ecology of South-West Nigeria. African Journal of Biotechnology.
- Gissa DW (2008) Genotypic variability and combining ability of quality protein maize inbred lines under stress and optimal conditions (Doctoral Dissertation, University of the Free State).
- Pfunde CN (2012) Parent characterization of quality protein maize (Zea mays L.) and combining ability for tolerance to drought stress (Doctoral Dissertation, University of Fort Hare).
- Lucchin M, Barcaccia G, Parrini P (2003) Characterization of a flint maize (Zea mays L. convar. mays) Italian landrace: I. Morpho-phenological and agronomic traits. Genetic Resources and Crop Evolution 50: 315-327.
- Gerdes JT, Tracy WF (1994) Diversity of historically important sweet corn inbreds as estimated by RFLPs, morphology, isozymes, and pedigree. Crop Science 34: 26-33.
- Kamara AY, Kling JG, Menkir A, Ibikunle O (2003) Agronomic performance of maize (Zea mays L.) breeding lines derived from a low nitrogen maize population. J Agr Sci 141: 221-230.
- Warburton ML, Xianchun X, Crossa J, Franco J, Melchinger AE, et al. (2002) Genetic characterization of CIMMYT inbred maize lines and open pollinated populations using large scale fingerprinting methods. Crop Science 42: 1832-1840.
- Patto MV, Satovic Z, Pego S, Fevereiro P (2004) Assessing the genetic diversity of Portuguese maize germplasm using microsatellite markers. Euphytica 137: 63-72.
- Legesse WB, Myburg AA, Pixley VK, Botha MA (2007) Genetic diversity of African maize inbred lines revealed by SSR markers. Hereditas 144: 10-17.
- Karanja J, Amugune NO, Ininda J, Kimatu JN, Danson JW (2009) Microsatellite analysis of the correlation between molecular and morphological traits in assorted maize inbred lines. ‎Afr Crop Sci 2: 1.
- Amelework B, Shimelis H, Laing M (2017) Genetic variation in sorghum as revealed by phenotypic and SSR markers: Implications for combining ability and heterosis for grain yield. Plant Genetic Resources 15: 335-347.
- Powell W, Morgante M, Andre C, Hanafey M, Vogel J, et al. (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Molecular Breeding 2: 225-238.
Citation: Terefe W, Teklewold A, Tesfaye K (2019) Characterization of Selected Maize Inbred Lines Adapted to Highland Agro-Ecologies of Ethiopia Using Morphological and Molecular Genetic Distances. Adv Crop Sci Tech 7: 421. DOI: 10.4172/2329-8863.1000421
Copyright: © 2019 Terefe W, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 5591
- [From(publication date): 0-2019 - Dec 13, 2025]
- Breakdown by view type
- HTML page views: 4482
- PDF downloads: 1109