Comparison of Extremely Halophilic Archaea Isolated from Three Salt Lakes in China
Received: 15-Dec-2017 / Accepted Date: 30-Jan-2018 / Published Date: 07-Feb-2018 DOI: 10.4172/2168-9652.1000227
Abstract
There’re many kinds of extremely halophilic archaea living in salt lakes all over the world and the ecological environment of lakes can affect their archaeal species. To illustrate the relationships between archaeal species and salt lake environment, we isolated and cultured 25 extremely halophilic archaeal strains from three salt lakes in China (Gou Salt Lake, Huama Salt Lake and Yuncheng Salt Lake). According to the 16S rRNA gene sequence analysis and the construction of Neighbor-Joining phylogenetic trees, the isolates were closely related to each other. The most dominant archaea was genus halorubrum (88.0%), followed by genus haloarcula (8.0%) and a small amount of genus halovivax (4.0%). Yuncheng salt lake (YC) had the most abundant number of species. Through the study of the basic information of the three sampling sites, we considered the species of extremely halophilic archaea were related to their living natural conditions of brine. Warm weather and high mineralization degree are likely to be functional components to enrich the species diversity.
Keywords: Salt lakes; Halophilic archaea; 16S rDNA; PCR; Phylogenetic tree
Introduction
Hypersaline salt lakes represent an extreme type of habitat where halophilic prokaryotes are the dominant forms of life [1]. Salt lakes are important mineral resources which contain a variety of salts (mainly NaCl). The formation of salt lake is closely related with natural conditions. In arid or semi-arid climate, when the evaporation of the lake is more than its resupply, the salts constantly concentrate and various elements in water become saturated or supersaturated [2]. Under strong evaporation, water disappears while salts are piling up, which creates salt lakes.
Extremely halophilic archaea (order Halobacteriales, family Halobacteriaceae, phylum Euryarchaeota, domain archaea) inhabit hypersaline regions [3,4]. They could survive extreme conditions such as high salinity, high temperatures, and neutral to alkaline pH, low water activity, and extreme gamma radiations [5]. People have proven that Na+ can maintain cell membrane, cell wall structure and functions to prevent lysis [6]. Na+ can also support the active transport system of amino acid and sugar [7]. Halophilic archaea mainly grow in salt lakes, salt marshes, Dead Sea, the saltern and other environments. They play an important role in the biogeochemistry of carbon and phosphorus in salt lake environments [8] and some halophiles have been shown to degrade organic compounds [9,10] such as pesticides and crude oil [11] for potential use in bioremediation studies and applications [12]. Moreover, halophiles may also have potential uses as biocontrol agents against certain pathogenic fungi [13,14]. Salt lakes are interesting model systems for studies on microbial diversity and ecosystem functions in extreme environments which are highly desirable due to their potential applications [15].
In this study, the extremely halophilic archaea were cultured from three salt lakes of China that were collected: Gou salt lake (GC), Huama salt lake (HMC) and Yuncheng salt lake (YC) respectively. We used phylogenetic analysis to research their compositions and the dominant halophilic archaea. The aim of this study is to find how the different ecological environment of the salt lakes affects the extremely halophilic archaeal species.
Methods
Sampling sites
The water samples were collected of the top 10 cm layer from three Chinese salt lakes in June 2012: Gou salt lake (GC) and Huama salt lake (HMC) are located in Shanxi province. Yuncheng salt lake (YC) is located in Shanxi province, China. After being sieved through microporous membranes of whose pore size is 0.22 μm. The sample was stored at 4°C before the microbial analysis.
Isolation and purification
In order to select extremely halophilic archaea, we chose high salt medium. The ingredients are: Yeast extract (Oxoid) 10 g, Peptone 7.5 g, Trisodium citrate 3 g, NaCl 200 g, KCl 2 g, MgSO4.7H2O 10 g, H2O 1000 ml, agar powder 20 g (solid medium), pH 7.5, 121°C, sterilized 25 min, for using later.
