Research Article Open Access
Degradation of Profenofos and λ-Cyhalothrin Using Endogenous Bacterial Isolates and Detection of the Responsible Genes
Reda R Abdullah1,2*, Sherif B Abdel Ghani3,4 and Noha A Sukar5
1Plant Protection Research Institute, Agriculture Research Center, Giza, Egypt
2Promising Research Center in Biological Control and Agricultural Information, Qassim University, Saudi Arabia
3Plant Production and Protection Department, College of Agriculture and Veterinary medicine, Qassim University, 51452 Buraydah, PO Box 6622, Al-Qassim, Saudi Arabia
4Department of Plant Protection, Faculty of Agriculture, Ain Shams University, PO Box 68 Hadayek Shoubra, 11241 Cairo, Egypt
5Department of Biological and Environmental Science, Al Azhar University, Tanta, Egypt
- *Corresponding Author:
- Reda R Abdullah
Promising Research Center in Biological Control and
Agricultural Information, Qassim University, Saudi Arabia
Tel: +966533800146
E-mail: redakenany@yahoo.com
Received date: April 04, 2016; Accepted date: July 14, 2016; Published date: July 15, 2016
Citation: Abdullah RR, Ghani SBA, Sukar NA (2016) Degradation of Profenofos and λ-Cyhalothrin Using Endogenous Bacterial Isolates and Detection of the Responsible Genes. J Bioremed Biodeg 7:360. doi:10.4172/2155-6199.1000360
Copyright: © 2016 Abdullah RR, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
An effective profenofos and λ-cyhalothrin degrading bacterium (strain DB17) was isolated from soil samples collected from Qassim region, Saudi Arabia. Based on the results of phylogenetic similarity of 16SrDNA gene sequences, strain DB17 was identified to be Pseudomonas putida. The isolate utilized profenofos and λ-cyhalothrin as the sole source of carbon for its growth. Analytical method was developed and validated for the determination of profenofos and λ-cyhalothrin in bacterial medium to monitor the intended biodegradation of both compounds. The inoculation of isolate DB17 109 and 1011 cells/ml to mineral salt medium supplemented by 100 mg/liter of profenofos and λ-cyhalothrin resulted in a higher degradation rate than in non-inoculated medium. The genes encoding organophosphorus hydrolase (mpd and opd) and pyrethroid-degrading esterase gene (pytY) were cloned using a PCR cloning strategy. The obtained results highlight the potential of this bacterium to be used in the cleanup of pesticide contaminated sites.
Keywords
Biodegradation; Bacteria; Profenofos; λ - Cyhalothrin; Responsible genes for degradation
Introduction
Crop production of Saudi Arabia is mainly cultivated under greenhouses because of the dominating arid climate of Saudi desert. Plants grown under greenhouse are more prone to infestation and infection by different types of pests. Pesticides usage is a pivotal strategy for pest control. Crop production under greenhouse conditions requires more pesticide usage. The excessive use of pesticides has led to accumulation of these pesticides' residues in the environment, presenting a substantial health hazard to human [1]. Using bacteria for the degradation of pesticides is an efficient approach. Isolation of indigenous bacteria capable of degrading pesticides provides an environmentally friendly way for pesticide detoxification [2].
Degradation by microbes depends not only on the presence of degradative enzymes, but also on a wide range of environmental parameters such as ambient temperature, nutrients status and pH. Also, pesticide concentration is a limiting factor [3]. All the above mentioned factors require investigation to assess its impact on activity and microbial population.
"Catabolic plasmids" are the plasmids of bacteria that carry the responsible genes for pesticide degradation [4]. Depending on the carried genes, the organism is able to use particular compound, hence degrade it [5]. Many catabolic plasmids have been reported in numerous species of Pseudomonas, Arthrobacter, Actinobacter, Alcaligenes and Flavobacterium [4].