Saltern samples were diluted with dilution gradients of 10-1, 10-2, 10-3, 10-4, 10-5 and 10-6. Then extracted 100 μl diluted liquid to solid MGM medium plate for a 10 to 15 days spread plate cultivation under the temperature of 37°C. The cultivated archaea were then streak cultivated to get a single colony. Archaea from the single colony were streak cultivated again to obtain purified strains [16,17].
DNA extraction, PCR and phylogenetic tree construction
DNA were extracted according to the instruction of Bacterial genomic DNA Extraction Kit by Shanghai Sangon Biological Engineering Technology & Services Co., Ltd. Sequences of primers were the consensus primer of Halophilic archaea--H21F/H1540R [18]. Forward primer: 5'-ATTCCGGTTGATCCTGCCGG-3' and reverse primer: 5'-AGGAGGTGATCCAGCCGCAG-3' were used for amplifying the archaeal 16S rRNA genes. The template 3 μl, every primer 1 μl, dNTP 1.5 μl, 10x PCR buffer 5 μl, MgCl2 4 μl, Taq enzyme 0.3 μl, steam sterilization water to 50 μl by using the PCR Gene Amp 9700 (Applied Biosystems, Foster City, CA, USA). The PCR thermal cycling conditions were as follows: 95°C predenaturation for 5 min, 95°C denaturation for 1 min, 50°C annealing for 1 min, 72°C extension for 1 min 30 s, 35 cycles at 72°C for 10 min, 4°C for preservation. The PCR product was purified by UNIQ-10 column DNA gel extraction kit (Shanghai Sangon Biological Engineering Technology & Services Co., Ltd.) and sequenced by Shanghai Sangon Biological Engineering Technology & Services Co., Ltd.
The 16S rRNA gene sequences were compared with the other strains from the EzTaxon Server database. ClustalX software was used to determine the matching alignment of multiple sequences. Phylogenetic trees were constructed using the Neighbor-Joining method under MEGA5.0 and appraised by using bootstrap value with 1,000 repeats [19].
Results
Descriptions of sampling sites
The basic information about three salt lakes was obtained from ‘Chinese Salt Lakes’ writen by Zheng [20] and we listed them in Table 1. It illustrated that HMC, GC and YC are located in Chinese inland and their climates were dry. What’s more, HMC was an alkaline lake while YC was acidic lake. HMC and GC had the highest altitude and the lowest temperature.
| Sites | pH | Annual Average Temperature(°) | Mineralization degree (g/L) | Altitude (m) | Coordinates |
|---|---|---|---|---|---|
| GC | 7.3 | 7 | 156.77 | 1303 | 107°31′25″E, 37°45′30″N |
| HMC | 8 | 7 | 108.5 | 1310 | 107°30′15″E, 37°41′20″N |
| YC | 5.5 | 14 | 236.4 | 311.77 | 107°31′-111°03′E, 34°56′15″-35°02′30″N |
Table 1: Basic information for the sampling sites.
Also, we listed the selected general physical and chemical properties of sampling sites according to reference book in Table 2 [20]. The cationic detection method is atomic absorption, and the anion’s is ion chromatography. The three study sites showed obvious differences with regard to the inorganic ions concentrations (Table 2). In general, Na+ was the most abundant cation among the four samples followed by Mg2+. The most abundant anion of the samples was Cl- except YC. It’s worth noting that the concentration of SO4 2- was the highest anion in YC.
| Sites | Na+ (mg/L) | K+ (mg/L) | Mg2+ (mg/L) | Ca2+ (mg/L) | Cl- (mg/L) | SO42- (mg/L) |
|---|---|---|---|---|---|---|
| GC | 55025 | 189 | 3731 | 434 | 96309 | 20 |
| HMC | 28740 | 157 | 8710 | 248 | 69998 | 13 |
| YC | 32671 | 835 | 28284 | 218 | 53220 | 119364 |
Table 2: General physical and chemical properties for the sampling sites.