Contaminated environments have participated with time in the evolution of endogenous microbial populations. Therefore, contaminated sites are the most appropriate locations for the isolation of degradative bacterial strains which are able to degrade contaminating compounds and also the related ones [6]. Decontamination of polluted areas is a need of the hour. The conventional means (physio chemical methods) for soil decontamination are rather expensive, labor intensive less efficient and may affect soil properties as well. Thus, biodegradation is becoming a method of choice for the remediation of polluted sites.
The Objective of this study was investigating the biodegradation of profenofos and λ-cyhalothrin using isolated indigenous bacteria from Qassim region, Saudi Arabia. Also, studying the effect of temperature, incubation time, bacterial inoculum and pesticide concentrations on the efficiency of degradation. Also, a special focus was given to development and validation of the analytical method. In addition, detection of the genes which are responsible for the degradation of tested compounds.
Materials and Methods
Chemicals and reagents
Two insecticides were selected for the current study. Log P, chemical group, toxicity and chromatographic parameters of the tested pesticides are summarized in Table 1. Chemical structure of profenofos and λ-cyhalothrin is illustrated in Figure 1. Profenofos and λ-cyhalothrin standards were obtained from Sigma–Aldrich (Germany). The stock solution was prepared in toluene and working solutions were prepared in acetonitrile, HPLC grade, obtained from BDH, UK. MgSO4 (anhydrous) and NaCl were purchased from Merck, Germany. Mineral salt medium (MSM) components salts (K2HPO4, K2HPO4, (NH4)2SO4, MgSO4.7H2O, CaCO3, FeCl2.4H2O) were all purchased from LOBA Chemie Pvt. Ltd. (India).
| Pesticide | LogP | Chemical group | Toxicity class (WHO) | Retention time (Rt) | Monitored ions (m/z) |
|---|---|---|---|---|---|
| Profenofos | 4.44 | Organophosphorus | II | 6.16 | 139, 206, 339 |
| λ-Cyhalothrin | 7 | Pyrethroid | II | 8.48 | 181, 197, 208 |
Table 1: Log P, chemical group, toxicity and chromatographic parameters of the tested pesticides.
Soil sample collection for enrichment studies
Five soil samples were utilized in this study for the isolation of profenofos and λ- cyhalothrin degrading microorganisms. Soil samples were collected from different sites with a history of both pesticides applications in Qassim region, Saudi Arabia. The collected soil samples were taken and stored in plastic bags at 4°C until isolation process [7].
Isolation of profenofos and λ- cyhalothrin degrading bacteria
Ten grams of soil sample was placed in a 250 ml conical flask containing 100 ml mineral salt medium MSM ((g L-1 deionized water) K2HPO4 0.225; K2HPO4 0.225; (NH4)2SO4 0.225; MgSO4 7H2O 0.05; CaCO3 0.005; and FeCl2 4H2O 0.005; pH- 7.2) and incubated at 30°C in a rotary shaker at 150 rpm for two days. The flasks were then left for a few hours to allow the soil particles to settle, and the suspension containing microorganisms was then used to inoculate fresh sterilized MSM containing 100 mg L-1 of one of the tested pesticides and incubated for 48 hr. at 28°C. In order to obtain pure cultures of bacteria, 5 ml aliquots of enrichment cultures were centrifuged at 3000 rpm for 5 min and the cell pellets were re-suspended in 2 ml sterile medium. Aliquots of this suspension were spread on MSM profenofos and λ- cyhalothrin agar plates. Inoculated plates were incubated at 28°C and discrete colonies were isolated. Strong resulting colonies in growth were repeatedly subcultured using the same procedures above to confirm their utilizing ability. All colonies were transferred into fresh sterile medium to obtain a pure culture [7]. Eighteen isolates of strong growth on the pesticide agar medium were selected for further investigation. Each isolate was cultured in carbon deficient broth medium containing 100 mg L-1 of one of the tested pesticides. Cultures were stirred for 3 days at 30°C before a sample was taken from each treatment. Samples were subjected for pesticide analysis.
Analytical method development and validation
An analytical method was developed using modified QuEChERS method for sample preparation followed by GC/MS for the determination step.