Diversity of halophilic archaea
We isolated and enriched 25 extremely halophilic archaeal strains from three samples, 4 of GC, 2 of HMC and 19 of YC respectively. After amplifying of PCR, there’re 1400 bp-1500 bp lengths of each sample’s 16S rDNA for phylogenetic analyses. Three phylogenetic trees were constructed (Figures 1-3) and Methanosarcina lacustera was chosen as the out-group.
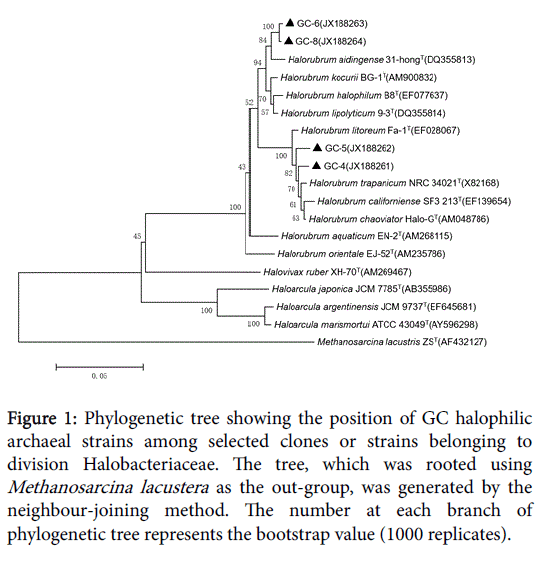
Figure 1: Phylogenetic tree showing the position of GC halophilic archaeal strains among selected clones or strains belonging to division Halobacteriaceae. The tree, which was rooted using Methanosarcina lacustera as the out-group, was generated by the neighbour-joining method. The number at each branch of phylogenetic tree represents the bootstrap value (1000 replicates).
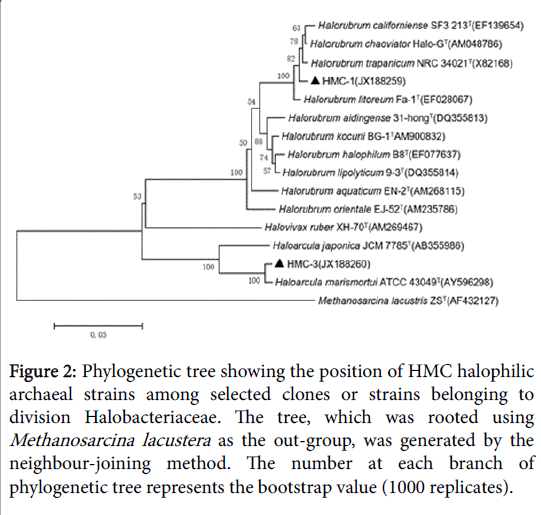
Figure 2: Phylogenetic tree showing the position of HMC halophilic archaeal strains among selected clones or strains belonging to division Halobacteriaceae. The tree, which was rooted using Methanosarcina lacustera as the out-group, was generated by the neighbour-joining method. The number at each branch of phylogenetic tree represents the bootstrap value (1000 replicates).
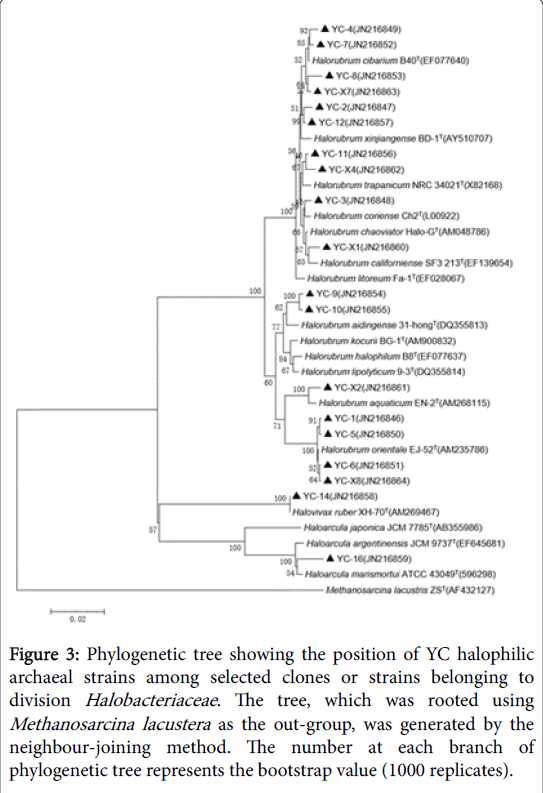
Figure 3: Phylogenetic tree showing the position of YC halophilic archaeal strains among selected clones or strains belonging to division Halobacteriaceae. The tree, which was rooted using Methanosarcina lacustera as the out-group, was generated by the neighbour-joining method. The number at each branch of phylogenetic tree represents the bootstrap value (1000 replicates).
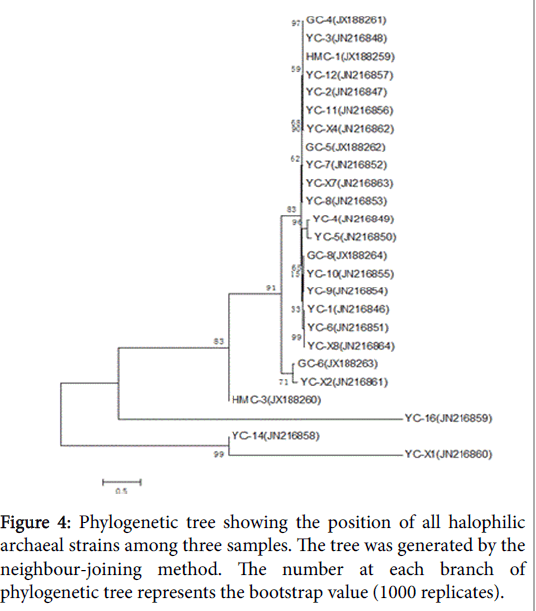
There is another phylogenetic tree that compared the species of three samples (Figure 4). Comparison of the sample sequences with other halobacterial sequences in the database indicated that they were most closely related to the other species of the genus Halorubrum.
GC-6 was highly homologous to GC-8which belonged to genus Halorubrum and clustered into one branch. There were two different strains in HMC but they belonged to different genus. HMC-1 was one species of genus Halorubrum while HMC-3 was confirmed to Haloarcula marismortui . YC had the most abundant diversity of strains, which harbored representatives of all the three genera. Much of them belonged to genus Halorubrum and they’re scattered throughout the phylogenetic tree, representing different species. In addition, YC-14 was same with Halovivax ruber and YC-16 was a species of genus Haloarcula.
When comparing the three samples together, most of them are homologous with each other, except for the following four: HMC-3, YC-14, YC-16 and YC-X1. YC-14 was more homologous to YC-16 by contrast.
From the perspective of all 25 samples, there were 22 of them belonged to genus Halorubrum (88.0%), suggesting that it may be the dominant genus in salt lakes. Moreover, genus Haloarcula (8.0%) and Halovivax (4.0%) also appeared in our study, but only a few, so they did not account for the advantage in salt lakes. This study didn’t found any unknown or novel archaea, but the species diversity in the salt lake is great and needed to be further investigated and studied.
Discussion
YC had the most abundant strains among the three. Previous studies have shown that pH correlated with prokaryotic distributions in highly saline environments [21]. We found that Yuncheng salt lake was an acidic lake, but alkaline environments were more suitable for the survival of halophilic bacteria [22]. The high archaeal diversity in YC could be due to the fact that some kinds of halorubrum can tolerate the relative low pH and there have been researches reported that some halorubrum species can live in pH 5.5-9.5 [23]. The contrary discovery indicated that there could be no direct correlation between pH and halophilic archaeal diversity and there were a lot of factors that could affect the results.