A house made QuEChERS method was followed in sample extraction [8]. Two and half ml sample of bacterial cultures were filtered through a micropore filter 0.2 μm (Whatman®, Inc., USA) into 15 ml centrifuge tubes. Five ml acetonitrile (1% acetic acid) was added and the tubes were shaken for mixing. Sample tubes were kept in the freezer for 15 min before adding 1 g MgSO4 and 0.25 g NaCl and tubes were tightly capped and shaken vigorously by hand for 1 min. then tubes were centrifuged (Centra MP4R centrifuge, IEC, MA USA) under cooling for 5 min at 4400 rpm. 1 ml aliquot was taken into 1.5 ml screw capped vial contains 150 mg MgSO4 and 25 mg PSA (primary secondary amine) and hand shaken for 1 min before centrifugation for 5 min at 4400 rpm. 500 μl of the extract was transferred to 1.5 ml vial for GC/MS residue analysis. Extracts were diluted when required using blank matrix extracts.
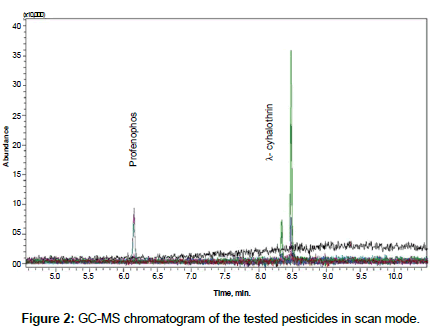
A Shimadzu GC/MS-Q2010 Ultra (Shimadzu Corporation, Japan) was used for determination of pesticide residues. The instrument is equipped with Shimadzu AOC-20i autosampler and split/splitless injector in the split mode at 260°C. RXI-SIL 5 MS fused silica column, 30 m length × 0.25 mm i.d. × 0.25 μm film thickness, sourced from Restek, USA, was used. Oven temperature program started at 200°C with hold time 2 minutes, than raised at 15°C min-1 to 290°C and held at final temperature for 4 minutes. Injected volume was 1 μl. The MS detector was run in the scan mode (from 50 m/z to 500 m/z). After that SIM mode was utilized for residue determination. Three ions were selected for each pesticide. 139 m/z and 181 m/z were utilized as quantification ions for profenofos and λ- cyhalothrin, respectively. 206 and 339 m/z were used as qualifiers for profenofos; 197 and 208 m/z were use as qualifier ions for λ- cyhalothrin. Injector was in the split mode. Injector temperature was 260°C, transfer line was 285°C and ion source was set at 235°C. Carrier gas, helium, flow was constant at 1 ml/ min. GC-MS chromatogram of the tested pesticide is shown in Figure 2.
Validation parameters of the analytical method were investigated to assure the quality of the analytical results. Method accuracy was assessed by extraction and determination of tested compounds concentrations in spiked matrix of bacterial media at different levels i.e., 0.1, 1.0 and 10 mgL-1 of the bacterial media. Method repeatability was calculated as RSD % of the recovery studies. Concerning linearity (R2), 5 concentrations of each compound i.e., 0.05, 0.1, 0.25, 0.5 and 1.0 mg L-1 were prepared. Each concentration was injected three times (n=3) and average area was used for calibration curve development. Limit of detection (LOD) and limit of quantification (LOQ) were calculated from the calibration curve data. In order to compensate for matrix effect, matrix matched standard approach was utilized in pesticide residues quantification.
Biodegradation studies
The bacterial cells of best isolate in degradation process were precultured in LB medium (g L-1 deionized water; NaCl 10.0; Tryptone 10.0; Yeast extract 5.0; pH- 7.2) at 30°C with 150 rpm shaking, harvested by centrifugation at 6000 rpm for 10 min and was washed three times with sterilized mineral salts medium then was quantified by the dilution plate count technique. The obtained bacterial cells were used in biodegradation studies according to the modified method of Ref. [9].
Degradation studies were carried out in liquid mineral salts medium (MSM) supplemented with profenofos and λ- cyhalothrin. These were assayed with batch cultures in MSM by the best isolate which was selected from the first screening of degradation. Effects of incubation temperature, incubation period, pesticide concentration and inoculums density also were studied. Cultures were run in triplicate to ensure accuracy [9]. Degradation of tested pesticides in broth media without inoculation was also investigated (Control treatment). Degradation percent was calculated as the reduction of pesticide concentration after subtraction of the control degradation percent.
Identification of the bacterial isolates by 16S rDNA
The genomic DNA was extracted by phenol-chloroform method according to Ref. [10] and amplified with the 16S rDNA specific primer; P1F (5'-AGA GTT TGA TCC TGG CTC AG-3') and P2R (5'- TGA CTG ACT GAG GCT ACC TG-3') (Macrogen Company, Korea). The thermal cycling conditions were as follows: one cycle of 95°C for 5 min followed by 35 cycles of denaturation at 94°C for 1 min., annealing at 55°C for 1 min. and extension at 72°C for 1 min, followed by final cycle of extension of 72°C for 7 min. The purified PCR product was analyzed for nucleotide sequence determination by using ABI PRISM® 3500 XL DNA Sequencer (Applied Biosystems) at Macrogen Company, Korea. The homology of partial sequences obtained were compared with the sequences from the DNA databases and similarity showing above 95% were retrieved by nucleotide Basic Local Alignment Search Tool (BLAST) program at the National Center for Biotechnology Information (NCBI) BLAST server [11].
Detection of organophosphorus hydrolase and pyrethroid esterase genes
The presence of three genes; methyl parathion- degradation gene (mpd) and Organophosphate-degradation gene (opd) as well as pyrethroid-degrading esterase gene (pytY) in the bacterial genome was assessed by polymerase chain reaction (PCR) with specific primers of three genes as shown in Table 2. PCR was performed as following conditions: initial denaturation at 94°C for 5 min, 30 cycles consisting of denaturation at 94°C for 0.5 min, annealing at 48 to 55°C for 0.5 min, and extension at 72°C for 1 min, and final chain elongation at 72°C for 7 min. PCR products were analyzed by electrophoresis in a 2% agarose gel with 1 × TBE as the running buffer.
| Primer ID | Target | Sequence (5' to 3') | References |
|---|---|---|---|
| F | mpd gene | GAATTCATATGCCCCTGAAGAAC | Yang et al. [9] |
| R | mpd gene | GAATTCTCGAGCTTGGGGTTGACGACCG | Yang et al. [9] |
| F196 | opd gene | CGCGGTCCTATCACAATCTC | Iyer et al. [12] |
| R840 | opd gene | CTTCTAGACCAATCGCACTG | Iyer et al. [12] |
| ZF | pytY gene | CGGGATCCATGACCACTCAAACCTATGAGC | Ruan et al. [17] |
| ZR | pytY gene | CGGAATTCTCAGTATGCGAGAAGCGACTG | Ruan et al. [17] |
Table 2: Sequences of specific primers to detect mpd, opd, pyty genes.
Results
Isolation and identification of profenofos and λ- cyhalothrin degrading bacteria
The selected eighteen bacterial isolates were found to possess ability to use the tested pesticides (profenofos and λ- cyhalothrin) as a carbon source with different efficiencies (Table 3). Strain coded DB17 was the most efficient isolate to degraded both pesticides. DB17 was subjected for further studies. This strain was a rod-shaped, gram negative bacterium. By sequencing the 16SrDNA gene of DB17 and comparing it with previously published 16S rDNA gene sequences, the strain was classified as a member of the genus Pseudomonas. The sequence of the strain displayed the highest identity (98%) with the 16S rDNA gene of a Pseudomonas putida.