What’s more, mineralization degree, which means the amount of salts in water, can also have impact on species. It had been proved that high level of mineralization degree was necessary for the survival of halobacteria [24]. As the amount of salt increases, the diversity enriches too. Among the three sample sites, the mineralization degree of YC was much higher than the others, which may result in their abundant species.
The locations of Huama salt lake and Gou salt lake are very close. They have the same climate and natural environment. In this study, we only collected and cultured 4 strains in GC and 2 in HMC. These four strains in GC were clustered into one branch, which meant they had very close genetic relationships. However, the two strains in HMC belonged to different genus. We didn’t culture many clones in our media due to the high salt concentration (NaCl 20%). Some studies considered that optimal salinity for moderately halophilic bacteria growth was 3%-15% NaCl [25,26]. Also, their diversity was affected by temperature. The temperature of the two samples was relatively low, which is not conducive to the growth of microorganisms.
The extreme halophilic archaea of the three salt lakes have been shown to be phylogenetically distinct, a result which is consistent with both the natural environmental conditions and the brine physicochemical properties. It’s clear that the warm weather and high mineralization degree are likely to be functional components to enrich the species diversity. In addition, halorubrum was the dominant genus in salt lakes. Although the temperatures were low at these three sample sites, archaea grew at 37°C, they are mostly resistant to high temperatures due to the properties of halophilic archaea and some extreme halophilic archaea are lightly thermophilic, so we don’t think this temperature could eliminate some archival species in isolated samples and that couldn’t reduce diversity of species in samples. However, other factors that can affect the diversity of halophilic archaea as well, and the specific mechanism of these factors are needed to be further investigated and studied.
Conclusion
There are many kinds of extremely halophilic archaea living in salt lakes all over the world and the ecological environment of lakes can affect their archaeal species. Through the study of the basic information of the three sampling sites, we considered the species of extremely halophilic archaea were related to their living natural conditions of brine. Warm weather and high mineralization degree are likely to be functional components to enrich the species diversity. In addition, Halorubrum was the dominant genus in salt lakes.
Acknowledgement
The authors gratefully acknowledge the financial support given by Priority Academic Program Development (PAPD). The authors are also grateful to Shanghai Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China) for their help in sample sequencing.
References
- Oren A (2002) Halophilic microorganisms and their environments. In: Joseph Seckbach (5th eds) Kluwer Academic Publisher, New York, pp: 151-152.
- Wang Q, Zhang WZ, Yue J, Lee CG (2002) Tremendous change of the earth surface system and tectonic setting of salt-lake formation in Yuncheng Basin since 7.1 Ma. Science China 45: 110-122.
- Ma Y, Galinski EA, Grant WD, Oren A (2010) Halophiles 2010: Life in saline environment. Appl Environ Microbiol 76: 6971-6981.
- Mani K, Salgaonkar BB, Braganca JM (2012) Culturable halophilic archaeal at the initial and crystallization stages of salt production in a natural solar saltern of Goa, India. Aquatic Biosystems 8: 15.
- Kottemann M, Kish A, Iloanusi C, Bjork S (2005) Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation. Extremophiles 9: 219-227.
- Oren A (1994) The ecology of the extremely halophilic archaeal. FEMS Microbiol Rev  13: 415-439.
- Von Weymarn N, Nyyssölä A, Reinikainen T, Leisola M (2001) Improved osmotolerance of recombinant Escherichia coli by de novo glycine betaine biosynthesis. Appl Microbiol Biotechnol 55: 214-218.
- López-López A, Yarza P, Richter M, Suárez-Suárez A (2010) Extremely halophilic microbial communities in anaerobic sediments from a solar saltern. Environ Microbiol Rep 2: 258-271.