| Bacterial isolate codes | Profenofos | λ- cyhalothrin | ||
|---|---|---|---|---|
| Conc. | Degradation % | Conc. | Degradation % | |
| Control | 97.21 | 2.79 | 98.45 | 1.55 |
| DB1 | 76.43 | 23.57 | 74.17 | 25.83 |
| DB2 | 84.49 | 15.51 | 73.55 | 26.45 |
| DB3 | 86.43 | 13.57 | 77.02 | 22.98 |
| DB4 | 61.43 | 38.57 | 74.94 | 25.06 |
| DB5 | 77.04 | 22.96 | 80.15 | 19.85 |
| DB6 | 67.65 | 32.35 | 61.64 | 38.36 |
| DB7 | 65.61 | 34.39 | 63.49 | 36.51 |
| DB8 | 56.63 | 43.37 | 64.49 | 35.51 |
| DB9 | 60.82 | 39.18 | 78.64 | 21.36 |
| DB10 | 75.71 | 24.29 | 73.4 | 26.6 |
| DB11 | 59.29 | 40.71 | 68.23 | 31.77 |
| DB12 | 48.06 | 50.92 | 65.88 | 34.12 |
| DB13 | 49.08 | 51.94 | 60.06 | 39.94 |
| DB14 | 71.43 | 28.57 | 64.23 | 35.77 |
| DB15 | 54.29 | 45.71 | 78.18 | 21.82 |
| DB16 | 62.45 | 37.55 | 65.34 | 34.66 |
| DB17 | 32.35 | 67.65 | 54.43 | 45.57 |
| DB18 | 67.76 | 32.24 | 74.79 | 25.21 |
Table 3: Evaluation of the degradation efficiency of the tested bacterial isolates.
Validation of the analytical method
Method validation process is the required proof to ensure the ability of an analytical method and the capability of the analyst to provide results of accepted quality. Profenofos and λ- cyhalothrin are widely used insecticides. Polarities of the compounds, expressed as Kow, are 4.44 and 7.0 for profenofos and λ- cyhalothrin, respectively. Analytical method was developed and validated for monitoring of profenofos and λ- cyhalothrin concentrations in liquid bacterial media as affected by the tested bacterial isolates. Validation parameters according to European guidelines i.e., accuracy, repeatability, linearity, limit of detection LOD and limit of quantitation LOQ, were investigated. Chromatographic conditions were optimized to ensure the absence of interferences during the resolution time of the target compound.
Calibration curves of profenofos and λ- cyhalothrin were constructed as a plot of the injected concentrations and the resulting absolute area under peak. Results showed that determination coefficients (R2) were 0.9997 and 0.9991 for profenofos and λ- cyhalothrin, respectively. LOD and LOQ were calculated from the standard calibration data. LOD was calculated at signal to noise ratio of 3 and LOQ was 3.3 folds of LOD. LODs were 0.02 and 0.04 mg L-1 while LOQs were 0.08 and 0.14 mg L-1 for profenofos and λ- cyhalothrin, respectively. Table 4 shows calibration curve parameters, LOD and LOQ values of both pesticides also recovery and repeatability of the method. The obtained results are within the norms reported by European guidelines to consider a method reliable and accurate.
| Pesticides | Intercept | Slope | R2 | LOD | LOQ | Recovery | Repeatability |
|---|---|---|---|---|---|---|---|
| (mg L-1) | (mg L-1) | (%) | RSD % | ||||
| Profenofos | 4.27 | 470.02 | 0.999 | 0.02 | 0.08 | 97.45 | 7.37 |
| λ- cyhalothrin | 1.26 | 274.17 | 0.999 | 0.04 | 0.14 | 99.95 | 6.86 |
Table 4: Intercept, slope, correlation coefficient (R2) of the calibration curves and LOD and LOQ of the tested pesticides.
Biodegradation of profenofos and λ- cyhalothrin in liquid culture media
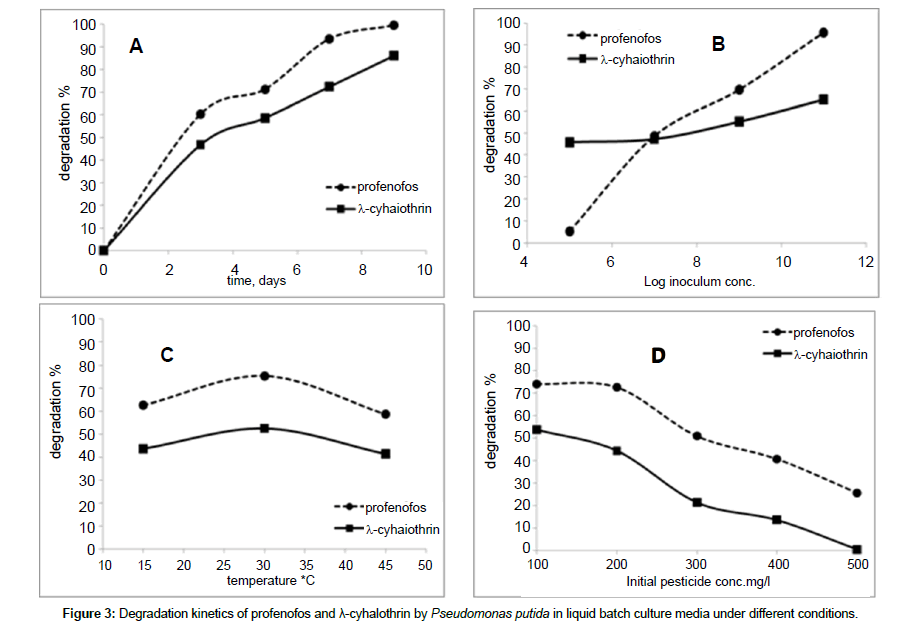
The degradation of profenofos and λ- cyhalothrin by DB17 isolate (Pseudomonas putida) under different conditions of incubation period, inoculum, temperature and pesticide concentration in liquid culture media was analyzed by GC/MS system. Degradation of profenofos and λ- cyhalothrin was monitored for 9 days after incubation. Degradation percentage reached 99.57% and 86.11% for profenofos and λ- cyhalothrin, respectively at the end of the experiment. Rapid degradation rate was observed for first 3 days after incubation. Slower rate was noted during further days after incubation.
The effect of consecutive initial inoculum concentrations of bacterial cells i.e., 105, 107, 109, 1011 on the degradation of the tested pesticides was investigated. Samples were taken after 3 days of incubation. A proportional relation was witnessed in profenofos degradation with inoculum concentration (slope 14.63) in comparison to λ- cyhalothrin where there was a little effect of inoculum concentration on the degradation (slope 3.30). The degradation percentages of profenofos were 5.31% and 95.81% with 105 to 1011 CFU, respectively. While, the degradation percentages of λ- cyhalothrin were 45.82% and 65.22% with 105 to 1011 CFU, respectively. The bacterial strain, DB17 (Pseudomonas putida) was more active under 30°C than at 15°C or 45°C as it appears from the degradation percent of both pesticides after 3 days of incubation. The ability of the isolate DB17 to tolerate an increasing concentration of the tested pesticides was a concern to investigate. Broth media augmented with successive concentrations of profenofos and λ- cyhalothrin i.e., 100, 200, 300, 400, 500 were cultured with 109 of DB17 bacterial cells as the initial concentration and kept stirred at 30°C for 3 days before a sample was taken. The degradation percentage decreased with pesticide concentration increase. Degradation percent were 74.15% and 25.65% at 100 and 500 ppm of profenofos, respectively while in case of λ- cyhalothrin were 53.74% and 0.53% at 100 and 500 ppm, respectively. Also, it can be inferred that the strain DB17 (Pseudomonas putida) was able to degrade profenofos more efficiently than λ- cyhalothrin. Degradation kinetic as affected with different conditions is illustrated in Figure 3.