- Le Borgne S, Paniagua D, Vazquez-Duhalt R (2008) Biodegradation of organic pollutants by halophilic bacteria and archaea. J Mol Microbiol Biotechnol 15: 74-92.
- Oncescu T, Oancea P, Enache M, Popescu G (2007) Halophilic bacteria are able to decontaminate dichlorvos, a pesticide, from saline environments. Cent Eur J Biol 2: 563-573.
- Al-Mailem DM, Sorkhoh NA, Al-Awadhi H, Eliyas M (2010) Biodegradation of crude oil and pure hydrocarbons by extreme halophilic archaea from hypersaline coasts of the Arabian Gulf. Extremophiles 14: 321-328.
- Zhao BS, Wang H, Mao XW (2009) Biodegradation of phenanthrene by a halophilic bacterial consortium under aerobic conditions. Curr Microbiol 58: 205-210.
- Chen L, Wang G, Bu T, Zhang Y (2010) Phylogenetic analysis and screening of antimicrobial and cytotoxic activities of moderately halophilic bacteria isolated from the Weihai Solar Saltern (China). World J Microbiol Biotechnol 26: 879-888.
- Sadfiz N, Essghaier B, Hajlaoui MR, Fardeau ML (2008) Ability of moderately halophilic bacteria to control grey mould disease on tomato fruits. J Phytopathol 156: 42-52.
- Jose PA, Santhi VS, Jebakumar SRD (2011)Phylogenetic-affiliation, antimicrobial potential and PKS gene sequence analysis of moderately halophilic Streptomyces sp. inhabiting an Indian saltpan. J Basic Microbiol 51: 348-356.
- Xu XW, Wu M, Huang WD (2005) Isolation and characterization of a novel strain of Natrinema containing a bop gene. J Zhejiang Univ Sci 6B: 142-146.
- Gerber GE, Anderegg RJ, Herlihy WC (1982) Partial primary structure of bacteriorhodopsin: Sequencing methods for membrane proteins. Proc Natl Acad Sci U S A 88: 56-74.
- Ventosa A, Gutierrez MC, Kamekura M, Oren A (2004) Taxonomic study of Halorubrum distributum and proposal of Halorubrum terrestre sp. nov. Int J Syst Evol Microbiol 54: 389-392.
- Felsenstein J (1985) Contidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783-791.
- Salgaonkar BB, Das D, Bragança JM (2015) Resistance of extremely halophilic archaeal to zinc and zinc oxide nanoparticles. Appl Nanosci 6: 251-258.
- Zhang WJ, Cui HL (2014) Halorubrum salinum sp. Nov., isolated from a marine solar saltern. Arch Microbiol. 196: 395-400.
- Yin S, Wang Z, Xu JQ, Xu WM (2015) Halorubrum rutilum sp. Nov. isolated from a marine solar saltern. Arch Microbiol 197: 1159-1164.
- Rivadeneyra MA, Delgado R, Párraga J, Ramos-Cormenza A (2006) Precipitation of minerals by 22 species of moderately halophilic bacteria in artificial marine salts media: Influence of salt concentration. Folia Microbiol (Praha) 51: 445-453.
- Van de Vossenberg JL, Driessen AJ, Grant WD (1999)Lipid membranes from halophilic and alkali-halophilic archaeal have a low H+ and Na+ permeability at high salt concentration. Extremophiles 3: 253-257.
- Kushner DJ (1978) Life in high salt and solute concentrations: Halophilic bacteria. microbial life in extreme environments 1978: 317-368.
Citation: Wen H, Yin X, Gao B, Su S, Xu X (2018) Comparison of Extremely Halophilic Archaea Isolated from Three Salt Lakes in China. Biochem Physiol 7: 227 DOI: 10.4172/2168-9652.1000227
Copyright: ©2018 Wen H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 5444
- [From(publication date): 0-2018 - Sep 01, 2025]
- Breakdown by view type
- HTML page views: 4514
- PDF downloads: 930