Detection of the pesticide degrading genes
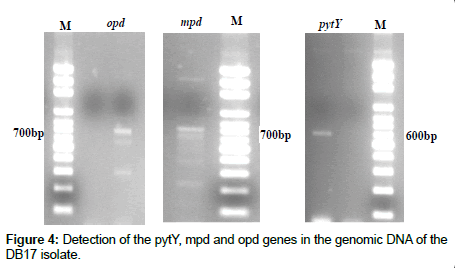
Detection of pesticide degrading genes (pytY, mpd and opd) in genomic DNA of DB17 isolate was performed according to an established protocol that relies on analytical PCR with their specific primers [12]. The amplified fragments are shown in Figure 4. The sequence BLAST result showed that the mpd gene was 99% similar to mpd gene of Plesiomonas sp., whereas the opd gene was 99% similar to opd gene of Sphingobium fuliginis and the pytY gene was 99% similar to pytY gene of Ochrobactrum anthropi at the nucleotide level. The expected lengths of PCR fragments are 700, 700 and 600 bp, to mpd, opd and pytY genes, respectively as shown in Figure 4.
Discussion
In the present study eighteen profenofos and λ- cyhalothrin degrading bacterial strains were enriched and isolated from soil samples collected from different agricultural sites in Qassim, Saudi Arabia. The isolate DB17 was found more efficient in comparison to other isolates. It was identified to be Pseudomonas putida using 16S rDNA gene detection strategy. Ref. [13] mentioned that isolation of bacterial strain through enrichment technique has been extensively used for biodegradation studies of pesticides. Ref. [7] isolated soil bacterial strains able to biodegrade profenofos in soil. They found that Pseudomonas putida was efficient to degrade profenofos. Also, Ref. [14] determined the degradation of the pyrethroid insecticide cypermethrin by Pseudomonas strains. Their study revealed that Pseudomonas strain degraded cypermethrin up to 100% within two days. Many of previous researches mentioned that the use of live biocatalysts for organophosphate degradation was demonstrated with Pseudomonas putida [15] and Pseudomonas sp [16].
In the present study rapid degradation was found with profenofos compared with λ- cyhalothrin in liquid culture. Also, the suitable conditions of pesticide degradation by Pseudomonas putida were 30°C for incubation temperature, 109 to 1011 cells / ml of bacterial inoculum concentrations and 100 to 200 ppm of pesticide concentrations in liquid media. Ref. [7] found that the pesticide profenofos was degraded rapidly in liquid culture of Pseudomonas putida where the degradation percentage reached to 27% after 6 hrs and 92.37% after 96 hrs. They used 100 ppm of pesticide concentration and 30°C of incubation temperature in degradation process. In this work some responsible genes for the degradation of organophosphorus and pyrethroid pesticides (mpd, opd and pyty) were detected in the genomic DNA of Pseudomonas putida (DB17) isolate. Ref. [9] cloned the gene encoding for organophosphorus hydrolase enzyme using PCR strategy based on the known methyl parathion degrading (mpd) gene of Plesiomonas sp. Our results were in agreement with Ref. [9] Sequence BLAST result indicated that this gene was 99% similar to mpd. Also, they mentioned that the mpd and opd genes appear to have evolved from different sources. Ref. [17] isolated pyrethroid-degrading gene pytY by screening the genomic library of Ochrobactrum anthropi YZ-1, and expressed it in Escherichia coli BL21 (DE3). Also, they reported estP, pytH, pye3, and pytZ as the pyrethroid degrading genes.
In conclusion, several samples of contaminated soils, with a history of profenofos and λ-cyhalothrin insecticides usage, were collected. Eighteen isolates were isolated from the collected soil samples. All the bacterial isolates were subjected to preliminary degradation test to investigate their ability to degrade profenofos and λ-cyhalothrin in liquid broth medium. Isolate DB17 showed the highest efficiency to degrade both pesticides among other isolates. An analytical method using QuEChERS followed by GC/MS was validated and used for samples' analysis. DB17 isolate was identified to be Pseudomonas putida. Genes responsible for organophosphorus and pyrethroid pesticides were detected in DB 17 isolate. The obtained results confirmed that the isolate DB17 P. putida can be successfully used for bioremediation of contaminated aquatic environments. Further investigation is required to confirm the ability of DB17 to degrade the tested pesticides in soil.
Acknowledgements
The authors wish to thank Promising Research Center in Biological Control and Agricultural Information (BCARC), Qassim University, Saudi Arabia for funding this work.
References
- Mosleh YY, Mofeed J, Almagrabi OA, Mousa T,Kadasa NMS (2014)Dietary Intake of Pesticides Based on Vegetable Consumption: A Case Study, Jeddah, Kingdom of Saudi Arabia. Life Science Journal 11: 680-688.
- MervatSM (2009)Degradation of methomyl by the novel bacterial strain StenotrophomonasmaltophiliaM1. Elect. J. Biotech12: 1-6.
- Singh DK (2008)Biodegradation and bioremediation of pesticide in soil: concept, method and recent developments. Indian J. Microbiol48:35-40.
- Sayler GS, Hooper SW,Layton AC,King JMH (1990)Catabolic plasmids of environmental and ecological significance. Microbial. Ecol19: 1-20.
- Laemmli CM,Leveau JHJ, Zehnder AJB,Vandermeer JR (2000)Characterization of a second tfdgene cluster for chlorophenol and chlorocatechol metabolism on plasmid pJP4 in RalstoniaeutrophaJMP134 (pJP4). J. Bacteriol182: 4165-4172.
- Horne I, Harcourt RL,Sutherland TD, RusselRJ,OakeshottJG(2002)Isolation of a Pseudomonasmonteilistrain with a novel phosphotroiesterase. FEMS Microbial Letters 206: 51-55.
- MalghaniS, Chatterjee N, Hu Xue Yu,Luo Z (2009)Isolation and identification of profenofos degrading bacteria. Brazilian Journal of Microbiology 40: 893-900.
- Abdel Ghani SB, Hanafi H (2016)QuEChERS Method Combined with GC–MS for Pesticide Residues Determination in Water . Journal of Analytical Chemistry 71: 508-512.
- Yang C, Liu N,Guo X,Qiao C(2006)Cloning of mpd gene from a chlorpyrifos-degrading bacterium and use of this strain in bioremediation of contaminated soil. FEMS MicrobiolLett26:118–125.
- Abed ThikraA,(2013)Evaluation of methods for the extraction and purification of DNA of cultured Lactobacillus colony isolated from dairy products. IJAMBR 1: 20-25.
- RamosSB, Pereyra de la IglesiaM T, ProbanzaA, Lucas GarciaJA, MegiasM, et al.(2006)Screening for PGPR to improve growth of Cistusladanifer seedlings for reforestation of degraded mediterranean ecosystem. Plant and Soil 287: 59-68.
- IyerR, StepanovVG, BrianI(2013)Isolation and molecular characterization of a novel Pseudomonasputidastrain capable of degrading organophosphate and aromatic compounds.Advances in Biological Chemistry 3: 564-578.
- Tatiane MS, MariaIS, AndreMM, FabianaDA, SoniaAVP, et al. (2007)Degradation of 2, 4 D herbicide by microorganisms isolated from Brazilian contaminated soil.Brazil. J. Microbiol38: 522-525.
- Malik D, SinghM, Bhatia P (2008)Biodegradation of cypermethrin by aPseudomonasstrainCyp19and its use in bioremediation of contaminated soil. Internet Journal of Microbiology 6: 2.
- Ramanathan MP,LalithakumariS(1996)Short communication: Methylparathion degradation by Pseudomonassp.A3 immobilized in sodium alginate beads. World Journal of Microbiology 12: 107-108.
- Rani NL,LalithakumariD (1994)Degradation of methyl parathion by Pseudomonasputida . Canadian Journal of Microbiology 40: 1000-1006.
- RuanZ,Zhai Y, Song JSY, LiK, Zhao B,et al. (2013)Molecular Cloning and Characterization of a Newly Isolated Pyrethroid-Degrading Esterase Gene from a Genomic Library of Ochrobactrumanthropi YZ-1. PLoS ONE 8: 1-7.
--
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 12645
- [From(publication date):
July-2016 - Aug 20, 2025] - Breakdown by view type
- HTML page views : 11442
- PDF downloads : 1203